INTRODUCTION
The dengue virus belonging to Flaviridae genus of the Flaviridae family is a common disease that affects about 400 million humans per year worldwide (Guzman et al., 2010; Kuno et al., 1998; Westaway et al., 1985). In 2020, India reported 16,439 cases, including 12 deaths, while the Lao PDR reported 6,015 cases of dengue and 11 deaths in 2020. 78,303 cases and 127 deaths have been reported in Malaysia (ECDC, 2020). 494 cases were reported from Bangladesh and 9,108 cases and 14 deaths from Cambodia, respectively. Nepal has reported 315 cases of dengue and Pakistan has reported a total of 743 cases of dengue by 2020 (ECDC, 2020). A record dengue fever epidemic in Latin America was reported in 2019, with over 2.7 million cases and 1,206 deaths during 10 months of 2019 (WHO, 2019). The genome of the Dengue virus consists of 10,696 nucleotides, of which 10,173 nucleotides encode 3,391 amino acids for a single open reading frame (Osatomi and Sumiyoshi, 1990). The viral genome of DEN encodes structural proteins as polyprotein, 2K peptide, Membrane glycoprotein, Envelope protein, Membrane Envelope protein, Anchored capsid protein, Capsid protein, and non-structural protein NS1, NS2a, NS2b, NS3, NS4A, NS4B, and NS5. A variety of human disorders are caused by dengue infection, ranging from asymptomatic, diarrhea, rash, knee pain and other mild symptoms to dengue hemorrhagic fever, and dengue shock syndrome. An important role is played by the immune system in embodying defensive responses as the first barrier to defense (Sai et al., 2013). During the acute phase of the febrile condition, a drop in platelet counts also occurs. After defervescence, most patients are resurrected, but extreme complications arise at this point as the fever subsides in a few patients, and this can potentially be lethal.
Dengvaxia, the world’s first authorized dengue vaccine, is a tetravalent live attenuated dengue vaccine officially approved in 19 nations (Scott, 2016; WHO, 2017). Its overall effectiveness against DENV was low, with approximately 50% against DENV1 and 39% against DENV2 in particular. However, recent clinical trials have found that CYDTDV vaccination has induced a high risk of hospitalization for children under the age of nine (Hadinegoro et al., 2015). Clinical trials of Dengvaxia have shown that the DENV2 strain is not very successful. Present vaccination testing findings suggest that multiple people have had severe potential allergic responses, such as allergies, headache, and urticaria. The outcomes of clinical trials of Dengvaxia demonstrate adverse consequences for a pregnant woman resulting in miscarriage, stillbirth, elective termination, and uterine mortality.
Lymphocytes are one group, and lymphocytes include T, B, and NK cells. Cell activation, antigen detection, and signal transduction are the most important external structures of B-cells responsible for (Sathe and Cusick, 2020) the antigen receptor is the functionally part of the cell surface multi molecular protein complexes (Justiz Vaillant et al., 2020). The sequencing of the repertoire of B cells and the growing structural characterization of protective antigens and epitopes currently provide molecular and mechanical expertise to guide the production of new vaccines that were previously impossible. A transmembrane receptor that reaches into the cytoplasm is the B-cell anti gen receptor. In transmitting the signals and triggering B cells, these tails are ineffective (Tanaka and Baba, 2020). The B cell receptor is a complex of multi molecular proteins non covalently bound to other proteins. B cell epitopes are intended for the production of diagnostic reagents and vaccines, not just for pathogenesis and immunological research (Jiang et al., 2010). Consequently, because of its potential to be bound by antibodies, the B cell epitope is antigenic (detected by immune system, potentially). If it is also capable of activating the production of such antibodies (e.g., by binding them to B cell surface immunoglobulins), it is immunoglobulinic. The characteristics of being antigenic and immunogenic are also antigenic and immunogenic.
