INTRODUCTION
Cardiovascular diseases (CVDs) constitute one of the most significant public health challenges worldwide, exerting a profound impact on morbidity, mortality, and healthcare systems. The World Health Organization (WHO) reports that CVDs are responsible for a staggering 17.9 million deaths annually, making them the leading cause of mortality globally [1]. The spectrum of CVDs encompasses a diverse array of conditions that affect the heart and blood vessels, each with its unique pathophysiological mechanisms and clinical manifestations [2,3]. Among the most prevalent CVDs are coronary artery disease, hypertension, stroke, heart failure, and peripheral artery disease. These conditions arise from multifaceted pathological processes, including endothelial dysfunction, atherosclerosis, inflammation, thrombosis, and vascular remodeling, which collectively contribute to the disruption of cardiovascular homeostasis [4,5]. Despite advancements in medical science and healthcare delivery, traditional treatment modalities for CVDs primarily focus on managing risk factors, alleviating symptoms, and modifying disease progression [6]. Pharmacological agents such as statins, antiplatelet drugs, and antihypertensive medications have demonstrated efficacy in reducing cardiovascular morbidity and mortality. However, these therapies are not without limitations, as they may be associated with adverse effects, drug interactions, and suboptimal response rates in certain patient populations [7,8]. Moreover, the emergence of drug resistance poses a significant challenge to the long-term effectiveness of existing treatments, highlighting the need for alternative therapeutic approaches [9,10].
Targeting neural precursor cells expressed developmentally downregulated 4 (NEDD4) represents a promising avenue for innovative cardiovascular therapeutics. NEDD4 is a ubiquitin ligase enzyme that plays a pivotal role in the regulation of protein degradation and cellular signaling pathways [11]. Its intricate involvement in these fundamental cellular processes positions it as a key player in maintaining cardiovascular homeostasis [12]. Mounting evidence from preclinical and clinical studies suggests that dysregulation of NEDD4 expression and activity is implicated in the pathogenesis of various cardiovascular disorders. One such disorder is cardiac hypertrophy, a condition characterized by an abnormal enlargement of the heart muscle cells, often in response to chronic stressors such as hypertension or myocardial injury. Dysregulated NEDD4 expression has been linked to aberrant signaling cascades involved in cardiac hypertrophy, leading to adverse structural and functional changes in the heart [13]. Additionally, NEDD4 has been implicated in the development of heart failure, a progressive condition marked by the heart’s inability to pump blood effectively, resulting in symptoms such as fatigue, shortness of breath, and fluid retention [14,15]. By modulating key signaling pathways implicated in heart failure pathogenesis, targeting NEDD4 holds promise for mitigating disease progression and improving cardiac function. Moreover, NEDD4 has emerged as a potential therapeutic target for addressing arrhythmias, which are abnormal heart rhythms that can significantly impair cardiovascular function and increase the risk of adverse cardiac events. Dysregulated NEDD4 activity has been implicated in the modulation of ion channels and cardiac electrical conduction, contributing to the development and maintenance of arrhythmias [16,17]. By targeting NEDD4-mediated signaling pathways, novel therapeutic interventions may offer the potential to restore normal cardiac rhythm and reduce the burden of arrhythmic events in affected individuals.
In addition to targeting NEDD4, natural products have garnered increasing attention as potential sources of bioactive compounds with therapeutic potential for CVDs. Earthworms belonging to the Lumbricus genus have emerged as particularly intriguing sources of bioactive molecules due to their rich biochemical composition and pharmacological properties [18]. With a history of use in traditional medicine systems spanning centuries and across cultures, earthworms have been employed for various ailments, including inflammatory conditions, wounds, and gastrointestinal disorders [19]. This historical precedent underscores the potential of earthworm-derived bioactive compounds in addressing contemporary healthcare challenges, including CVDs. Notably, earthworms have been shown to possess potent fibrinolytic effects attributed to their bioactive component, Lumbrokinase. Lumbrokinase has been reported to exhibit thrombolytic activity, facilitating the breakdown of fibrin clots, and thereby potentially preventing or overcoming cardiac fibrosis, a hallmark of several cardiovascular pathologies [20,21]. The bioactive compounds found in earthworms may exert beneficial effects on cardiovascular health through a variety of mechanisms, including anti-inflammatory, antioxidant, and vasodilatory actions [22,23]. By modulating these pathways, earthworm-derived compounds have the potential to mitigate the underlying pathological processes driving CVDs and improve clinical outcomes for affected individuals. Furthermore, the exploration of earthworm-derived bioactive compounds represents a novel and promising approach to drug discovery and development, offering the potential for the identification of new therapeutic agents with improved efficacy and safety profiles for the treatment of CVDs.
Molecular simulation approaches, including molecular docking and molecular dynamics (MD) simulations, play a crucial role in modern drug discovery, particularly in elucidating protein-protein interactions and facilitating the identification of potential therapeutic agents [24,25]. In the context of CVDs, where the molecular mechanisms underlying pathogenesis are often intricate and multifactorial [26,27], molecular simulation techniques offer valuable insights into the interactions between bioactive molecules and their target proteins. The aims of this study encompassed a comprehensive investigation into the potential therapeutic benefits of targeting NEDD4 in CVDs through interactions with bioactive compounds derived from earthworms (Lumbricus genus). First, we aimed to elucidate the intricate protein-protein interactions between bioactive compounds sourced from earthworms and NEDD4 utilizing molecular docking techniques. By simulating the binding of these compounds to specific regions within the NEDD4 protein structure, we sought to identify potential lead molecules that exhibited high binding affinity and favorable interactions, thereby guiding the selection and optimization of promising therapeutic candidates. Subsequently, our study endeavored to predict the binding affinities and modes of interaction between the identified earthworm-derived compounds and distinct binding sites on the NEDD4 protein. Through molecular docking simulations, we aimed to characterize the molecular recognition events that underlay the formation of stable protein-protein complexes, providing valuable insights into the structural basis of compound-target interactions. Furthermore, we sought to explore the dynamic behavior and structural stability of protein-protein complexes formed between earthworm-derived compounds and NEDD4 using MD simulations. By simulating the movements and interactions of atoms within the protein-protein complexes over time, we aimed to elucidate the conformational dynamics and energetics governing the binding process, thus enhancing our understanding of the underlying mechanisms of action. Moreover, our study aimed to investigate the therapeutic potential of the identified earthworm-derived compounds in the context of CVDs by targeting NEDD4. Through in silico analyses and computational modeling, we aimed to assess the efficacy and selectivity of these compounds in modulating NEDD4 function and mitigating the pathological processes associated with CVDs, thereby laying the groundwork for future preclinical and clinical studies.
MATERIALS AND METHODS
Materials
The protein and peptide sequences sourced from earthworms belonging to the Lumbricus genus were retrieved. To acquire these sequences, a systematic search strategy was employed using UniProt, a comprehensive database of protein sequences and functional information. The search strategy was based on UniProtKB with the MeSH term “Lumbricus”, which allowed for the specific targeting of proteins and peptides associated with the Lumbricus genus. Following the search, the data were meticulously curated to eliminate any duplicate entries, ensuring the integrity and accuracy of the dataset used for subsequent analyses. This process of data curation was crucial for minimizing redundancy and potential biases, thereby enhancing the reliability and validity of the study outcomes. Through rigorous data retrieval and curation procedures, a comprehensive and high-quality dataset was obtained, serving as the foundation for investigations into the bioactive components derived from earthworms and their potential therapeutic applications in CVDs.
Methods
3D structure modeling
Various computational tools were employed to construct the 3D structures of the protein and peptide sequences obtained. This process resulted in the retrieval of a substantial dataset comprising 979 records. The application of rigorous criteria during the selection process of peptide and protein sequences from earthworms (Lumbricus genus) aimed to ensure the quality and reliability of the dataset used for subsequent analyses. Several key criteria were employed to filter and refine the initial dataset obtained, thereby minimizing the likelihood of generating incomplete or inaccurate structural models [28]. Additionally, stringent quality control measures were applied to screen for sequencing errors, ambiguous residues, or other anomalies that could compromise the reliability of subsequent analyses. Sequences failing to meet quality standards were excluded from upholding the overall accuracy and robustness of the dataset [29]. Furthermore, duplicate entries within the dataset were identified and removed to prevent redundancy and potential biases in subsequent analyses. Moreover, ensuring the taxonomic identity of the earthworm species represented in the dataset was crucial for maintaining specificity and relevance to the Lumbricus genus. Taxonomic verification procedures [30] were employed to confirm that all sequences originated from earthworms of the Lumbricus genus, thereby preventing the inclusion of irrelevant or misclassified data.
I-TASSER (Iterative Threading ASSEmbly Refinement) [31] was employed for proteins and peptides that possessed templates in the protein data bank (PDB). I-TASSER is particularly adept at constructing 3D structures when template structures are available in the PDB, leveraging these templates to predict the tertiary structure of the query sequences. For proteins and peptides lacking templates in the PDB, AlphaFold [32] was utilized to generate their 3D structures. It predicted the spatial arrangement of amino acids by iteratively refining its predictions based on attention mechanisms and multiple sequence alignments. Subsequently, the active sites of the proteins and peptides from Lumbricus were analyzed using CASTp 3.0 [33]. Accessing the CASTp 3.0 server through a web interface, protein structures were uploaded and the analysis was initiated. CASTp 3.0 employed algorithms to analyze the protein surfaces and identify potential active sites based on geometric and analytical criteria, generating quantitative measurements such as surface area and volume of identified cavities. Furthermore, the NEDD4 receptor, serving as the target for subsequent docking studies, was retrieved from the PDB with the identifier 2XBB (resolution of 2.68 Å) [34]. This step ensured the availability of the target receptor structure for subsequent molecular docking simulations, facilitating the investigation of protein-protein interactions between the bioactive compounds from Lumbricus and NEDD4.
