INTRODUCTION
Zika virus (ZIKV) is the mosquito-borne Flavivirus that identified for the first time in 1947 (Basile et al., 2017). There was the first large outbreak of ZIKV infection on Yap Island (Micronesia) in 2007 (WHO, 2016). The regions of ZIKV transmissions have increased in many countries including South and Central America, the Caribbean, and the United States (Hendrixson and Newland, 2018). In 2016, WHO declared that there was an association between ZIKV infection and the cluster of microcephaly and other neurological disorders (WHO, 2016). ZIKV infection during pregnancy causes both symptomatic and asymptomatic congenital infection that associates with central nervous system abnormalities, fetal death, and fetal growth restriction (Brasil et al., 2016; Hendrixson and Newland, 2018). In addition, ZIKV infection can result in life-threatening neurologic illness in adults, including encephalitis, and Guillain-Barré syndrome (Barbi et al., 2018; Krow-lucal et al., 2018; Miller et al., 2017; Salinas et al., 2017; Sebastián et al., 2017).
ZIKV is a single stranded, positive-sense RNA virus with a 10.7 kb genome encoding a single polyprotein that is cleaved into three structural proteins (C, prM/M, and E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4BM, and NS1). The envelope (E) glycoprotein is the major antigenic determinant on ZIKV that mediates the binding and fusion during virus entry (Lindenbach and Rice, 2003; Zhang et al., 2017). ZIKV glycoprotein binds Axl receptor in the microglia and astrocyte through Gas6 bridging and promotes ZIKV infection such as microcephaly (Meertens et al., 2017; Retallack et al., 2016). Therefore, this glycoprotein becomes an important target for the research to develop vaccine candidates and antiviral therapeutics. Many pharmacological inhibitors of ZIKV infection have been investigated in an in silico study and in the experimental models (Munjal et al., 2017). But, at present, there is no antiviral drug, vaccine, or other modality available to prevent or treat multiple strain of ZIKV infection (Poland et al., 2018). Therefore, in this study, our attempt is to identify the conserved region of ZIKV envelope glycoprotein that could act as B cell epitope by immunoinformatics, thus could be developed to the multi-strain vaccine.
MATERIAL AND METHODS
Protein sequence preparation
The ZIKV polyprotein sequences from all strains were retrieved from the National Center of Biotechnology Information (https://www.ncbi.nlm.nih.gov/) using the keyword “Flavi_glycoprot”.
Conserved region identification
Multiple alignment methods were used to analyze the conserved regions. All ZIKV sequences were aligned using molecular evolutionary genetics analysis (MEGA) 5 software to identify the conserved region of the ZIKV glycoprotein. The 3D structure of the conserved region was modeled using the Swiss Model (www.swissmodel.expasy.org) (Bienert et al., 2018; Waterhouse et al., 2018). Then, this 3D structure was aligned using PyMol software with the template obtained from homology modeling to confirm the conserved region of ZIKV glycoprotein. Homology modeling was used to visualize the B cell epitope in the 3D structure (Benkert et al., 2018; Bertoni et al,. 2017; Guex et al., 2009). Cluspro web server (https://cluspro.bu.edu/login.php) was used to stimulate the binding of the ZIKV glycoprotein envelope and Axl receptor tyrosine kinase, a receptor that mediates the ZIKV entry to the host cell. The position of interaction between amino acid residues was evaluated using knowledge-based fast atomic density evaluation (FADE) and contact (KFC) web server (https://mitchell-lab.biochem.wisc.edu/KFC_Server/index.php).
B cell epitope analysis
B cell epitope is a specific antigen region that has the high interaction with B cell lymphocyte. As a result, B cell could produce the antigen-specific antibody and memory cell. B cell epitope is divided based on hydrophilicity, accessibility, and β turn region (Hasan et al., 2013). To predict the B cell epitope, we used the methods provided by the immune epitope database (IEDB) web server (http://tools.iedb.org/main/bcell/) with default thresholds: BepiPred (Erik et al., 2006) with threshold 0.229, Emini surface accessibility (Emini et al., 1985) with threshold 1,000, Kolaskar–Tongaonkar antigenicity (Kolaskar and Tongaonkar, 1990) with threshold 1.015, Parker hydrophilicity (Parker et al., 1986) with threshold 2.068, Karplus and Schulz flexibility (Karplus and Schulz, 1985) with threshold 1.002, and Chou and Fasman beta turn (Chou and Fasman, 1978) with threshold 1.017.