Predicting linear B-cell epitopes with high accuracy is of key importance for epitope-based immunotherapy. For predicting B cell epitopes, several bioinformatics servers and algorithms are available. Prediction algorithms for B cell epitopes rely on linear epitopes since it is known that linear epitopes can evoke an antibody response that can interact with the parental antigen (Saha and Raghava, 2007a, 2007b). B-cell epitope can be more immunogenic than another if both are present on the same molecule and in particular, if they overlap physically with each other so that, compared to the other, the more immunogenic one thus described as immunodominant will induce the production of antibodies on its own while effectively suppressing productivity.
MATERIALS AND METHODS
Collection of source data
Resource gene database of structural protein amino acid sequence polyprotein, 2K peptide, membrane glycoprotein, protein envelope, membrane protein envelope, capsid protein anchor, capsid protein and nonstructural protein NS1, NS2a, NS2b, NS3, NS4A, NS4B, and NS5 Dengue virus (https:/www.viprbrc.org/brc/vipr proteinserch.spg). For the prediction of B cell epitopes, proteomes of DV-2 serotypes were evaluated.
Prediction of linear B-cell epitopes
Using multiple epitope properties, three separate methods were used to predict B cell epitopes.
ABCpred (http://www.imtech.res.in/raghava/abcpred/) and BCPREDS (http://ailab.ist.psu.edu/bcpred/predict.html), were used to predict potential 12, 16, and 20 mer B-cell epitopes from dengue virus 2 serotype proteins. ABCPred is a tool based on a neural network for predicting continuous B-cell epitopes by means of a fixed length pattern (Saha and Raghava, 2006). The ABCPred dataset includes data from virus epitopes contained in the BciPep database with 65.9% prediction accuracy. In both the B cell epitope prediction servers, fixed duration patterns are common. BCPREDS comprises two fixed length methods BCPred and AAP methods. For B-cell epitope prediction, BCPred and AAP methods were chosen in this analysis. The default parameter given by the servers was used to calculate the B-cell epitope prediction.
Antigenicity prediction
Antigenicity is a vaccine property that specifies whether an antigen or epitope will react with antibodies or not and induces the immune system from subsequent challenge by DENV to establish a protective mechanism. Using VaxiJen (http://www.ddgpharmfac.net/vaxijen/VaxiJen/VaxiJen.html) to ascertain the existence of antigen in its sequence, the proteins acquired undergo antigenicity prediction. The peptide sequence with a VaxiJen value of 0.4 is anticipated to have antigenicity properties above the viral threshold level. The expected B-cell epitopes should be theoretically antigenic, so that lymphocytes can elcitate optimum immune response upon parental antigen presentation.
Predictions the allergenicity of protein
Allergen FP (http://ddg-pharmfac.net/Allergen FP) was used to predict allergenicity of proteins. Non allergenic protein picked on a similarity index basis.
Toxicity prediction
The anticipated putative candidates were unable to cause any adverse effects on humans during administration. The toxic nature of the epitopes anticipated was then analyzed using a web-based server Toxinpred (http://www.imtech.res.in/raghava/toxinpred/design.php).
Consensus epitope prediction
For the preparation of Dengue Virus vaccines, the prediction between or within current serotypes of common epitopes can be used. The results of the predicted dengue virus 2 serotype epitopes (12, 16, and 20 mer) were compared to each other and the typical peptides were found to be consensus epitopes. The key explanation for the use of the consensus epitope strategy was to classify putative candidates with a higher likelihood of Dengue Virus immune response.
Conservancy analysis
Conservancy study is used to determine the degree of distribution of the epitope in a homologous protein sample in the in silico vaccinology technique. We used the epitope conservancy research method (http:/tools.iedb.org/conservancy/) at the IEDB (Immune Epitope DataBase) for the prediction of the conservancy trend of the desired epitopes (Bui et al., 2007). We used a method to calculate the variability of epitopes within a given range of protein sequences to aid with the optimal degree of conservation in the collection of epitopes.