Protein-protein docking simulation
In this part, we conducted a detailed exploration of the intermolecular interactions between the proteins and peptides sourced from Lumbricus and NEDD4 (target receptor). Utilizing protein-protein docking simulations, our objective was to elucidate key aspects of the binding process, including the identification of crucial residues responsible for forming protein-protein complexes, elucidation of the types of intermolecular interactions involved, determination of binding affinities, assessment of binding modes, and analysis of orientations. To delineate the binding sites of the target receptor, we analyzed protein-protein interactions using PDBSum [35]. Initially, the 3D structures of the target receptor and interacting proteins were retrieved from the PDB and prepared for analysis. Accessing the PDBSum web interface, protein structures were uploaded for detailed analysis. PDBSum employed its algorithms to identify and characterize binding sites between the target receptor and interacting proteins, providing insights into residues involved in binding interfaces, their interactions including hydrogen bonds and van der Waals contacts, and any structural motifs or domains contributing to interaction specificity. This analysis allowed us to gain insights into the spatial arrangement of key residues within the binding site of NEDD4 and their potential interactions with the proteins and peptides from Lumbricus. To ensure the accuracy of our analysis, the receptor was refined using Swiss-PdbViewer v4.1.1 [36] before proceeding with protein-protein docking analysis, ensuring a reliable foundation for our subsequent investigations. To further characterize the potential agonistic or antagonistic properties of the Lumbricus-derived proteins and peptides towards NEDD4, we employed a standard agonist (polyubiquitin, PDB ID: 2XBB) [34] and antagonist (name PDB ID: 8T48) [37] molecules for comparative analysis. This approach enabled us to evaluate whether the proteins and peptides from Lumbricus could potentially act as agonists or antagonists to NEDD4, providing valuable insights into their functional roles in modulating NEDD4 activity.
Subsequently, protein-protein docking calculations were performed using the advanced interface option within the high ambiguity driven protein-protein docking (HADDOCK) stand-alone version [38]. By leveraging this approach, we were able to assess the interactions between the proteins and peptides from Lumbricus and NEDD4, facilitating the prediction of potential binding modes and affinity. The selection of optimal protein-protein docking results for each resultant complex was based on two critical criteria: first, the highest number of clusters or populations observed, indicating the robustness of the predicted interactions, and secondly, the highest docking score (HADDOCK score), which serves as a measure of the strength of the binding affinity between the proteins and peptides from Lumbricus and NEDD4 within the protein-protein complex. The molecular docking results provide a comprehensive overview of the binding affinity, structural parameters, and cluster characteristics of the various NEDD4 complexes formed with earthworm-derived proteins and peptides. Notably, comparisons were drawn with standard agonist and antagonist complexes, NEDD4:Polyubiquitin and NEDD4:N4BP1, respectively, serving as benchmarks for evaluating the efficacy of the Lumbricus-derived compounds. One of the primary indicators of the strength of interaction between molecules is the binding affinity, represented by the ΔG (Gibbs free energy) value. Lower ΔG values indicate stronger binding, implying more stable and favorable interactions between the proteins or peptides and NEDD4.
MD simulation
MD simulations were employed to analyze the dynamics and stability of the protein-protein complexes formed between the proteins and peptides from Lumbricus and NEDD4. The simulations were conducted using GROMACS 2022.5 [39]. To set up the simulations, the optimized potentials for liquid simulations all-atom force field was employed, providing accurate descriptions of molecular interactions within the system [40,40]. The simulation box dimensions were defined according to default cubic box parameters, ensuring adequate space for the biomolecular complexes within the simulation environment. Standard procedures were followed to prepare the input files for the simulations, including the addition of water molecules using the single point charge extended model and the incorporation of counterions to maintain system neutrality [41]. Energy minimization was carried out using the steepest-descent approach to remove steric clashes and relax the system to a stable state. Following energy minimization, a two-phase equilibration process was conducted. In phase 1, the system was equilibrated in the number of particles, volume, and temperature ensemble to control temperature fluctuations and stabilize the system. Subsequently, in phase 2, equilibration was performed in the number of particles, pressure, and temperature ensemble to maintain constant pressure and temperature conditions. Once equilibration was achieved, production MD simulations were carried out for a total of 100 nanoseconds to capture the dynamics of the protein-protein complexes over an extended period. Throughout the simulations, various parameters such as root mean square deviations (RMSD), root mean square fluctuation (RMSF), radius of gyration (RoG), potential energies, and intermolecular hydrogen bonding interactions were monitored and analyzed to assess the stability and conformational dynamics of the complexes. To visualize critical residues and intermolecular interactions within the anticipated protein-protein complexes, manual inspection was conducted using molecular visualization software such as PyMOL [42] and UCSF Chimera [43]. These tools facilitated the visualization and analysis of key structural features and interactions within the simulated complexes, providing valuable insights into the mechanisms underlying the binding and stability of the Lumbricus-derived proteins and peptides with NEDD4.
Molecular mechanics/Poisson–Boltzmann surface area (MM/PBSA) calculations
The MM/PBSA calculation method, which relies on MD simulations, was utilized to explore the protein-protein interactions between proteins and peptides sourced from Lumbricus and NEDD4. MD simulations generated a diverse ensemble of protein conformations, from which representative snapshots were selected for further analysis [44]. The MM/PBSA calculations served as a powerful tool for assessing the thermodynamic stability and binding affinity of the NEDD4 protein-protein complexes, offering valuable insights into their molecular interactions [45]. Each snapshot underwent comprehensive energy calculations, including gas-phase energy calculations, solvation energy estimations employing a continuum solvent model, and entropy calculations. These individual energy contributions were then combined to compute the binding free energy of the protein-protein complex [46,47]47]. To perform these calculations, the gmx_MMPBSA module within the GROMACS simulation package was employed [48,49]49] enabling the efficient and accurate computation of binding free energies for biomolecular complexes. The MM/PBSA method is widely recognized for its capability to predict the binding free energy of protein-protein interactions, making it a valuable tool in elucidating the energetics of biomolecular interactions [50]. The calculation of the MM/PBSA binding free energy is based on the following equation:
ΔG_binding = ΔG_complex – ΔG_proteinX1 - ΔG_proteinX2
where,
ΔG_binding: the binding free energy associated with the formation of the protein-protein complex.
ΔG_complex: the free energy of the fully solvated protein-protein complex.
ΔG_proteinX1: the free energy of protein 1 in its solvated state when unbound.
ΔG_proteinX2: the free energy of protein 2 in its solvated state when unbound.
The binding free energy is determined by computing the difference between the free energy of the complex and the combined free energies of the unbound proteins. This calculation provides insights into the energetic alterations that occur during the formation of the protein-protein complex, thereby elucidating the strength and stability of the interaction.
Statistical analysis
In this study, statistical analysis was conducted to ascertain the relationships among all parameters derived from both molecular docking and MD simulations. The data obtained from these computational experiments were subjected to comprehensive statistical analysis and interpretation using SPSS 25 statistical software [51] and OriginLab Pro 2022 [52]. This analytical approach enabled the exploration of correlations, trends, and patterns within the dataset, providing valuable insights into the relationships between different variables and parameters. Additionally, statistical analysis facilitated the validation of computational results and the identification of significant findings, contributing to a more robust understanding of the molecular interactions under investigation.
RESULTS
3D structure modeling
In the 3D structure modeling phase of the study, 979 substantial datasets were retrieved and evaluated for completeness, with sufficient information for accurate 3D modeling. Thorough comparison and validation of sequence data ensured that each unique protein or peptide was represented only once in the final dataset. The obtained database of protein sequences from earthworms (Lumbricus genus) can be seen in Supplementary Data 1. Subsequently, the 3D structures of these proteins and peptides were generated using advanced computational tools tailored to their structural characteristics. For proteins and peptides with available templates in the PDB, the I-TASSER algorithm was employed for the accurate prediction of tertiary structures based on sequence similarity and structural homology accuracy, even for sequences with no homologous structures available in the PDB. By employing advanced machine learning algorithms trained on vast protein sequence and structure databases, AlphaFold can accurately predict the 3D structures of proteins based solely on their amino acid sequences [32]. The 3D structure modeling phase of the study utilized cutting-edge computational methodologies to generate accurate and reliable structural models for the identified proteins and peptides from earthworms of the Lumbricus genus (Fig. 1).
