INTRODUCTION
The continuous increase in cancer cases and the limitations of current treatments have prompted ongoing efforts to discover new drugs. A significant issue in cancer treatment is drug retention, and the complications associated with cancer often require patients to consume multiple drugs that can act synergistically to kill cancer cells. Polypharmacology phenomena involve either (a) a single drug acting on multiple targets in one disease pathway or (b) a single drug acting on multiple targets related to various disease pathways [1]. One strategy in polypharmacology is hybridization. Hybridization of multitarget drug compounds is a popular approach among researchers. This strategy combines compounds already possessing therapeutic potential to create opposing or synergistic functions on different receptor targets, including enhancing synergy, reducing side effects, and preventing drug resistance [2,3]. Compound hybridization comes in various forms, one of which is linked hybridization. Linked hybridization occurs when a compound acts as a bridge between two pharmacophores. Linked hybridization can work in a cleavable manner, where the drug compound will later separate in the body and interact with the target receptors [4,5]. A previous study of linked hybridization compounds with anticancer potential demonstrates the potential of amide-linked compounds as anti-proliferation agents against cancer, with inhibition effectiveness four times better than that of its pharmacophore compounds. Furthermore, the study explains that forming ester-linked compounds also exhibits anti-proliferative properties and can reduce toxicity, making them safe for human hepatocytes [6].
Natural compound preparations as pharmacophores, such as volatile compounds, are influenced by chemical structures such as terpenes and polyphenols with substituent groups like hydroxyl, aromatic rings, esters, aldehydes, methoxy, methylenedioxy, and so on. Essential oil compounds and their derivatives, such as eugenol and salicylic acid, have been reported to possess antibacterial, antifungal, antioxidant, anti-inflammatory, and anticancer capabilities [7–9]. Previous research also reported that salicylic acid and its derivatives, including acetylsalicylic acid, can induce endoplasmic reticulum stress and promote cancer cell death, making them a potential candidate for anticancer drugs, suppressing the epidermal growth factor receptor (EGFR) [10,11]. In addition to the constituents of essential oils, amino acids have been identified for their potential as anticancer agents in recent studies. When combined with certain compounds, amino acid derivatives demonstrate a notable reduction in the cytotoxicity associated with drug compounds. Nonetheless, their efficacy in treating various cancer types [12].
The supportive predictive methods for drug discovery have become increasingly popular lately, one of which is by integrating ADMET information and Lipinski’s principles. This approach allows the research on new drugs to lead to the design of molecules that are not only biologically active but also meet the physicochemical requirements necessary for clinical success. This initiative provides a strong foundation for optimizing the therapeutic potential, bioavailability, and safety of new drugs throughout the drug development stages [13]. Molecular docking is also one of the in-silico structure-based methods used to predict interactions between molecules and biological targets and estimate their complementarity through screening functions. Drug design has also evolved due to natural compounds with good bioactivity, although only some can meet the drug’s capabilities. Natural compounds can be optimized through structural modifications with principles such as enhancing binding potential and selectivity, improving physicochemical properties distribution, enhancing biochemical properties, pharmacokinetics, and reducing side effects, among others [14,15].
The application of computational analysis to evaluate the anticancer potential of both linked and nonlinked hybrid compounds through forming amides and esters. Therefore, the influence of hybridization on cancer drug development can be understood. The formation of ester and amide compounds can be achieved through stieglich and amidation reactions. The stieglich reaction involves esterification formation with the assistance of catalysts such as DCC and DMAP, and both reactions have the same mechanism. The steglich reaction is performed with carboxylic acid (COOH) compounds and alcohols or amines with high selectivity and yield percentages [16]. Meanwhile, amidation reactions using DCC as a catalyst have been reported by George A. Kraus. Therefore, considering the availability of functional groups such as alcohol (OH), COOH, and amine (NH2) in eugenol, salicylic acid, and amino acids, it is highly feasible to synthesize hybrid linker compounds, both esters, and amides, for preparing anticancer drug compounds.