Similarity analysis
The conserved region that was predicted as epitopes need to be analyzed the similarity to the receptor on the cell surface to prevent the autoimmune. The similarity was analyzed using the basic local alignment search tool protein (BLASTp) and the resulted score must be under 70%.
RESULTS AND DISCUSSION
Conserved region of ZIKV glycoprotein
In this study, we retrieved 363 strains of ZIKV polyprotein sequences from 34 different locations. Previously study only retrieved 50 strains of ZIKV polyprotein sequences to propose the conserved epitopes of ZIKV envelope glycoprotein (Badawi et al., 2016). The identified regions in this study were Uganda, Cuba, Brazil, Dominican Republic, USA, Colombia, Honduras, China, Senegal, Cambodia, Philippines, Thailand, South Korea, Haiti, Taiwan, Suriname, Russia, Singapore, Asian, Mexico, Nicaragua, Italy, Japan, French, UK, Martinique, Guatemala, Puerto Rico, Micronesia, Ecuador, Peru, Panama, and Central African Republic (Fig. 1), but there was one unknown location found. The conserved sequence obtained from the alignment results using MEGA 5 software has 266-mer length with the sequence as follows: “RCIGVSNRDFVEGMSGGTWVDVLEHGGCVTMAQDKPVDIELVTTVSNMAERCYEAISDSDSRCPTQGEALKQSDTQYVCKRTLVDRGWGNGCGLFGKGSLVTCAKFCSKKMTGKSIQPENLEYRIMLSVHGSQHSGMGETDERAKVETPNSPRAEATLGFGSLGLDCEPRTDFSDLYYLTMNNKHWLHKEWFHDIPLPWHGTGTPHWNNKEALVEFDAHAKRQTVLGSQEGAVHALAGALEAEMDGKGLSGLKCRLKMDKRLKG.”
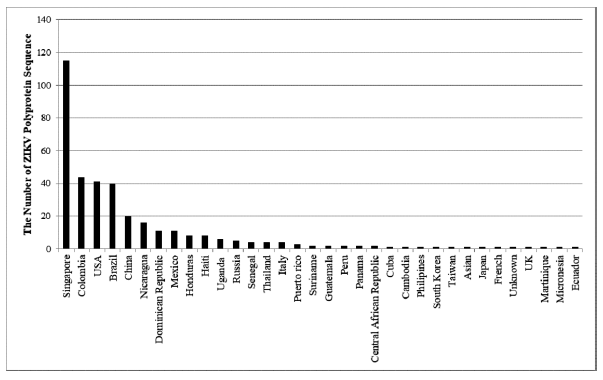 | Figure 1. The source of ZIKV strains retrieved from NCBI. ZIKV polyprotein sequences were retrieved from 34 countries which Singapore has the highest number of ZIKV polyprotein sequences (151 from 343 sequences or 31.68%). [Click here to view] |
After obtaining the conserved region, we modeled the protein using the homology modeling method. The result showed that the conserved region was 98.5% similar to the 5JHM template identified as a ZIKV envelope protein. In order to obtain the highest quality model, the resulted model must have similarity more than 20% to the template (Bienert et al., 2018; Waterhouse et al., 2018). The modeled protein was shown in the cartoon and colored based on the secondary protein structures (Fig. 2a). To determine the coverage level of the modeled protein on the protein template (Protein Data Bank ID: 5JHM), we did the 3D alignment using PyMol software. The result revealed that the modeled protein matched with the A chain on the template (Fig. 2b).
The molecular docking is not only could be used to optimize the shape of the protein conformation and ligand orientation, but also to identify the low energy needed for the protein-ligand binding (Sitompul et al., 2012). After the 3D alignment process, we confirmed the binding ability of the ZIKV model protein with the Axl receptor tyrosine kinase using the molecular docking method (Badawi et al., 2016; Nowakowski et al., 2016). The Axl receptor was retrieved from RSCB PDB (ID: 5U6B) that play a role as the target protein receptor of ZIKV envelope glycoprotein. The binding energy of the ZIKV envelope glycoprotein-Axl receptor complex was −841.0 kJ/mol. The binding interaction was evaluated using the KFC web server. The interaction between two proteins is essential for studying the properties of a protein when experiencing combinations in complex structures (Sitompul et al., 2012). The result showed that approximately 36 amino acid residues of ZIKV envelope glycoprotein were involved in the binding interaction. These amino acid residues that positively bound to the Axl receptor were Asp9, Glu12, Glu40, Asp177, Glu243, and Glu245 (Fig. 3).