Protein-protein docking simulation
The protein-protein docking simulations conducted in this study aimed to unravel the intricate interactions between NEDD4 and proteins or peptides sourced from earthworms (Lumbricus genus), with a specific focus on identifying potential therapeutic candidates for CVDs. The best binding pose of standard agonist, standard antagonist, and top-performing protein from Lumbricus was presented in Figure 2. The comparison between the top-performing proteins and peptides derived from Lumbricus and the standard agonist (NEDD4:Polyubiquitin) and antagonist (NEDD4:N4BP1) complexes, based on binding affinity, provides valuable insights into the potential efficacy of Lumbricus-derived bioactive compounds in modulating NEDD4 activity compared to established proteins. First, the standard agonist, NEDD4:Polyubiquitin, demonstrated a binding affinity with a ΔG value of −10.7 kcal/mol and a dissociation constant (Kd) of 3.00e-8 M. This indicates a strong interaction between NEDD4 and Polyubiquitin, highlighting its efficacy as an agonist for NEDD4 activity modulation. In contrast, the standard antagonist, NEDD4:N4BP1, exhibited a slightly lower binding affinity, with a ΔG value of −9.4 kcal/mol and a Kd of 2.20e-7 M. Despite its lower binding affinity compared to the agonist, NEDD4:N4BP1 still displayed a notable interaction with NEDD4, indicative of its efficacy as an antagonist for NEDD4 activity inhibition. Among the Lumbricus-derived complexes, Actin-1, Heat shock protein 70, Lumbrokinase-7T1, and EF-1-alpha emerged as top-performing candidates based on their low values of binding affinity ΔG (kcal/mol). Actin-1 exhibited an ΔG value of −15.2 kcal/mol, surpassing the binding affinity of the standard agonist, NEDD4:Polyubiquitin. This suggests that Actin-1 may possess a stronger interaction with NEDD4, potentially surpassing the efficacy of Polyubiquitin and N4BP1 in modulating NEDD4 activity. Similarly, Heat shock protein 70, Lumbrokinase-7T1, and EF-1-alpha displayed notable binding affinities with ΔG values of −14.1 kcal/mol, −14.1 kcal/mol, and −13.4 kcal/mol, respectively (Table 1 and Fig. 3c). These values compare favorably with the binding affinity of the standard agonist and antagonist, further highlighting the potential of these Lumbricus-derived compounds as effective modulators of NEDD4 activity for cardiovascular disease intervention. The complete docking results can be seen in Supplementary Data 2.
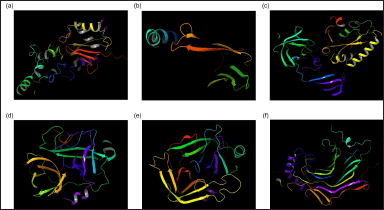 | Figure 1. 3D structure modeling outcomes depicting proteins and peptides derived from the earthworm (Lumbricus genus). (a) Actin-1. (b) Heat shock protein 70. (c) Elongation factor 1-alpha. (d) Lumbrokinase-7T1. (e) Fibrinolytic enzyme. (f) CCF-like protein. [Click here to view] |
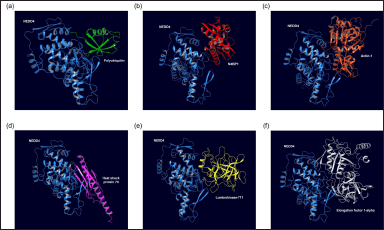 | Figure 2. Molecular docking simulations depict the best binding poses for interactions between proteins from the earthworm (Lumbricus genus) and NEDD4. (a) NEDD4:Polyubiquitin (agonist) complex. (b) NEDD4:N4BP1 (antagonist) complex. (c) NEDD4:Actin-1 complex. (d) NEDD4:Heat shock protein 70 complex. (e) NEDD4:Lumbrokinase-7T1 complex. (f) NEDD4:Elongation factor 1-alpha complex. [Click here to view] |
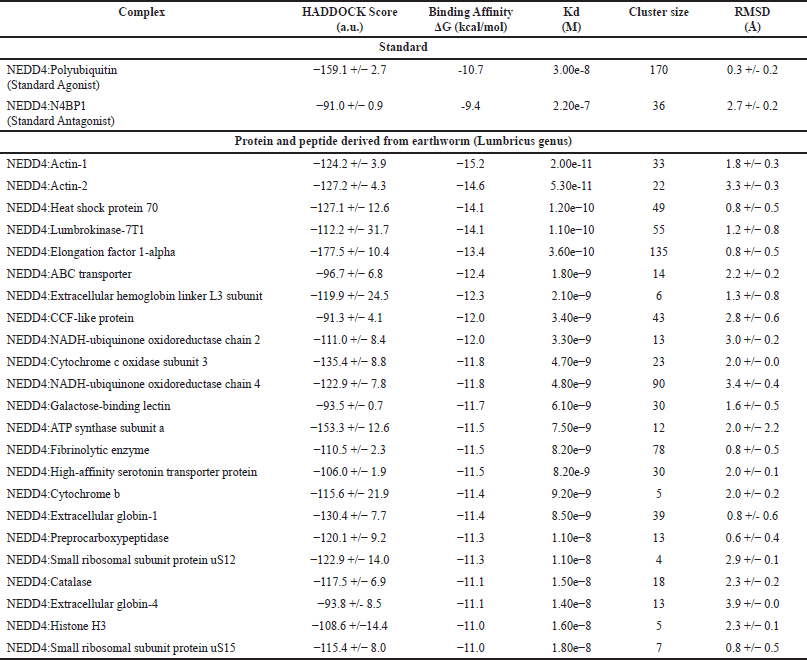 | Table 1. Molecular docking results: binding affinity and structural parameters of NEDD4 interactions with proteins and peptides derived from earthworm (Lumbricus genus). [Click here to view] |
Furthermore, the cluster size and RMSD values offer valuable structural information about the protein-protein complexes. Cluster size refers to the number of distinct conformations or poses observed within the generated ensemble of protein-protein complexes [53]. A larger cluster size suggests a greater structural diversity, indicating the presence of multiple binding modes or orientations between the proteins and peptides from Lumbricus and NEDD4. This structural diversity within the complexes reflects the potential for various interaction configurations, which may influence the functional properties and biological effects of the complexes [54,54]54]. For instance, the standard agonist NEDD4:Polyubiquitin complex exhibits a notably large cluster size of 170, indicating the presence of diverse conformations or binding modes between NEDD4 and polyubiquitin. This diversity suggests the potential for versatile interactions, which could play crucial roles in various cellular processes regulated by ubiquitination. Conversely, the standard antagonist NEDD4:N4BP1 complex has a smaller cluster size of 36, implying fewer distinct binding configurations compared to the agonist complex. This difference in cluster size may reflect the specific nature of the antagonist interaction and its regulatory role in modulating NEDD4 activity. Analyzing the Lumbricus-derived protein/peptide complexes, it is evident that the cluster sizes vary across different interactions. For instance, complexes involving proteins like EF-1-alpha and Fibrinolytic enzyme exhibit relatively larger cluster sizes (135 and 78, respectively), suggesting structural diversity and potential functional versatility in their interactions with NEDD4. On the other hand, complexes such as NEDD4:Extracellular hemoglobin linker L3 subunit and NEDD4:Small ribosomal subunit protein uS12 have smaller cluster sizes (6 and 4, respectively), indicating a more limited range of binding configurations.
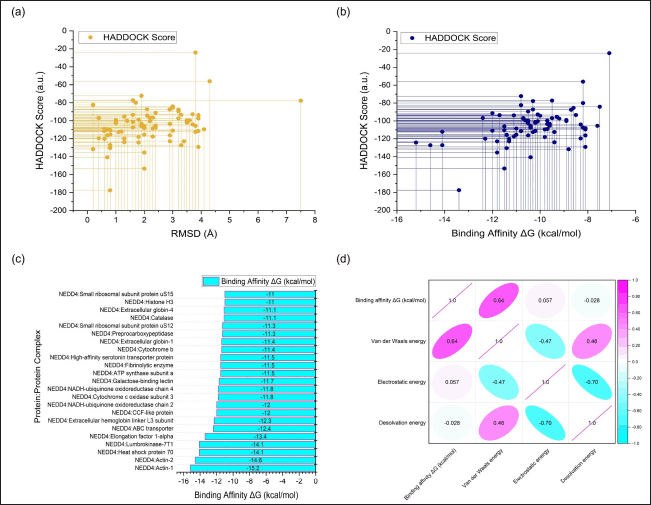 | Figure 3. Overview of molecular docking results. (a) Relationship between HADDOCK score and RMSD. (b) Correlation between HADDOCK score and binding affinity. (c) Binding affinity values of the top-performing protein derived from the earthworm (Lumbricus genus), with a threshold of −11.0 kcal/mol. (d) Correlation matrix depicting the relationship of binding energy (kcal/mol) with individual energy components. [Click here to view] |
On the other hand, RMSD values quantify the structural deviations or differences between individual conformations within the same cluster. Lower RMSD values indicate minimal deviation or closer structural resemblance between different conformations, suggesting a higher degree of stability and consistency in the binding interactions over time [55]. This stability is indicative of the robustness of the protein-protein complexes and their ability to maintain specific structural configurations despite fluctuations or perturbations in the surrounding environment [56]. For instance, the standard agonist NEDD4:Polyubiquitin complex exhibits a low RMSD value of 0.3 Å, indicating minimal structural deviation among its conformations and suggesting a stable and well-defined binding mode. Similarly, Lumbricus-derived complexes like NEDD4:Heat shock protein 70 and NEDD4:Fibrinolytic enzyme show low RMSD values (0.8 Å), indicating stable binding interactions and consistent structural configurations. In contrast, complexes with higher RMSD values, such as NEDD4:Actin-2 (RMSD = 3.3 Å), suggest greater structural variability among their conformations, potentially reflecting dynamic binding interactions with NEDD4. These higher RMSD values could indicate conformational flexibility or transient interactions, which may have implications for the functional roles of these complexes in cellular processes [57].