RESULTS
B-cell epitopes prediction
The DENV2 proteome generated 1458 B-cell epitopes in total. The ABCpred server predicted the largest number of B-cell epitopes 873, followed by 330 and 255 epitopes predicted by the BCpred and AAP methods of the BCPREDS server as shown in Figure 1. Out of 1458 epitopes examined by fixed length pattern (12, 16, and 20 mer), 681 epitopes (46.70%) were 12 mer in length, 330 (22.63%), 16 mer and 447 (30.65%) 20 mer length, respectively. Less than one epitope score predicted by every server was discarded. On the basis of score value 1 or more, a total of 66 epitopes were chosen for further analysis.
Antigenicity prediction
VaxiJen 2.0 was used to estimate the vaccine construction antigenicity for 66 epitopes and resulted in the antigenic score of more than 0.8 chosen for further study of all 66 epitopes as shown in Table 1. BCpred was chosen among the largest numbers of antigenic epitopes (55), followed by AAppred (11) and no epitopes were selected from ABCpred. The epitopes expected by each approach displayed different degrees of antigenicity: 0.8–1.3 for BCpred, 0.8–1.7. For AApred, 83.33% (55 out of 66) of the epitopes in the analysis showed an antigenic score greater than 0.8 and 16.66% of the BCpred analysis showed an antigenic score greater than 0.8 (11 out of 36) epitopes.
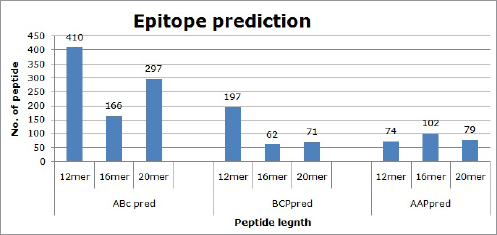 | Figure 1. Prediction of the epitope by methods of ABCpred, BCpred, and AAP. The maximum epitope numbers were found in the study of ABCpred. [Click here to view] |
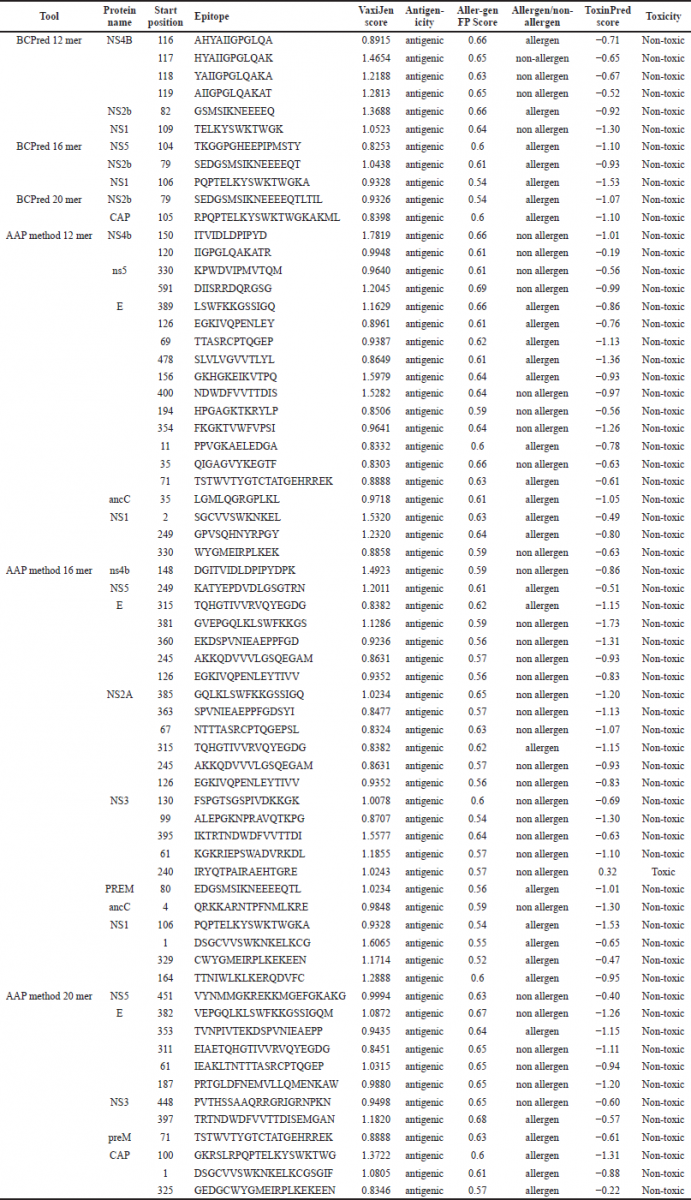 | Table 1. Predictions of antigenicity, allergenicity and toxicity index of epitopes. [Click here to view] |
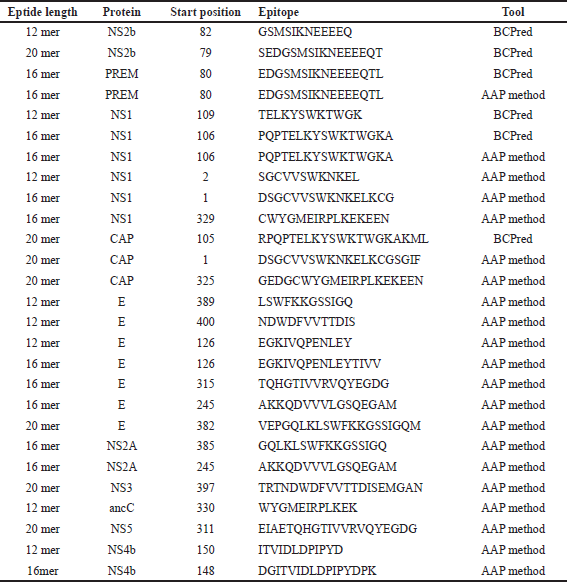 | Table 2. Predictions of consensus epitope. [Click here to view] |
