Molecular construction of NADH-cytochrome b5 reductase inhibition by flavonoids and chemical basis of difference in inhibition potential: Molecular dynamics simulation study
Sharad Verma, Amit Singh, Abha Mishra
DOI: 10.7324/JAPS.2012.2804Pages: 33-39

QSAR, Molecular Docking and Dynamics Studies of Quinazoline Derivatives as Inhibitor of Phosphatidylinositol 3-Kinase
Muhammad Arba, Ruslin, Waode Umi Kalsum, Armid Alroem, Muhammad Zakir Muzakkar, Ida Usman, Daryono Hadi Tjahjono
DOI: 10.7324/JAPS.2018.8501Pages: 001-009

Computational approach toward targeting the interaction of porphyrin derivatives with Bcl-2
Muhammad Arba, Yamin, Sunandar Ihsan, Daryono Hadi Tjahjono
DOI: 10.7324/JAPS.2018.81208Pages: 060-066
Identification of polyketide synthase 13 inhibitor: Pharmacophore-based virtual screening and molecular dynamics simulation
Muhammad Arba, Andy-Nur Hidayat, Henny Kasmawati, Yamin Yamin, Ida Usman
DOI: 10.7324/JAPS.2019.90702Pages: 012-017
Computational study of the potential molecular target for antibreast cancer activity of limonoid derivatives from chisocheton sp
Nurlelasari Nurlelasari, Ade Rizqi Ridwan Firdaus, Desi Harneti, Nenden Indrayati, Umi Baroroh, Muhammad Yusuf
DOI: 10.7324/JAPS.2020.102002Pages: 007-012

Designing of next-generation dihydropyridine-based calcium channel blockers: An in silico study
Sujoy Karmakar, Hriday Kumar Basak, Uttam Paswan, Arun Kumar Pramanik, Abhik Chatterjee
DOI: 10.7324/JAPS.2022.120414Pages: 127-135

Identification of potential bioactive compounds from Azadirachta indica (Neem) as inhibitors of SARS-CoV-2 main protease: Molecular docking and molecular dynamics simulation studies
Donny Ramadhan, Firdayani Firdayani, Nihayatul Karimah, Elpri Eka Permadi, Sjaikhurrizal El Muttaqien, Agus Supriyono
DOI: 10.7324/JAPS.2023.139799Pages: 040-049

Therapeutic potential of earthworm-derived proteins: Targeting NEDD4 for cardiovascular disease intervention
Doni Dermawan, Faisal Alsenani, Nasr Eldin Elwali, Nasser Alotaiq
DOI: 10.7324/JAPS.2024.204162Pages: 216-232
_.webp)
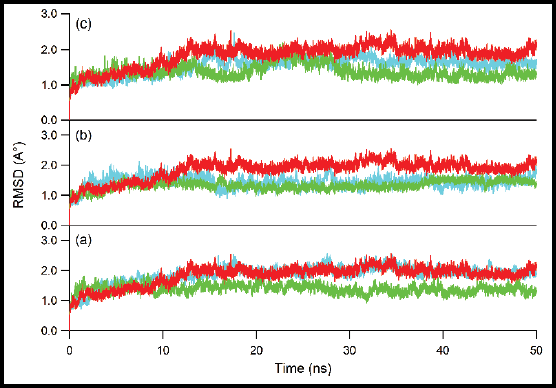
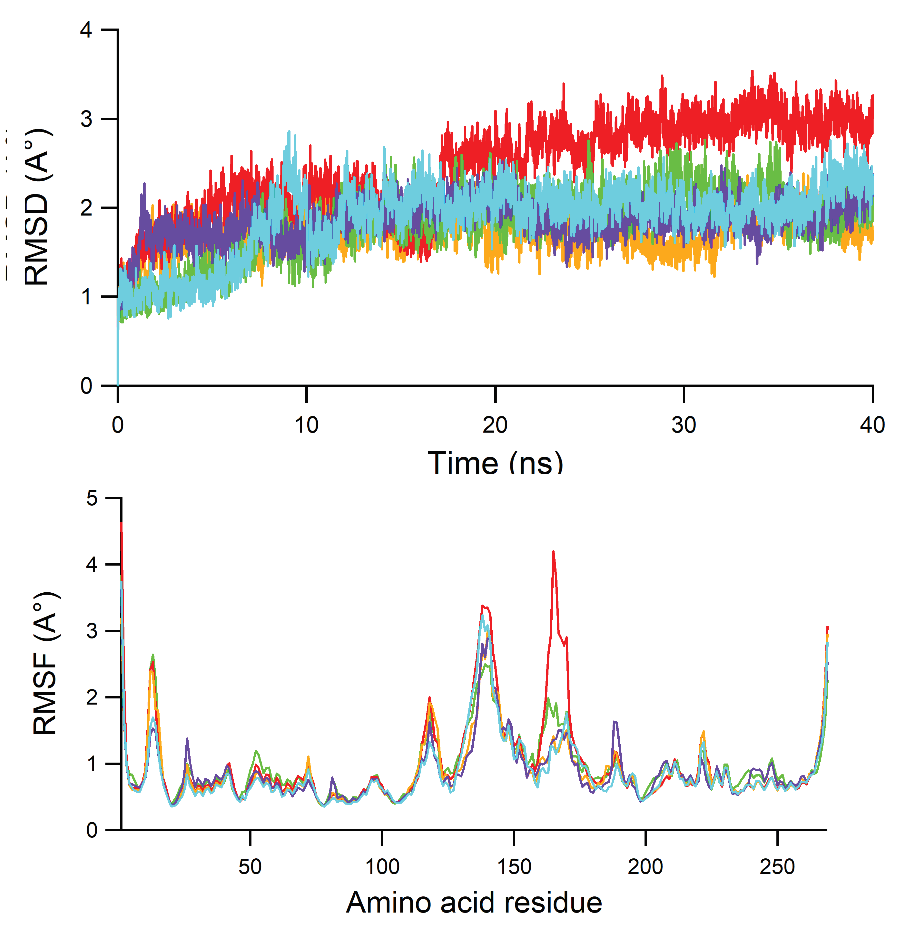
Molecular docking and molecular dynamics simulation of glycyrrhizic acid in multitarget agents as potential inhibitors of respiratory influenza viruses
Aarati Ramesh Supekar, Santosh Bhujbal, Rashmi Yadav
DOI: 10.7324/JAPS.2025.197023Pages: 190-204