The statistical analysis conducted to investigate the relationship between the HADDOCK score and RMSD revealed a Pearson correlation coefficient (r) of 0.234. This coefficient indicates the strength and direction of the linear relationship between the two variables. A value of 0.234 suggests a weak positive correlation, meaning that as the HADDOCK score tends to decrease (indicating better docking quality), the RMSD tends to decrease slightly as well. However, it is important to note that the correlation is relatively weak, indicating that other factors beyond the HADDOCK score contribute to the variability in RMSD values. Further analysis revealed that approximately 51.94% of the proteins and peptides derived from Lumbricus demonstrated favorable RMSD values, defined as RMSD values equal to or less than 2.00 Å (Fig. 3a). These favorable RMSD values suggest that a significant portion of the predicted protein-protein complexes exhibit structural conformations that closely resemble experimental or reference structures. This finding underscores the overall success of the docking simulations in accurately predicting the structural arrangements of the complexes. However, despite the majority of proteins and peptides exhibiting favorable RMSD values, the analysis also identified three outlier points characterized by very large RMSD values. These outlier points indicate instances where the predicted structures deviate significantly from the experimental or reference structures. Such deviations may result from various factors, including inaccuracies in the docking algorithm, limitations in the experimental data used as input, or inherent complexities in the protein-protein interactions being studied [58].
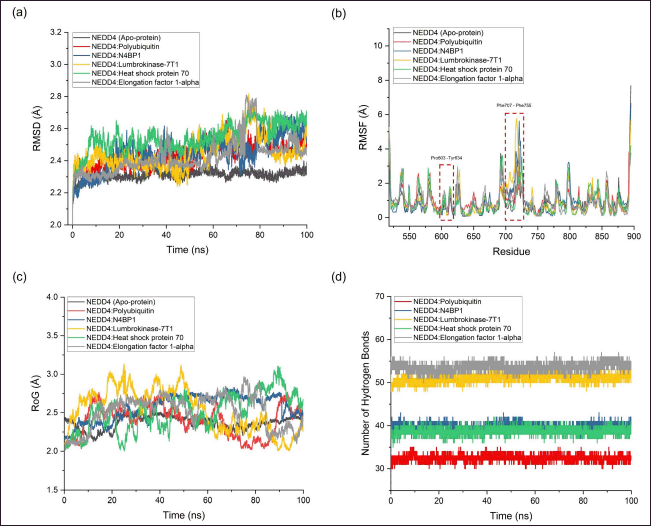 | Figure 4. Analysis of molecular dynamics (MD) simulations for complexes between proteins derived from the earthworm (Lumbricus genus) and NEDD4. (a) Root mean square deviation (RMSD) indicating structural stability, (b) Root mean square fluctuation (RMSF) depicting residue flexibility, (c) Radius of gyration (RoG) illustrating structural compactness, and (d) Number of hydrogen bonds highlighting intermolecular interactions. [Click here to view] |
The statistical analysis was also conducted to examine the relationship between the HADDOCK score and binding affinity (ΔG), revealing a Pearson correlation coefficient (r) of 0.4499 (Fig. 3b). This coefficient indicates the strength and direction of the linear relationship between the two variables. A value of 0.4499 suggests a moderate positive correlation, implying that as the HADDOCK score decreases (reflecting improved docking quality), the binding affinity tends to decrease as well. A moderate positive correlation coefficient like 0.4499 indicates that there is a discernible trend in the relationship between the HADDOCK score and binding affinity. In this case, as the predicted quality of the protein-protein docking improves (reflected by lower HADDOCK scores), there is a tendency for the binding affinity to decrease, indicating stronger binding between the proteins involved in the complex formation. Conversely, higher HADDOCK scores are associated with weaker binding affinity. This finding suggests that the HADDOCK scoring system effectively captures aspects of the protein-protein interaction that influence binding affinity, such as the complementarity of molecular surfaces, electrostatic interactions, and van der Waals forces. The correlation matrix depicted in Figure 3d provided further insight into the relationship between binding energy (kcal/mol) and individual energy components. Specifically, positive correlation scores were observed between binding affinity and both van der Waals energy (correlation score: 0.64) and electrostatic energy (correlation score: 0.057). This positive correlation indicated that as the van der Waals and electrostatic energy components increased, the binding affinity also tended to increase, suggesting a stronger interaction between the proteins involved in the complex formation. Conversely, the correlation between binding affinity and desolvation energy showed a negative value (−0.028), indicating an inverse relationship. This suggested that the binding affinity tended to decrease as the desolvation energy increased. Desolvation energy refers to the energy required to remove solvent molecules from the binding interface, and a higher value implies greater disruption to the solvent molecules surrounding the interacting proteins, potentially weakening the binding affinity [59,60]60]. However, it is essential to interpret this correlation cautiously, as other factors beyond the HADDOCK score may also contribute to variations in binding affinity, such as the specific amino acid residues involved in the binding interface, post-translational modifications, or environmental conditions [61,62]62].
The comprehensive analysis of hydrogen bond interactions between NEDD4 and the top-performing proteins derived from the earthworm (Lumbricus genus) sheds light on the molecular mechanisms underpinning their binding processes (Table 2). These interactions play a crucial role in stabilizing protein-protein complexes and mediating specific recognition between the proteins involved [63]. For instance, in the NEDD4:Polyubiquitin complex, notable hydrogen bond interactions include Glu554(OE2)-Arg74(N), Tyr604(O)-Leu71(N), and Asn628(OD1)-Leu73(N), with interaction distances ranging from 2.68 to 3.08 Å. These interactions are indicative of the complementarity and specificity between the receptor and interacting protein residues, contributing to the overall stability of the complex. Similarly, in the NEDD4:N4BP1 complex, hydrogen bond interactions such as Glu554(OE2)-Lys177(NZ), Glu559(OE2)-Gln144(NE2), and Tyr604(O)-Ser143(OG) are observed, with distances ranging from 2.69 to 3.17 Å. These interactions highlight the key residues involved in mediating the binding between NEDD4 and N4BP1, underscoring the specificity of their interaction. Moreover, in the NEDD4:Actin-1 complex, hydrogen bond interactions between Ala550(O)-Asp180(N), Asp578(O)-Lys285(NZ), and Tyr605(OH)-His74(NE2) are identified, with distances ranging from 2.74 to 3.13 Å. These interactions suggest the involvement of specific residues in facilitating the binding between NEDD4 and Actin-1, potentially influencing the structural conformation and functional properties of the complex. In the NEDD4:Heat shock protein 70 complex, hydrogen bond interactions such as Tyr605(OH)-Lys85(NZ), Gly625(O)-Asn78(ND2), and Asp630(OD1)-Lys80(NZ) are observed, with distances ranging from 2.59 to 3.06 Å. These interactions highlight the role of key residues in mediating the binding between NEDD4 and Heat shock protein 70, providing insights into the molecular basis of their interaction. Furthermore, in the NEDD4:Lumbrokinase-7T1 complex, hydrogen bond interactions between Thr551(OG1)-Arg17(NH1), Cys627(OD1)-Asp23(N), and Tyr634(OH)-Lys20(NZ) are identified, with distances ranging from 2.72 to 3.14 Å. These interactions underscore the importance of specific residues in facilitating the binding between NEDD4 and Lumbrokinase-7T1, potentially modulating their biological functions.
MD simulation
The MD simulation results offer a comprehensive understanding of the behavior of protein-protein complexes formed between Lumbricus-derived proteins and NEDD4. Throughout the 100 ns simulation, NEDD4 maintained a relatively stable conformation, as evidenced by the average RMSD values ranging from 2.310 to 2.741 Å without significant spikes (Fig. 4a). This stability suggests that NEDD4 interactions with both standard agonist and antagonist, as well as Lumbricus-derived proteins, were dynamically consistent over the simulation period. When analyzing the RMSD values, which indicate the deviation of protein structures from their initial conformations, notable differences emerge among the complexes. For instance, the NEDD4:Polyubiquitin (Standard Agonist) complex displays a slightly higher average RMSD (2.439 Å) compared to apo-protein NEDD4 (2.310 Å). This observation suggests a moderate increase in structural flexibility upon the binding of the standard agonist, indicating potential conformational adjustments required for effective binding. Conversely, the RMSD value for the NEDD4:N4BP1 (Standard Antagonist) complex (2.442 Å) remains similar to that of the apo-protein, indicating that the binding of the antagonist may not significantly perturb the structural stability of NEDD4. Furthermore, the Lumbricus-derived protein complexes, including Actin-1, Actin-2, Heat shock protein 70, Lumbrokinase-7T1, and EF-1-alpha, exhibit slightly higher average RMSD values ranging from 2.424 to 2.741 Å compared to the standard complexes. These differences suggest potential variations in the dynamic behavior and conformational changes induced by the binding of Lumbricus-derived proteins. The higher RMSD values imply that these Lumbricus-derived proteins may interact with NEDD4 in a manner that elicits different structural adjustments or conformational dynamics compared to the standard agonist and antagonist.