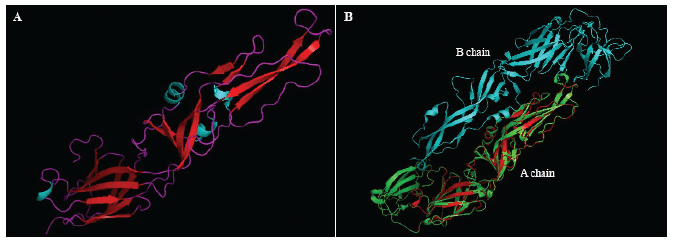 | Figure 2. Visualization and 3D alignment of conserved ZIKV envelope glycoprotein. (A) The conserved sequence structure of ZIKV envelope glycoprotein. The cyan, red, and purple color were the structure of α-helix, β-sheet, and coil. (B) 3D alignment between conserved ZIKV envelope glycoprotein and template (ID 5JHM). The conserved sequence was superimposed with the A chain of the template. The green and the cyan color were A and B chain of the template, the red color was the conserved ZIKV envelope glycoprotein. [Click here to view] |
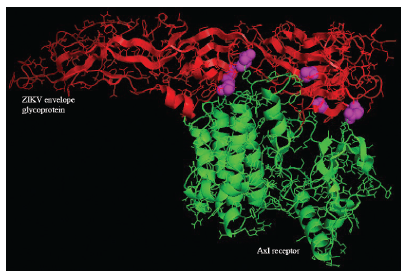 | Figure 3. The interaction between a conserved region on ZIKV envelope glycoprotein and Axl receptor. The Axl receptor domain (green) bond to the conserved region of ZIKV envelope glycoprotein (red). The purple color was the amino acid residues of ZIKV envelope glycoprotein that interacted with the Axl receptor. [Click here to view] |
Prediction of B-cell epitope
Epitope-related studies have been previously done in several viruses such as dengue virus (DENV), human papillomavirus (HPV), Epstein–Barr virus (EBV), and ZIKV (Badawi et al., 2016; Baidya et al., 2017; Himmah et al., 2016; Nusrat et al., 2018; Sitompul et al., 2012). A study conducted by Himmah et al. (2016) aimed to develop DENV vaccine by designing a polytope protein from the conserved DENV serotypes 1–4, to analyze antigenicity, and to know the interaction and the binding energy between the antibody and the polytope protein. In order to predict the epitope, they used epitope prediction parameters such as antigenicity, flexibility, and hydrophilicity (Himmah et al., 2016). B cell epitope prediction also used in the epitope mapping study of gp350 conserved domain of EBV for the development of the nasopharyngeal vaccine. They used the Kolaskar–Tongaonkar and Welling scale antigenicity parameters and BepipPred predictions on the IEDB web server (Sitompul et al., 2012). In addition, the immunoinformatic epitope B cell prediction of L1 protein HPV types 16 and 18 derived from all strains was performed on the IEDB web server by simply using the Kolaskar and Tongaonkar scale antigenicity parameter which is a standard feature for epitope prediction (Baidya et al., 2017). From the previous research, we can conclude that there are several parameters for the B cell epitope prediction on the IEDB web server, such as hydrophilicity, accessibility, antigenicity, turn, flexibility, and polarity. All predictions have a calculation based on a trend scale for every 20 amino acids. In the graphic result, the Y-axis defines for each amino acid residual score, whereas X is the position of the amino acid sequence residue. The yellow region is a positive prediction of B cell epitope while green is negative (IEDB, 2018).