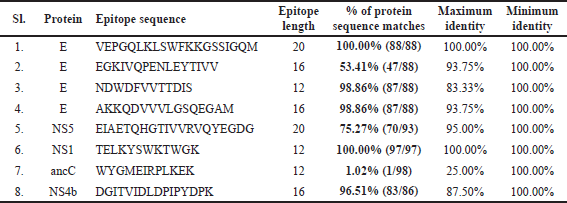 | Table 3. Predictions of conservancy analysis. [Click here to view] |
Toxicity prediction of B cell
Toxicity analysis was conducted on the ToxinPred web server. Anything expected by the ToxinPred web server as toxic has been discarded. The negative value of the peptide showed that it was non toxic. On the other hand, positive value of the peptide showed that it was toxic. To analyze toxicity of the 66 epitopes, it was expected that 65 were nontoxic (Table 1). Just 1 of the toxic peptides and the majority of the 65 non-toxic epitopes were omitted for further study.
Prediction of the allergenicity
The Allergen FP server was used to forecast the final vaccine construct’s non-allergic actions. Forecasting is based on SVM method. 31 allergenic epitopes and 35 non-allergic activity prediction epitopes were seen out of 66 epitopes by Allergen FPP, using the amino acid sequence for prediction. The BC pred server reveals 7 allergenic and 4 non allergic predictions, while the AApred server shows 24 allergenic and 31 non allergic predictions (Table 1). For further research, only non-allergenic epitopes were used.
Consensus epitopes prediction
In this analysis, a total of 27 consensus epitopes were predicted as shown in Table 2. The AAA method predicted 24 epitopes, while the BCpred method predicted only three. The majority of the consensus 12 epitopes were found to be 16 mer long. The numbers 8 and 7 were 12 and 20 mer, respectively. The E protein has four overlap epitopes, like VEPGQLKLSWFKKGSSIGQM, EGKIVQPENLEYTIV, NDWDFVVTTDIS, and AKKQDVVVLGSQEGAM. Epitopes EIAETQHGTIVVRVQYEGDG, TELKYSWKTWGK, WYGMEIRPLKEK, and DGITVIDLDPIPYDPK from NS5, NS1, ancC, and NS4b proteins were also found overlapped.