During MD simulations, the RMSF analysis was conducted to evaluate the flexibility profile of individual amino acid residues within NEDD4. The obtained average RMSF values ranged from 0.310 to 0.577 Å, indicating moderate flexibility across different regions of the protein. RMSF analysis offers crucial insights into the mobility and flexibility of specific residues within protein structures, shedding light on their functional roles [64]. Upon investigating the interaction of the top-performing protein complex from Lumbricus with NEDD4, it became evident that this interaction led to the disruption of hydrogen bonds with specific residues, particularly in the region spanning amino acid residues Pro603 to Tyr634 and Phe707 to Phe755 (Fig. 4b). Notably, these regions represent the active binding site of NEDD4 as a target receptor [34]. The disruption of hydrogen bonds in these critical areas resulted in higher residue fluctuations compared to the NEDD4 agonist and apo-protein complex. The observed increase in residue fluctuations suggests enhanced mobility and flexibility of these residues upon binding of the top-performing protein from Lumbricus to NEDD4. Interestingly, this pattern of increased flexibility mirrors that of N4BP1, a standard antagonist. The resemblance in flexibility patterns at these specific residues indicates that top-performing proteins derived from Lumbricus have the potential to act as inhibitors akin to standard antagonists. This finding holds significant implications, as it suggests that Lumbricus-derived proteins could modulate the activity of NEDD4 by acting as inhibitors, similar to known antagonists. By disrupting hydrogen bonds and inducing higher flexibility in crucial binding site residues, these proteins may interfere with the functional interactions of NEDD4, offering promising avenues for the development of therapeutic interventions targeting NEDD4-mediated cellular processes.
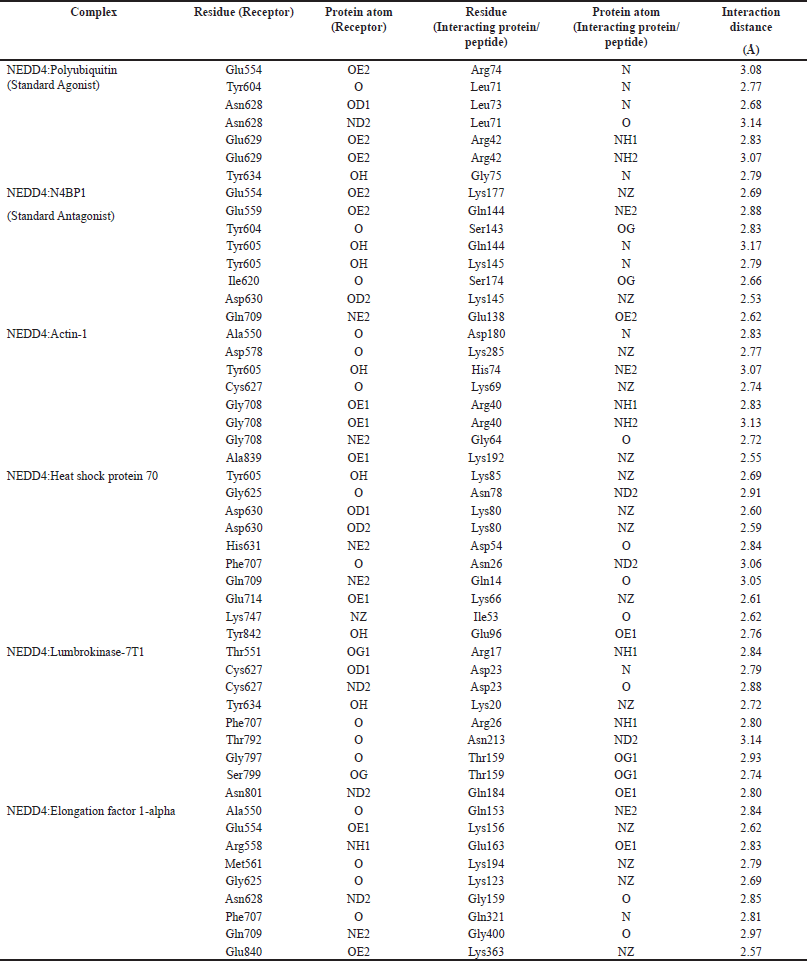 | Table 2. Comprehensive analysis of hydrogen bond interactions between NEDD4 (within the binding sites) and top-performing proteins derived from earthworm (Lumbricus genus), highlighting residues and atoms involved in the binding process. [Click here to view] |
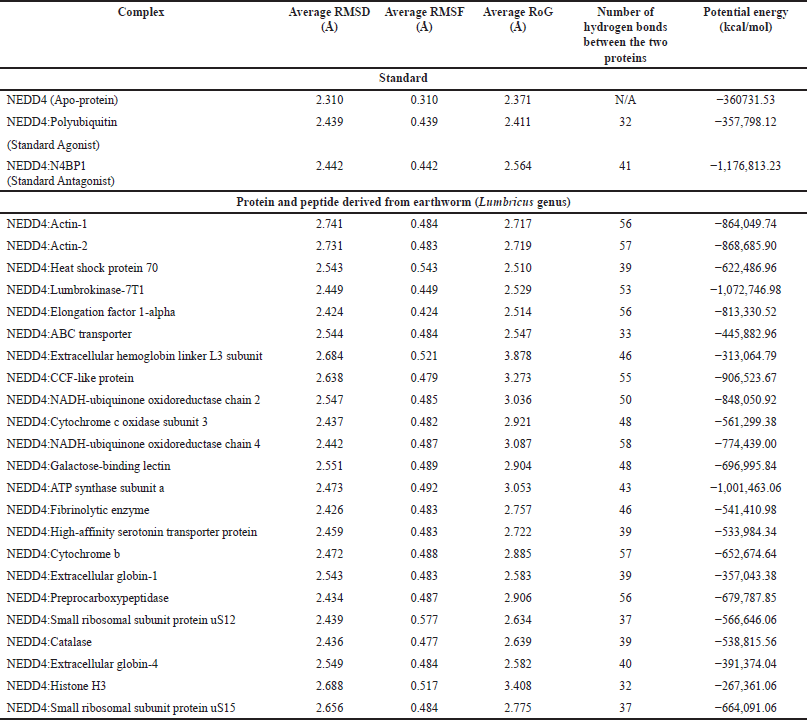 | Table 3. Time-averaged structural properties obtained from the MD simulations of NEDD4 protein-protein complexes. [Click here to view] |
The RoG provided insights into the compactness or the extent of the spreading of protein structures during MD simulations [65]. For the NEDD4 complexes, the average RoG values ranged from 2.371 to 2.564 Å. The RoG values for the standard NEDD4 agonist (Polyubiquitin) and antagonist (N4BP1) complexes were 2.411 Å and 2.564 Å, respectively. In contrast, for the Lumbricus-derived protein complexes, RoG values ranged from 2.510 to 2.529 Å, slightly higher than the standard agonist but comparable to the antagonist. The number of hydrogen bonds formed between the two interacting proteins was indicative of the strength and specificity of their interaction. For the standard complexes, the NEDD4:Polyubiquitin agonist complex formed 32 hydrogen bonds, while the NEDD4:N4BP1 antagonist complex formed 41 hydrogen bonds. Comparatively, the Lumbricus-derived protein complexes displayed similar or slightly higher numbers of hydrogen bonds, with values ranging from 39 to 56. Notably, the NEDD4:Lumbrokinase-7T1 complex formed the highest number of hydrogen bonds (53), followed closely by the NEDD4: EF-1-alpha complex (56) (Table 3). These results suggested that the interactions between NEDD4 and the Lumbricus-derived proteins were characterized by comparable or slightly higher numbers of hydrogen bonds compared to standard interactions. This indicated strong and specific binding between these proteins, potentially contributing to their functional relevance. Additionally, the RoG values suggested that the Lumbricus-derived protein complexes exhibited similar compactness or spreading compared to standard complexes, implying that they maintained stable structural conformations during the simulation period.
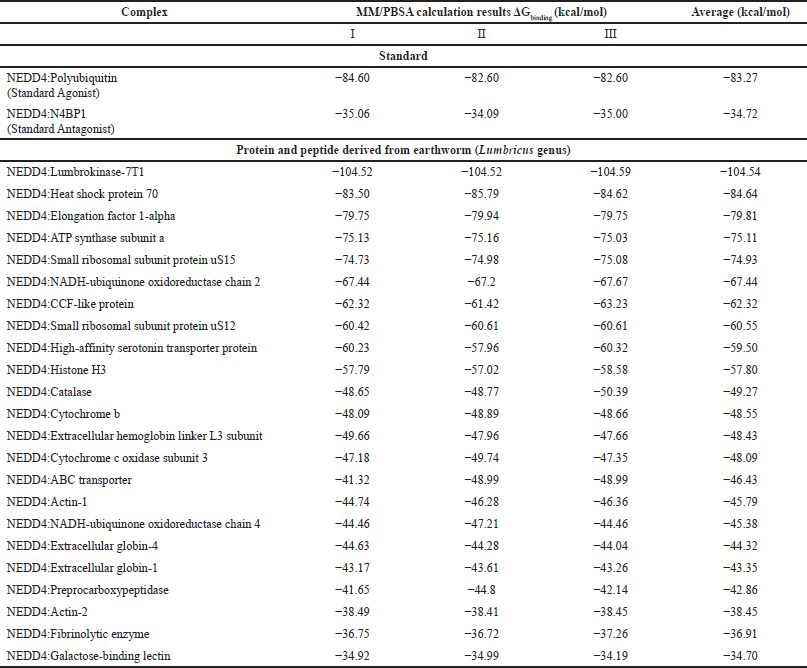 | Table 4. Result of the MM/PBSA calculations for the NEDD4 protein−protein complexes. The mean ΔGbinding is presented with standard deviation in units of kcal/mol. [Click here to view] |
MM/PBSA calculations
The strength of the protein-protein interactions was judged by determining the mean ΔGbinding values, which represent the binding free energy, for each complex. In the case of the standard complexes, NEDD4 bound to Polyubiquitin (Standard Agonist) demonstrated a mean ΔGbinding of −83.27 kcal/mol, indicating a substantial level of stability in the interaction. Conversely, when bound to N4BP1 (Standard Antagonist), NEDD4 exhibited a lower mean ΔGbinding of −34.72 kcal/mol, suggesting a comparatively weaker interaction. However, the Lumbricus-derived protein complexes presented intriguing findings, showcasing similar or even more negative mean ΔGbinding values compared to the standard complexes. Notably, NEDD4 complexed with Lumbrokinase-7T1 displayed a notably high mean ΔGbinding of −104.54 kcal/mol, indicative of a robust and energetically favorable interaction. This suggests that the Lumbricus-derived protein has a strong affinity for NEDD4, potentially surpassing the binding strength observed with the standard agonist and antagonist. Furthermore, the complexes formed between NEDD4 and Heat shock protein 70 and EF-1-alpha exhibited mean ΔGbinding values of −84.64 kcal/mol and −79.81 kcal/mol, respectively (Table 4). These values suggest strong binding affinities comparable to or even exceeding those observed in the standard complexes. This implies that the Lumbricus-derived proteins possess significant potential in modulating NEDD4 activity, potentially rivaling or surpassing the efficacy of standard proteins.