In our study, we predicted the B cell epitopes of the conserved region of the ZIKV envelope glycoprotein based on the BepiPred method, Emini surface accessibility, Kolaskar–Tongaonkar antigenicity, Parker hydrophilicity, Chou and Fasman beta-turn, and Karplus and Schulz flexibility on IEDB. These methods were used to predict specific areas in proteins that bind to the B cell receptor, and the area must be on the surface and immunogenic (Fig. 4). The BepiPred linear epitope prediction works based on the combination of Markov’s hidden method model and a trend scale which the positive predictions are characterized by “E” as B cell linear epitope (Erik et al., 2006). In this study, the average ZIKV envelope glycoprotein binding score in B cells was 0.299 with a maximum and minimum values were 2.140 and -1.559. All values equal to or greater than the threshold were predicted to bind to the B cell receptor. The Emini surface accessibility scale describes that every peptide having a surface probability value of 1.000 is located on the cell surface (Emini et al., 1985). In this study, the surface accessibility scores of the ZIKV envelope glycoprotein peptide were 1.000 with a maximum and minimum values were 4.401 and 0.084. All values equal to or greater than the threshold were located on the surface.
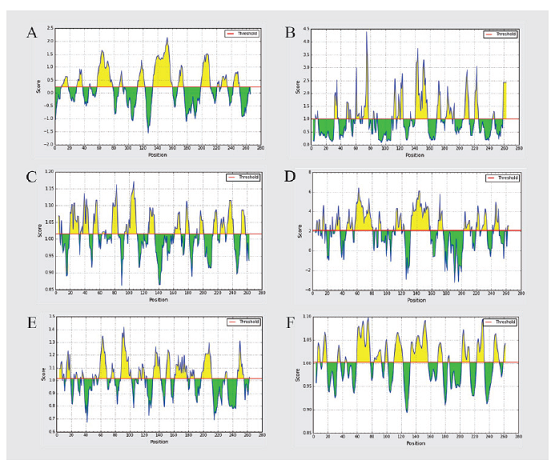 | Figure 4. Prediction of B cell epitopes by different parameters. (A) BepiPred, (B) Emini surface accessibility, (C) Kolaskar–Tongaonkar antigenicity, (D) Parker hydrophilicity, (E) Chou and Fasman beta-turn, and (F) Karplus and Schulz flexibility. B cell epitope prediction was done using the IEDB server. The yellow region was a positive prediction of B cell epitope, whereas the green region was negative. [Click here to view] |
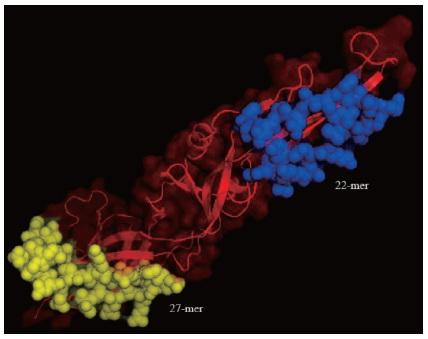 | Figure 5. The location of B cell epitope in the conserved region of ZIKV envelope glycoprotein domain. There were two sequences that potential as the B cell epitopes with 22-mer (blue) and 27-mer (yellow) sequence length. The red color was the non-epitope sequence of the conserved region of ZIKV envelope glycoprotein. [Click here to view] |
The Kolaskar and Tongaonkar antigenicity scale works based on the physicochemical properties of amino acid residues and the known tendency frequency as an experimental epitope. This method has 75% accuracy (Kolaskar and Tongaonkar, 1990). The default threshold of the Kolaskar and Tongaonkar antigenicity in this study was 1.015 with a maximum and minimum values were 1.172 and 0.861. The values that are equal to or above the default threshold were potential as antigenic determinants. The Parker hydrophilicity prediction works to predict epitopes based on peptide retention time during high-performance liquid chromatography (Parker et al., 1986). In Parker’s prediction of hydrophilicity, the average score was 2.068 with the maximum and minimum scores were 6.443 and −3.257. All values that are equal to and greater than the threshold were likely to be hydrophilic.
The Chou and Fasman beta-turn prediction predicts the epitope-based on the turn structure scale. It predicts that the epitope has been always in beta-shaped turn (Chou and Fasman, 1978). The Chou and Fasman Beta-Turn prediction, the average score was 1.017 with the maximum and minimum scores were 1.421 and 0.674. All values that are equal to and greater than the threshold are likely to be predicted as epitopes. The Karplus and Schulz flexibility scale works based on the flexibility scale in protein segment mobility. It is used as the base to know the B factors in the carbon chains of 31 known protein structures (Karplus and Schulz, 1985). The prediction of the flexibility of Karplus and Schulz has an average score of 1.002, a maximum score of 1.098 and 0.894 for the minimum score, all values that are equal to and higher than the threshold are likely to be predicted as highly flexible epitopes.