Conservancy analysis
Conservancy at 70% sequence identity threshold across consensus sequences, 2 out of 8 epitopes showed 100% conservancy across all DENV2 serotypes consensus sequences (Table 3). We found in this study that the epitope VEPGQLKLSWFKKGSSIGQM length 20 mer and TELKYSWKTWGK length 12 mer showed 100% preservation, while the epitope NDWDFVVTTDIS and AKKQDVVVLGSQEGAM length 12 mer and 16 mer showed 98.86% preservation. Other epitope length 16 mer DGITVIDLDPIPYDPK and length 20 mer EIAETQHGTIVVRVQYEGDG display 96.51% and 75.27% Conservancy. Remaining EGKIVQPENLEYTIVV length 16 mer and WYGMEIRPLKEK length 12 mer epitope reveals low Conservancy 53.41% and 1.02% discarded from the vaccine selection process, respectively. All the predicted B cell epitopes were then supplied with the IEDB conservancy analysis interface with a sequence identity threshold set at 70%. Both the DENV2 serotype consensus sequences envelope protein and NS5, NS1, ancC and NS4b were selected as homologous protein sets, respectively.
DISCUSSION
Every year, a large portion of the population is infected with one of the dengue virus serotypes, and a substantial number of these people experience serious DSS/DHF (Amorim et al., 2016). This illustrates the challenging task of developing a safe and efficient dengue virus vaccine. In contrast to conventional vaccine production, bioinformatics analysis can predict potent epitopes, making vaccine design simple and fast (Rappuoli et al., 2016). Within protection of viral infections, humoral immunity plays an essential function, alongside cell-mediated immunity. Recognition of B-cell epitopes is therefore essential in the understanding of viral pathogenesis and in the production of vaccines (Barlow et al., 1986). Antigenicity, allergenicity, and toxicity of predicted epitopes were evaluated by the VaxiJen v2.0 tool, Allergen FP server and ToxinPred web-server, respectively. The VaxiJen tool was used to analyze the defensive antigens based on the overall antigenicity score to further improve the prediction (Mehla and Ramana, 2016). Toxic peptide prediction is an important step in the production of a potent epitope-based vaccine. Using the Toxinpred tool, the toxic epitopes were omitted from further analysis. For consensus epitopes, the predicted B cell epitopes in each were also manually compared with each other. Epitope-based vaccines are a safe option for vaccine production since they can stimulate a particular immune response without causing any side effects (Oany et al., 2015).
CONCLUSION
VEPGQLKLSWFKKGSSIGQM, TELKYSWKTWGK, NDWDFVVTTDIS, AKKQDVVVLGSQEGAM, EIAETQHGTIVVRVQYEGDG, and DGITVIDLDPIPYDPK epitopes were selected for designing a vaccine against dengue virus. These epitopes had good antigenicity, allergicity, and conservancy. After experimental study, these epitopes may form the basis for the production of a Dengue vaccine.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FUNDING
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Amorim JH, dos Santos Alves RP, Bizerra R, Pereira SA, Pereira LR, Fabris DLN, Santos RA, Romano CM, de Souza Ferreira LC. Antibodies are not required to a protective immune response against dengue virus elicited in a mouse encephalitis model. Virology, 2016; 487:41–9. CrossRef
Barlow D, Edwards M, Thornton J. Continuous and discontinuous protein antigenic determinants. Nature, 1986; 322(6081):747 CrossRef
Bui HH, Sidney J, Li W, Fusseder N, Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinformatics, 2007; 8(1):361. CrossRef
ECDC. Geographical distribution of dengue cases reported worldwide, 2020. 2020. Available via https://www.ecdc.europa.eu/en/dengue-monthly
Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martínez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. Dengue: a continuing global threat. Nat Rev Microbiol, 2010; 8(12):S7–16. CrossRef
Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, Ismail HIHM, Reynales H, Limkittikul K, Rivera-Medina DM, Tran HN, Bouckenooghe A, Chansinghakul D, Cortés M, Fanouillere K, Forrat R, Frago C, Gailhardou S, Jackson N, Noriega F, Plennevaux E, Wartel TA, Zambrano B, Saville M, CYD-TDV Dengue Vaccine Working Group. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med, 2015; 373:1195–206. CrossRef
Jiang L, Zhou JM, Yin Y, Fang DY, Tang YX, Jiang LF. Selection and identification of B-cell epitope on NS1 protein of dengue virus type 2. Virus Res, 2010; 150(1–2):49–55. CrossRef
Justiz Vaillant AA, Jamal Z, Ramphul K. Immunoglobulin. StatPearls Publishing, Treasure Island, FL, 2020.
Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus flavivirus. J Virol, 1998; 72:73–83. CrossRef
Mehla K, Ramana J. Identification of epitope-based peptide vaccine candidates against enterotoxigenic Escherichia coli: a comparative genomics and immunoinformatics approach. Mol Biosyst, 2016; 12(3):890–901. CrossRef
Oany AR, Ahmad SAI, Hossain MU, Jyoti TP. Identification of highly conserved regions in L-segment of Crimean–Congo hemorrhagic fever virus and immunoinformatic prediction about potential novel vaccine. Adv Appl Bioinform Chem, 2015; 8:1. CrossRef
Osatomi K, Sumiyoshi H. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology, 1990; 176:643–7. CrossRef
Rappuoli R, Bottomley MJ, D’Oro U, Finco O, De Gregorio E. Reverse vaccinology 2.0: human immunology instructs vaccine antigen design. J Exp Med, 2016; 213(4):469–81. CrossRef
Saha S, Raghava GP. Prediction methods for B-cell epitopes. In: Schönbach C, Ranganathan S, Brusic V, (eds.). Immunoinformatics, Springer, Humana Press, Totowa, NJ, pp 387–94, 2007a. CrossRef
Saha S, Raghava GP. Searching and mapping of B-cell epitopes in BciPep database. In: Immunoinformatics, Springer, Humana press, New Jersey, USA, 2007b, 113–24. CrossRef
Saha S, Raghava GPS. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins, 2006; 65(1):40–8. CrossRef
Sai T, Lee CY, Lee IK, Yang KD, Liu JW. Comparisons of dengue illness classified based on the 1997 and 2009 World Health Organization dengue classification schemes. J Microbiol Immunol Infect, 2013; 46(4):271–81. CrossRef
Sathe A, Cusick JK. Biochemistry, immunoglobulin M. StatPearls Publishing, Treasure Island, FL, 2020. CrossRef
Scott LJ. Tetravalent dengue vaccine: a review in the prevention of dengue disease. Drugs, 2016; 76(13):1301–12.
Tanaka S, Baba Y. B cell receptor signaling. Adv Exp Med Biol, 2020; 1254:23–36. CrossRef
Westaway EG, Brinton MA, Gaidamovich S, Horzinek MC, Igarashi A, Kääriäinen L, Lvov DK, Porterfield JS, Russell PK, Trent DW. Flaviviridae. Intervirology, 1985; 24(4):183–92. CrossRef
WHO. WHO Region of the Americas records highest number of dengue cases in history- cases spike in other regions. 2019. Available via https://www.who.int/news/item/21-11-2019-who-region-of-the-americas-records-highest-number-of-dengue-cases-in-history-cases-spike-in-other-regions
World Health Organization. Updated questions and answers related to information presented in the Sanofi Pasteur press release on 30 November 2017 with regards to the dengue vaccine Dengvaxia®. World Health Organization, Geneva, Switzerland, 2017.