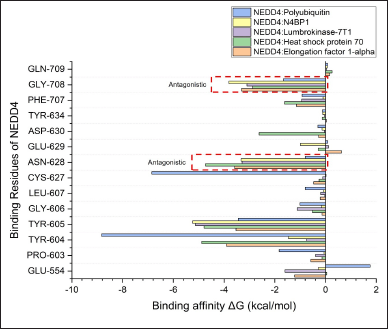 | Figure 5. Binding free energy analysis of individual amino acids in NEDD4 interactions with Polyubiquitin (standard agonist), N4BP1 (standard antagonist), and top three proteins derived from the earthworm (Lumbricus genus), as determined by MM/PBSA calculation. [Click here to view] |
Furthermore, a granular examination of the binding free energy of individual amino acid residues within NEDD4 provided deeper insights into the molecular mechanisms governing binding specificity and affinity. Among these residues, Gly708 and Asn628 emerged as particularly influential in dictating the antagonistic activity observed within the complexes. In the context of the interactions between NEDD4 and the top-performing proteins from Lumbricus, these specific residues exhibited notably high binding affinity values compared to the standard agonist complex. The heightened binding affinity of Gly708 and Asn628 of top-performing proteins suggests their pivotal roles in mediating the antagonistic effects observed in the protein-protein complexes (Fig. 5). These residues likely participate in critical interactions that govern the stability and specificity of the complexes, thereby influencing their overall functional outcomes. The elevated affinity observed in the Lumbricus-derived protein complexes underscores the significance of these interactions in modulating NEDD4 activity and highlights their potential as key determinants of therapeutic efficacy. By elucidating the contributions of individual amino acid residues to the binding energetics, this analysis provides valuable insights into the structural basis of protein-protein interactions. The identification of Gly708 and Asn628 as key contributors to the antagonistic activity enhances our understanding of the molecular determinants underlying the complex interplay between NEDD4 and its interacting partners. These findings pave the way for targeted manipulation of specific residues to modulate NEDD4 function effectively, offering promising avenues for the development of therapeutic interventions targeting NEDD4-associated pathways.
DISCUSSION
The study employed a multifaceted approach to investigate the interactions between NEDD4 and proteins and peptides derived from Lumbricus earthworms, with a specific focus on their potential therapeutic applications in CVDs. Building upon the foundation of robust data collection and stringent quality control measures, the study utilized advanced computational tools and methodologies to model 3D structures, conduct protein-protein docking simulations, perform MD simulations, and calculate binding free energies. In the 3D structure modeling phase, the study leveraged the power of bioinformatics and computational biology to generate accurate and reliable structural models of Lumbricus-derived proteins and peptides. By systematically collecting sequences from the UniProt database and applying rigorous selection criteria, the study ensured the quality and relevance of the dataset. This approach was consistent with previous studies that utilized bioinformatics tools to predict protein structures with high accuracy [66]. Moreover, the utilization of both I-TASSER and AlphaFold algorithms allowed for comprehensive coverage of proteins and peptides, whether they had homologous structures in the PDB or not, thereby maximizing the scope and depth of the structural modeling efforts [32].
The study delved into the intricate dynamics between NEDD4 and Lumbricus-derived compounds through protein-protein docking simulations to uncover potential therapeutic avenues for CVDs. By meticulously comparing the binding affinities of Lumbricus-derived proteins against standard agonist and antagonist complexes, the research pinpointed promising candidates that demonstrated comparable or heightened efficacy in modulating NEDD4 activity. This discovery resonated with prior investigations that underscored the therapeutic promise of natural compounds in the realm of cardiovascular disease management. Notably, previous literature has highlighted the medicinal potential of Lumbricus earthworm, particularly its primary constituent, Lumbrokinase, in the treatment of cardiovascular ailments [67]. By elucidating the molecular intricacies of Lumbrokinase’s interaction with the NEDD4 pathway, this study contributes novel insights into the therapeutic potential of Lumbrokinase for cardiovascular treatment. Additionally, the analysis of cluster size and RMSD values provided insights into the structural diversity and stability of protein-protein complexes, offering valuable information for rational drug design and optimization [68]. MD simulations further elucidated the dynamic behavior of protein-protein complexes over time, uncovering potential mechanisms for modulating NEDD4 activity. The study provided detailed insights into the structural dynamics and functional implications of NEDD4-therapeutic protein interactions by examining residue flexibility and hydrogen bond interactions. This aligned with previous studies that had used MD simulations to investigate protein-protein interactions and elucidate their dynamic behavior [69]. The study observed that Lumbrokinase, heat shock protein, and EF-1-alpha demonstrated sustained interactions with the NEDD4 protein throughout the simulation period, akin to the behavior exhibited by the standard antagonist complex. Furthermore, the study discussed the structural features and key interaction residues within these protein-protein complexes, elucidating the molecular basis underlying their binding affinity and activity. By analyzing the conformational changes and intermolecular forces at play during the MD simulations, the study highlighted the structural motifs and binding pockets crucial for stabilizing the interactions between the Lumbricus-derived compounds and NEDD4. The resemblance in activity between these Lumbricus-derived compounds and the standard antagonist suggests that they may function through similar mechanisms or binding modes, warranting further investigation into their therapeutic potential.
Our findings align with previous research indicating that heat shock proteins (Hsp), such as Hsp70, can modulate the ubiquitination process mediated by NEDD4. Specifically, it has been shown that Hsp70 plays a critical role in regulating the ubiquitination and degradation of p63 isoforms by CHIP (C-terminus of Hsc70-interacting protein), thereby influencing their stability in cells. By inhibiting CHIP-mediated ubiquitination, Hsp70 helps maintain the stability of TAp63 or ΔNp63 isoforms, which are implicated in various cellular processes, including cell proliferation, differentiation, and apoptosis [70]. This previous study provides valuable insights into the regulatory mechanisms governing protein ubiquitination and degradation pathways, highlighting the intricate interplay between Hsp and E3 ubiquitin ligases like NEDD4. Similarly, another study demonstrated that EF-1 can inhibit the ubiquitin ligase activity of SIAH-1 (Seven In Absentia Homolog 1), further corroborating the findings of the current study based on MD simulations. EF-1, a highly conserved protein involved in protein synthesis, exerts its inhibitory effect on SIAH-1-mediated ubiquitination by competing for binding to the E3 ligase substrate-binding site [71]. By blocking the interaction between SIAH-1 and its substrates, EF-1 interferes with the ubiquitination process, thereby modulating the stability and turnover of target proteins involved in cellular homeostasis and signaling pathways. This study’s findings complement the emerging evidence implicating protein elongation factors in the regulation of ubiquitin-mediated protein degradation pathways, highlighting their multifaceted roles beyond translation elongation. Together, these previous studies provide additional support for the current study’s findings regarding the inhibitory effects of Lumbricus-derived compounds, such as heat shock protein and EF-1-alpha, on NEDD4 ubiquitin ligase activity. By elucidating the molecular mechanisms underlying these interactions, the collective body of research contributes to our understanding of the intricate regulatory networks governing protein homeostasis and cellular function, with implications for therapeutic interventions in various diseases, including cancer and cardiovascular disorders. The comprehensive approach and integration of diverse computational techniques in this study represented a significant advancement in the field of drug discovery and protein engineering. By combining bioinformatics, molecular modeling, and simulation methodologies, the study provided a holistic understanding of NEDD4-therapeutic protein interactions and offered valuable insights into the development of novel therapeutics for CVDs. Moreover, the findings underscored the importance of computational approaches in accelerating the drug discovery process and optimizing therapeutic interventions.
