Computational modeling and analysis of prominent T-cell epitopes for assisting in designing vaccine of ZIKA virus
Garima Yadav, Richa Rao, Utkarsh Raj, Pritish Kumar Varadwaj
DOI: 10.7324/JAPS.2017.70816Pages: 116-122

Study of B cell epitope conserved region of the Zika virus envelope glycoprotein to develop multi-strain vaccine
Oktavia Rahayu Adianingsih, Viol Dhea Kharisma
DOI: 10.7324/JAPS.2019.90114Pages: 098-103

In silico prediction of b-cell epitopes of dengue virus – A reverse vaccinology approach
Manikandan Mohan, Prabu Shanmugaraja, Rajeswari Krishnan, Kamaraj Rajagopalan, Krishnan Sundar
DOI: 10.7324/JAPS.2020.10109Pages: 077-085

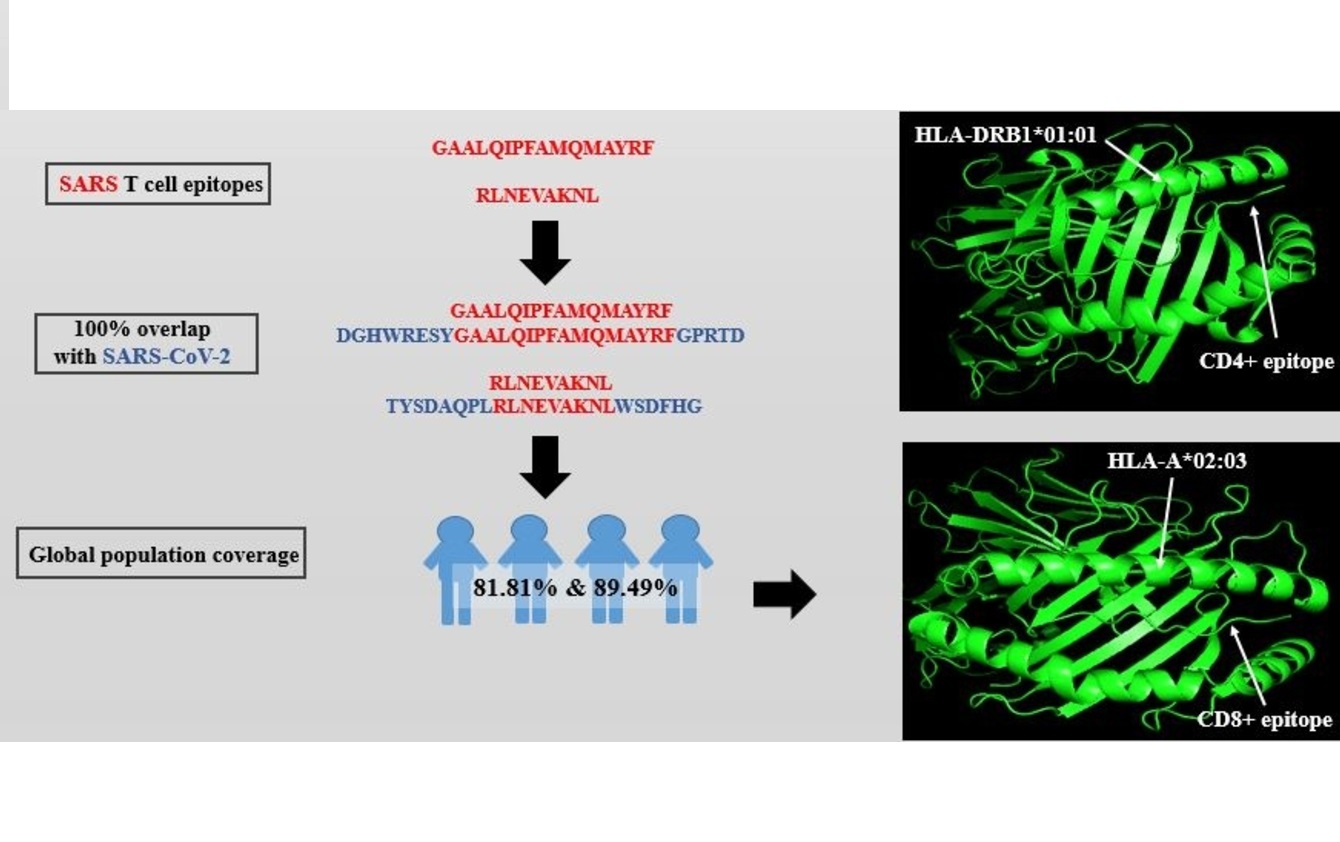
Immunoinformatics approach in designing SARS-CoV-2 vaccine from experimentally determined SARS-CoV T-cell epitopes
Leana Rich De Mesa Herrera
DOI: 10.7324/JAPS.2021.110303Pages: 029-036
Identification of dengue virus proteome B-cell epitopes using an immunoinformatic approach
Neeraj Kumar Dixit, Anup Kumar
DOI: 10.7324/JAPS.2021.1101209Pages: 107–113
Chimeric vaccine against multi-drug resistant Mycobacterium tuberculosis using in silico reverse vaccinology approach
Arpita Batta , Vineeta Singh, Bhartendu Nath Mishra, Tapankumar N. Dhole, Prahlad Kishore Seth
DOI: 10.7324/JAPS.2022.120609Pages: 086-114
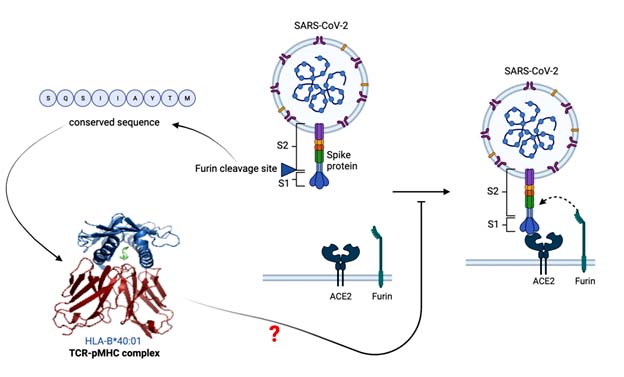
Mapping T-cell furin cleavage site epitopes in SARS-CoV-2 spike protein sequences from the Philippines and global variants
Maria Crischen Perono, Jewel Pasion, John Sylvester Nas, Francisco III Heralde
DOI: 10.7324/JAPS.2024.150608Pages: 197-211