The results of the epitope predictions indicate that there were the potential peptide sequences as the B cell epitopes. These sequences were “ISDSDSRCPTQGEALKQSDTQY” with 22-mer length and “SQH SGMGETDERAKVETPNSPRAEATL” with 27-mer length (Fig. 5). From the similarity analysis using the BLASTp, we confirmed that both peptides have a low similarity to the cell surface proteins in human. As previously reported that the inhibition of complement receptor (CR2) binding domains with gp350 using antibodies could prevent the spread of EBV virus and provoke the immune system to eliminate the virus in the patient’s body. The identification of the conserved domain residues is required to predict the peptide that has the high antigenicity. The epitope prediction tools could facilitate the development of vaccines and predict the epitopes (Sitompul et al., 2012).
CONCLUSIONS
The residues of a conserved region in ZIKV envelope glycoprotein could interact with the virus receptor on the host cell (Axl receptor tyrosine kinase) through amino acid residues Asp9, Glu12, Glu40, Asp177, Glu243, and Glu245. We proposed that sequence “ISDSDSRCPTQGEALKQSDTQY” (22-mer) and “SQHSGMGETDERAKVETPNSPRAEA TL” (27-mer) in ZIKV envelope glycoprotein could be used as B cell epitopes for the multi-strain ZIKV vaccine development. Further work required to clone the gene for these peptides on some carrier vectors in order to collect data on the extent of elicitation of immune response and to ensure that this vaccine will provide long-term protection. Further study is also needed to identify the interaction between these peptides with T cells and antibodies.
REFERENCES
Badawi MM, Osman MM, Abdelhamid A, Alla F. Highly conserved epitopes of ZIKA envelope glycoprotein may act as a novel peptide vaccine with high coverage : immunoinformatics approach. Am J Biomed Res, 2016; 4(3):46–60.
Baidya S, Das R, Kabir G. Epitope design of L1 protein for vaccine production against human papilloma virus types 16 and 18. Bioinformation, 2017; 13(3):86–93.
Barbi L, Coelho AVC, de Alencar CLA, Crovella S. Prevalence of Guillain-Barré syndrome among Zika virus infected cases: a systematic review and meta-analysis. Braz J Infect Dis, 2018; 22(2):137–41.
Basile K, Kok J, Dwyer DE. Zika Virus: What , Where from and Where To? Pathol, 2017; 49(7):698–706.
Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics, 2018; 27(3):343–50.
Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci Rep, 2017; 7:1–15.
Bienert S, Waterhouse A, de Beer TAP, Tauriello G, Studer G, Bordoli L, Schwede T. The SWISS-MODEL Repository—new features and functionality. Nucleic Acids Res, 2018; 45:313–9.
Brasil P, Pereira JP, Moreira ME, Nogueira RMR, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, Zin AA, Horovitz D, Daltro P, Boechat M, Gabaglia CR, Carvalho de Sequeira P, Pilotto JH, Medialdea-Carrera R, Cotrim da Cunha D, Abreu de Carvalho LM, Pone M, Machado Siqueira A, Calvet GA, Rodrigues Baião AE, Neves ES, Nassar de Carvalho PR, Hasue RH, Marschik PB, Einspieler C, Janzen C, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med, 2016; 24(375):2321–34.
Chou BPY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol, 1978; 47(1195):45–148.
Emini EA, Hughes JV, Perlow DS, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol, 1985; 55(3):836–9.
Erik J, Larsen P, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res, 2006; 7:1–7.
Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis, 2009; 30:162–73.
Hasan A, Hossain M, Alam M. A computational assay to design an epitope-based peptide vaccine against Saint Louis encephalitis virus. Bioinform Biol Insights, 2013; 7:347–55.
Hendrixson DT, Newland JG. Zika virus infection in children. Infect Dis Clin NA, 2018; 32:215–24.
Himmah K, Ardyati T, Deocaris C. Polytope prediction for dengue vaccine candidate based on conserved envelope glycoprotein of four serotypes of dengue virus and its antigenicity. J Pure App Chem Res, 2016; 5:101–7.
Immune Epitope Database. Overview: epitope prediction and analysis tools, 2018. Available via http://tools.iedb.org/main/.
Karplus PA, Schulz GE. Prediction of chain flexibility in proteins—a tool for the selection of peptide antigen. Naturwissenchaften, 1985; 72:2–3.
Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants protein antigens. FEBS Lett, 1990; 276(1):172–4.
Krow-lucal ER, de Andrade MR, Nunes J, Cananéa A, Moore CA, Leite PL, Biggerstaff BJ, Cabral CM, Itoh M, Percio J, Wada MY, Powers AM, Barbosa A, Abath RB, Staples JE, Coelho GE; the Paraíba Microcephaly Work Group. Association and birth prevalence of microcephaly attributable to Zika virus infection among infants in Paraíba, Brazil, in 2015–16 : a case-control study. Lancet, 2018; 4642(18):1–9.
Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res, 2003; 59:23–61.
Meertens L, Labeau A, Dejamac O, Gressens P, Schwartz O, Amara A. Axl mediates ZIKA virus entry in human glial cells and modulates innate immune responses. Cell Rep, 2017; 18:324–33.
Miller E, Becker Z, Shalev D, Lee CT, Cioroiu C, Thakur K. Probable Zika virus-associated Guillain-Barré syndrome: challenges with clinico-laboratory diagnosis. J Neurol Sci, 2017; 375:367–70.
Munjal A, Khandia R, Dhama K, Sachan S, Karthik K, Tiwari R, Malik YS, Kumar D, Singh RK, Iqbal HMN, Joshi SK. Advances in developing therapies to Combat Zika Virus: current knowledge and future perspectives. Front Micobiol, 2017; 8:1–19.
Nowakowski TJ, Pollen AA, Lullo ED, Sandoval-espinosa C, Bershteyn M. Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells: brief report. Cell Stem Cell, 2016; 18(5):591–96.
Nusrat S, Amran S, Rahvia G, Sthity A. Designing of epitope-focused vaccine by targeting E6 and E7 conserved protein sequences: an immuno-informatics approach in human papillomavirus 58 isolates. Interdiscip Sci Comput Life Sci, 2018; 10(2):251–60.
Parker J, Guo D, Hodges R. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites? Am Chem Soc, 1986; 25:5425–32.
Poland GA, Kennedy RB, Ovsyannikova IG, Palacios R, Ho PL, Kalil J. Development of vaccines against Zika virus. Lancet Infect Dis, 2018:1–9.
Retallack H, Lullo ED, Arias C, Knopp KA, Laurie MT, Sandoval-Espinosa C, Leon WRM, Krencik R, Ullian EM, Spatazza J, Pollen AA, Mandel-Brehm C, Nowakowski TJ, Kriegstein AR, DeRisi JL. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. PNAS, 2016; 113(5):14408–13.
Salinas JL, Walteros DM, Styczynski A, Garzón F, Quijada H, Bravo E, Chaparro P, Madero J, Acosta-Reyes J, Ledermann J, Arteta Z, Borland E, Burns P, Gonzalez M, Powers AM, Mercado M, Solano A, Sejvar JJ, Ospina ML. Zika virus disease-associated Guillain-Barré syndrome—Barranquilla, Colombia 2015–2016. J Neurol Sci, 2017; 381:272–7.
Sebastián UU, Villamizar A, Ricardo A, Alvarez BC, Cubides A, Luna AF, Arroyo-Parejo M, Acuña CE, Quintero AV, Villareal OC, Pinillos OS, Vieda E, Bello M, Peña S, Dueñas-Castell C, Rodriguez GMV, Ranero JLM, López RLM, Olaya SG, Vergara JC, Tandazo A, Ospina JPS, Soto IML, Fowler RA, Marshall JC; on behalf of LACCTIN Group. Zika virus-induced neurological critical illness in Latin America: severe Guillain-Barre syndrome and encephalitis. J Crit Care, 2017; 42:275–81.
Sitompul LS, Widodo N, Djati MS, Utomo DH. Epitope mapping of Gp350/220 conserved domain of epstein barr virus to develop nasopharyngeal carcinoma (Npc) vaccine. Bioinformation, 2012; 8(10):8–11.
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res, 2018; 46:296–303.
World Health Organization. Zika Situation Report. 2016. Available via http://www.who.int/emergencies/zika-virus/situation-report/5-may-2016/en/.
Zhang X, Jia R, Shen H, Wang M, Yin Z, Cheng A. Structure and functions of the envelope glycoprotein in flavivirus infections. Viruses, 2017; 9(338):1–14.