LIMITATIONS AND FUTURE WORKS
Despite the promising findings of this study, several limitations need to be acknowledged. First, while computational approaches like MD simulations provide valuable insights into protein interactions, they are inherently limited by simplifications and approximations in the underlying models. The accuracy of these simulations relies heavily on the parameters and force fields used, which may not fully capture the complexity of biological systems. Additionally, the predictive power of computational modeling depends on the availability and quality of experimental data for validation, which can be limited in the case of newly discovered or understudied proteins and peptides. Furthermore, the study primarily focused on in silico analyses, and the predicted interactions between Lumbricus-derived compounds and NEDD4 have yet to be experimentally validated. Experimental techniques such as protein-protein interaction assays, enzyme activity assays, and cellular studies are necessary to confirm the binding affinities and functional effects of these compounds on NEDD4 activity. Moreover, investigating the pharmacokinetic and pharmacodynamic properties of Lumbricus-derived compounds, including their stability, bioavailability, and efficacy in relevant disease models, is essential for translating these findings into clinical applications.
In terms of future directions, there are several avenues for further research. First, conducting comprehensive experimental validation studies to confirm the predicted interactions between Lumbricus-derived compounds and NEDD4 would strengthen the credibility of the computational findings. This could involve biochemical assays, structural biology techniques, and cell-based assays to characterize the binding kinetics, specificity, and functional consequences of these interactions. Additionally, exploring the therapeutic potential of Lumbricus-derived proteins in preclinical models of CVDs, such as animal models of hypertension, atherosclerosis, and myocardial infarction, would provide valuable insights into their efficacy and safety profiles in relevant physiological contexts. Moreover, investigating the mechanisms underlying the observed similarities between Lumbricus-derived compounds and standard antagonists in modulating NEDD4 activity could uncover novel regulatory pathways and therapeutic targets. This could involve elucidating the structural determinants of binding specificity and affinity, exploring the downstream signaling cascades affected by NEDD4 inhibition, and identifying potential synergistic interactions with existing pharmacological agents. Additionally, considering the multifaceted roles of NEDD4 in various cellular processes beyond CVDs, such as cancer, neurodegenerative disorders, and immune regulation, could expand the scope of potential therapeutic applications for Lumbricus-derived proteins.
CONCLUSION
In conclusion, this study offered a comprehensive exploration of the interactions between selected Lumbricus-derived proteins and NEDD4, shedding light on their potential therapeutic applications in CVDs. Through a multifaceted approach combining bioinformatics, computational modeling, and MD simulations, valuable insights were gained into the structural features and binding affinities of Lumbricus-derived proteins and peptides with NEDD4. The findings suggested that certain compounds, such as Lumbrokinase, heat shock protein, and EF-1-alpha, exhibited similar activities to standard antagonists in modulating NEDD4 activity, highlighting their potential as therapeutic candidates for CVDs. Moreover, the study underscored the importance of computational approaches in drug discovery and development, particularly in the context of natural products with diverse chemical structures and biological activities. By leveraging advanced computational tools and methodologies, the challenges associated with traditional drug discovery approaches were overcome, and a large number of candidate compounds were efficiently screened for their potential interactions with NEDD4. This approach not only accelerated the drug discovery process but also provided valuable mechanistic insights into the mode of action of these compounds at the molecular level. However, it is important to acknowledge the limitations of this study, including the reliance on computational predictions that require experimental validation. Future research should focus on corroborating the findings through biochemical assays, structural biology techniques, and preclinical studies in relevant disease models. Furthermore, exploring the therapeutic potential of Lumbricus-derived proteins beyond CVDs and investigating their synergistic interactions with existing pharmacological agents could open up new avenues for drug development and personalized medicine.
LIST OF ABBREVIATIONS
CVDs, Cardiovascular diseases; EF-1, Elongation factor 1; HADDOCK, High ambiguity-driven protein-protein docking; Hsp: Heat shock proteins; I-TASSER, Iterative threading assembly refinement; MD, Molecular dynamics; MM/PBSA, Molecular mechanics/poisson-boltzmann surface area; NEDD4, Neural precursor cell expressed developmentally downregulated 4; PDB, Protein data bank; RMSD, Root mean square deviations; RMSF, Root mean square fluctuation; RoG, Radius of gyration.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be authors as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
The authors extend their appreciation to the Deanship of Scientific Research, Imam Mohammad Ibn Saud Islamic University (IMSIU), Saudi Arabia, for funding this research work through Grant No. (221413005).
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors, and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declare that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. WHO. Cardiovascular diseases (CVDs): Geneva, Switzerland: World Health Organization; 2021 [cited 2024 April 1]. Available from: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds)
2. Roth Gregory A, Mensah George A, Johnson Catherine O, Addolorato G, Ammirati E, Baddour Larry M, et al. Global burden of cardiovascular diseases and risk factors, 1990–2019. J Am Coll Cardiol. 2020;76(25):2982–3021. CrossRef
3. Del Buono MG, Montone RA, Camilli M, Carbone S, Narula J, Lavie CJ, et al. Coronary microvascular dysfunction Across the Spectrum of Cardiovascular Diseases: JACC State-of-the-Art Review. J Am Coll Cardiol. 2021;78(13):1352–71. CrossRef
4. Gimbrone MA, García-Cardeña G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118(4):620–36. CrossRef
5. Thiriet M. Cardiovascular disease: an introduction. Vasculopathies. 2019;8(1):1–90. CrossRef
6. Ullah A, Kumar M, Sayyar M, Sapna F, John C, Memon S, et al. Revolutionizing cardiac care: a comprehensive narrative review of cardiac rehabilitation and the evolution of cardiovascular medicine. Cureus. 2023;15(10):1–11. CrossRef
7. Jansen-Chaparro S, López-Carmona MD, Cobos-Palacios L, Sanz-Cánovas J, Bernal-López MR, Gómez-Huelgas R. Statins and peripheral arterial disease: a narrative review. Front Cardiovasc Med. 2021;8:1–18. CrossRef
8. Rousan TA, Thadani U. Stable angina medical therapy management guidelines: a critical review of guidelines from the European Society of Cardiology and National Institute for Health and Care Excellence. Eur Cardiol. 2019;14(1):18–22. CrossRef
9. Kario K, Hoshide S, Narita K, Okawara Y, Kanegae H, Aoki K, et al. Cardiovascular prognosis in drug-resistant hypertension stratified by 24-hour ambulatory blood pressure: The JAMP Study. Hypertension. 2021;78(6):1781–90. CrossRef
10. Wijkman MO, Malachias MVB, Claggett BL, Cheng S, Matsushita K, Shah AM, et al. Resistance to antihypertensive treatment and long-term risk: the atherosclerosis risk in communities study. J Clin Hypertens (Greenwich). 2021;23(10):1887–96. CrossRef
11. Boase NA, Kumar S. NEDD4: the founding member of a family of ubiquitin-protein ligases. Gene. 2015;557(2):113–22. CrossRef
12. Li M, Sun G, Wang P, Wang W, Cao K, Song C, et al. Research progress of Nedd4L in cardiovascular diseases. Cell Death Discov. 2022;8(1):1–10. CrossRef
13. Goto J, Otaki Y, Watanabe T, Watanabe M. The role of HECT-Type E3 ligase in the development of cardiac disease. Int J Mol Sci. 2021;22(11):1–26. CrossRef
14. Hauck L, Stanley-Hasnain S, Fung A, Grothe D, Rao V, Duncan G, et al. Cardiac-specific ablation of the E3 ubiquitin ligase Mdm2 leads to oxidative stress, broad mitochondrial deficiency and early death. PLOS One. 2017;12(12):1–35. CrossRef
15. Kiryluk K, Isom R. Thiazolidinediones and fluid retention. Kidney Int. 2007;72(15):762–8. CrossRef
16. Reisqs JB, Qu YS, Boutjdir M. Ion channel trafficking implications in heart failure. Front Cardiovasc Med. 2024;11(1):1–16. CrossRef
17. Daimi H, Lozano-Velasco E, Aranega A, Franco D. Genomic and non-genomic regulatory mechanisms of the cardiac sodium channel in cardiac arrhythmias. Int J Mol Sci. 2022;23(3):1–45. CrossRef
18. Mustafa R, Saiqa A, Domínguez J, Jamil M, Manzoor S, Wazir S, et al. Therapeutic values of earthworm species extract from Azad Kashmir as anticoagulant, antibacterial, and antioxidant agents. Can J Infect Dis Med Microbiol. 2022;2022(1):1–20. CrossRef
19. Cooper EL, Balamurugan M, Huang CY, Tsao CR, Heredia J, Tommaseo-Ponzetta M, et al. Earthworms dilong: ancient, inexpensive, noncontroversial models may help clarify approaches to integrated medicine emphasizing neuroimmune systems. Evid Based Complement Alternat Med. 2012;2012(1):1–11. CrossRef
20. Dong Y, Woo YM, Lee YH, Ahn MY, Lee DG, Lee SH, et al. Data on the potent fibrinolytic effects of the Lumbricus rubellus earthworm and the Perinereis linea lugworm. Data Brief. 2019;26(1):1–7. CrossRef
21. Lai CH, Han CK, Shibu MA, Pai PY, Ho TJ, Day CH, et al. Lumbrokinase from earthworm extract ameliorates second-hand smoke-induced cardiac fibrosis. Environ Toxicol. 2015;30(10):1216–25. CrossRef
22. Chen M, Wang L, Zheng C, Ma A, Hu K, Xiang A, et al. Novel ACE inhibitory peptides derived from bighead carp (Aristichthys nobilis) hydrolysates: screening, inhibition mechanisms and the bioconjugation effect with graphene oxide. Food Biosci. 2023;52:1–14. CrossRef
23. Prakash M, Balamurugan M, Parthasarathi K, Gunasekaran G, Cooper E, Ranganathan L. Anti-ulceral and anti-oxidative properties of “earthworm paste” of Lampito mauritii (Kinberg) on Rattus Norvegicus. Eur Rev Med Pharmacol Sci. 2006;11:9–15.
24. Dermawan D, Prabowo BA, Rakhmadina CA. In silico study of medicinal plants with cyclodextrin inclusion complex as the potential inhibitors against SARS-CoV-2 main protease (Mpro) and spike (S) receptor. Inform Med Unlocked. 2021;25:1–18. CrossRef
25. Challapa-Mamani MR, Tomás-Alvarado E, Espinoza-Baigorria A, León-Figueroa DA, Sah R, Rodriguez-Morales AJ, et al. Molecular docking and molecular dynamics simulations in related to Leishmania donovani: an update and literature review. Trop Med Infect Dis. 2023;8(10):1–13. CrossRef
26. Sánchez-Gloria JL, Arellano-Buendía AS, Juárez-Rojas JG, García-Arroyo FE, Argüello-García R, Sánchez-Muñoz F, et al. Cellular mechanisms underlying the cardioprotective role of allicin on cardiovascular diseases. Int J Mol Sci. 2022;23(16):1–21. CrossRef
27. Choudhury TZ, Garg V. Molecular genetic mechanisms of congenital heart disease. Curr Opin Genet Dev. 2022;75:1–13. CrossRef
28. Chen S, Fan G, Li J. Improving completeness and accuracy of 3D point clouds by using deep learning for applications of digital twins to civil structures. Adv Eng Inform. 2023;58:1–13. CrossRef
29. Davis EM, Sun Y, Liu Y, Kolekar P, Shao Y, Szlachta K, et al. SequencErr: measuring and suppressing sequencer errors in next-generation sequencing data. Genome Biol. 2021;22(1):1–18. CrossRef
30. Odom AR, Faits T, Castro Nallar E, Crandall KA, Johnson WE. Metagenomic profiling pipelines improve taxonomic classification for 16S amplicon sequencing data. Sci Rep. 2023;13(1):1–12. CrossRef
31. Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(40):1–8. CrossRef
32. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–9. CrossRef
33. Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46(1):363–7. CrossRef
34. Maspero E, Mari S, Valentini E, Musacchio A, Fish A, Pasqualato S, et al. Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 2011;12(4):342–9. CrossRef
35. Laskowski RA, Jab?o?ska J, Pravda L, Va?eková RS, Thornton JM. PDBsum: structural summaries of PDB entries. Protein Sci. 2018;27(1):129–34. CrossRef
36. Johansson MU, Zoete V, Michielin O, Guex N. Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics. 2012;13(173):1–10. CrossRef
37. Gitlin AD, Maltzman A, Kanno Y, Heger K, Reja R, Schubert AF, et al. N4BP1 coordinates ubiquitin-dependent crosstalk within the IκB kinase family to limit Toll-like receptor signaling and inflammation. Immunity. 2024;57(5):973–86. CrossRef
38. Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: a protein−protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–7. CrossRef
39. Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–54. CrossRef
40. Robertson MJ, Tirado-Rives J, Jorgensen WL. Improved peptide and protein torsional energetics with the OPLSAA force field. J Chem Theory Comput. 2015;11(7):3499–509. CrossRef
41. Yuet P, Blankschtein D. Molecular dynamics simulation study of water surfaces: comparison of flexible water models. J Phys Chem B. 2010;114:13786–95. CrossRef
42. Schrödinger. The PyMOL Molecular Graphics System. 2.4 ed 2020.
43. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. CrossRef
44. Tian S, Sun H, Pan P, Li D, Zhen X, Li Y, et al. Assessing an ensemble docking-based virtual screening strategy for kinase targets by considering protein flexibility. J Chem Inf Model. 2014;54(10):2664–79. CrossRef
45. Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51(1):69–82. CrossRef
46. Yuan Z, Chen X, Fan S, Chang L, Chu L, Zhang Y, et al. Binding free energy calculation based on the fragment molecular orbital method and its application in designing novel SHP-2 allosteric inhibitors. Int J Mol Sci. 2024;25(1):1–24.
47. Rifai EA, Ferrario V, Pleiss J, Geerke DP. Combined linear interaction energy and alchemical solvation free-energy approach for protein-binding affinity computation. J Chem Theory Comput. 2020;16(2):1300–10. CrossRef
48. Valdés-Tresanco MS, Valdés-Tresanco ME, Valiente PA, Moreno E. gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J Chem Theory Comput. 2021;17(10):6281–91. CrossRef
49. Miller BR 3rd, McGee TD Jr, Swails JM, Homeyer N, Gohlke H, Roitberg AE. MMPBSA.py: an efficient program for end-state free energy calculations. J Chem Theory Comput. 2012;8(9):3314–21. CrossRef
50. Panday SK, Alexov E. Protein-protein binding free energy predictions with the MM/PBSA approach complemented with the gaussian-based method for entropy estimation. ACS Omega. 2022;7(13):11057–67. CrossRef
51. IBM. IBM SPSS Statistics for Windows. Version 25.0 ed. New York, NY: IBM Corp; 2017.
52. OriginLab. Origin(Pro). 2022 ed. Northampton, MA: OriginLab Corporation; 2022.
53. Mohammadi S, Narimani Z, Ashouri M, Firouzi R, Karimi-Jafari MH. Ensemble learning from ensemble docking: revisiting the optimum ensemble size problem. Sci Rep. 2022;12(410):1–15. CrossRef
54. Shyamal M, Mandal TK, Panja A, Saha A. Influence of anionic co-ligands on the structural diversity and catecholase activity of copper(II) complexes with 2-methoxy-6-(8-iminoquinolinylmethyl)phenol. RSC Advances. 2014;4:53520–30.
55. Kufareva I, Abagyan R. Methods of protein structure comparison. Methods Mol Biol. 2012;857:231–57. CrossRef
56. Russell R, Alber F, Aloy P, Davis F, Korkin D, Pichaud M, et al. A structural perspective on protein-protein interactions. Curr Opin Struct Biol. 2004;14:313–24. CrossRef
57. Dagliyan O, Proctor EA, D’Auria KM, Ding F, Dokholyan NV. Structural and dynamic determinants of protein-peptide recognition. Structure. 2011;19(12):1837–45. CrossRef
58. Guedes IA, Pereira FSS, Dardenne LE. Empirical scoring functions for structure-based virtual screening: applications, critical aspects, and challenges. Front Pharmacol. 2018;9:1–18. CrossRef
59. Zhou HX, Pang X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem Rev. 2018;118(4):1691–741. CrossRef
60. Misra VK, Hecht JL, Yang AS, Honig B. Electrostatic contributions to the binding free energy of the λcI repressor to DNA. Biophys J. 1998;75(5):2262–73. CrossRef
61. Kastritis PL, Rodrigues JPGLM, Bonvin AMJJ. HADDOCK2P2I: a biophysical model for predicting the binding affinity of protein–protein interaction inhibitors. J Chem Inf Model. 2014;54(3):826–36. CrossRef
62. Kastritis PL, Bonvin AM. On the binding affinity of macromolecular interactions: daring to ask why proteins interact. J R Soc Interface. 2013;10(79):1–27. CrossRef
63. Dermawan D, Sumirtanurdin R, Dewantisari D. Simulasi dinamika molekular reseptor estrogen alfa dengan andrografolid sebagai anti kanker payudara. Indones J Pharm Sci Technol. 2019;6(2):65–76.
64. Craveur P, Joseph AP, Esque J, Narwani TJ, Noël F, Shinada N, et al. Protein flexibility in the light of structural alphabets. Front Mol Biosci. 2015;2:1–20. CrossRef
65. Sanusi ZK, Lobb KA. Insights into the dynamics and binding of two polyprotein substrate cleavage points in the context of the SARS-CoV-2 main and papain-like proteases. Molecules. 2022;27(23):1–16. CrossRef
66. Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J. Critical assessment of methods of protein structure prediction (CASP)-Round XIII. Proteins. 2019;87(12):1011–20. CrossRef
67. Wang YH, Chen KM, Chiu PS, Lai SC, Su HH, Jan MS, et al. Lumbrokinase attenuates myocardial ischemia-reperfusion injury by inhibiting TLR4 signaling. J Mol Cell Cardiol. 2016;99:113–22. CrossRef
68. Lenselink EB, Louvel J, Forti AF, van Veldhoven JPD, de Vries H, Mulder-Krieger T, et al. Predicting binding affinities for GPCR ligands using free-energy perturbation. ACS Omega. 2016;1(2):293–304. CrossRef
69. Salsbury FR Jr. Molecular dynamics simulations of protein dynamics and their relevance to drug discovery. Curr Opin Pharmacol. 2010;10(6):738–44. CrossRef
70. Wu HH, Wang B, Armstrong SR, Abuetabh Y, Leng S, Roa WHY, et al. Hsp70 acts as a fine-switch that controls E3 ligase CHIP-mediated TAp63 and ΔNp63 ubiquitination and degradation. Nucleic Acids Res. 2021;49(5):2740–58. CrossRef
71. Wu H, Shi Y, Lin Y, Qian W, Yu Y, Huo K. Eukaryotic translation elongation factor 1 delta inhibits the ubiquitin ligase activity of SIAH-1. Mol Cell Biochem. 2011;357(1-2):209–15. CrossRef