EXPERIMENTAL SECTION
Materials and instrumentation
Eugenol (grade: 99%), alanine, salicylic acid, DCC and DMAP catalysts were obtained from Sigma-Aldrich. Ethyl acetate and n-hexane solvents, as well as reagents, were purchased from Merck. The ultrasonic cleaner used was the Krisbow DSA100-GLI-2.8L model KW1801033. thin-layer chromatography (TLC) analysis was performed using TLC Silica gel 60 F254 plates. FTIR analysis was performed using the SHIMADZU 8,400s instrument, LCMS analysis was conducted with an HP-5 capillary column on an LC C8 column. Molecular Docking was performed using Pyrx 9.0 software.
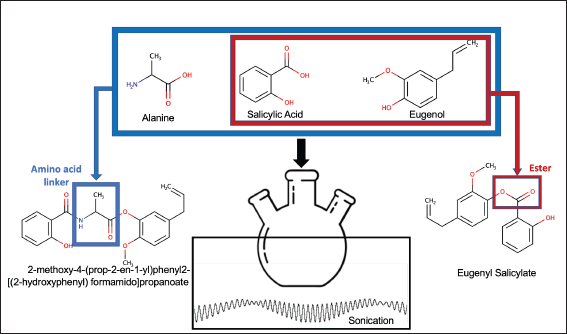 | Figure 1. Synthesis illustration. [Click here to view] |
Procedure
The research was conducted in two stages: 1. Anticancer analysis using ADMET, Lipinski rules, and in-silico docking; 2. Synthesis of linked compounds through amidation and esterification and synthesis of nonlinked compounds through esterification (Fig. 1).
Anticancer potential (Lipinski Rules, ADMET, in-silico Docking)
In this procedure, an analysis was conducted on two molecular hybrid compounds, with or without alanine linkage, to determine their drug similarity characteristics in accordance with Lipinski’s rule. The assessment utilized simplified molecular input line entry system (SMILES) representations of the active compounds, which were input into the swissADME web server (http://www.swissadme.ch/index.php). The examined compounds underwent ADMET analysis using either http://lmmd.ecust.edu.cn/admetsar1/ or http://lmmd.ecust.edu.cn/admetsar2/. This assessment required input of the SMILES representation of the drug candidate molecule. The ADMET analysis evaluated various parameters associated with absorption, distribution, excretion, metabolism, and drug toxicity, offering valuable insights into the safety and effectiveness of the drug candidates. In-silico analysis was conducted using Molecular Docking Pyrx 9.0. Table 1 provides comprehensive details concerning the active site grid box of the receptor. The docking results were selected based on criteria such as RMSD (<2) and binding affinity. These results were subsequently visualized in 3D using the BIOVIA Discovery Studio 2019 application, while their interactions were analyzed in 2D using Ligplot. This analysis aimed to identify optimal interactions compared to both the ligand compounds and the standard drug DOX (Doxorubicin).
Synthesis of eugenol and salicylic acid combination compounds with amino linker
The complete reaction for the synthesis of the combination compound of Eugenol and Salicylic Acid with an amino linker is shown in Figure 2. The first stage of amino acid-linked compound synthesis involved amidation, where alanine (0.09 g, 1 mmol) was reacted with salicylic acid (0.3 g, 2 mmol) using DCC Catalyst (0.412 g, 2 mmol) in THF solvent at 60°C under sonication for 1.5 hours. The resulting product was then separated through filtration using Whatman filter paper.
Subsequently, the synthesis proceeded to the second stage, esterification. In this stage, the compound from the first stage, 2-[(2-hydroxyphenyl)formamido] propanoic acid (414 mg, 3 mmol), was reacted with DCC catalyst (118 mg, 3 mmol) and DMAP catalyst (50 mg, 0.4 mmol) in a solvent-free reaction. Eugenol (985 mg, 6 mmol) was added dropwise, and the reaction was conducted for 1–3 hours with monitoring using TLC. Following the reaction, the mixture underwent filtration, and the liquid phase was washed successively with 5% HCl (2 × 5 ml), 5% NaHCO3 (3 × 5 ml), and H2O (3 × 5 ml). Anhydrous Na2SO4 was employed for drying the organic phase before the final compound, 2-methoxy-4-(prop-2-en-1-yl)phenyl 2-[(2-hydroxyphenyl)formamido] propanoate, was evaporated using nitrogen gas.
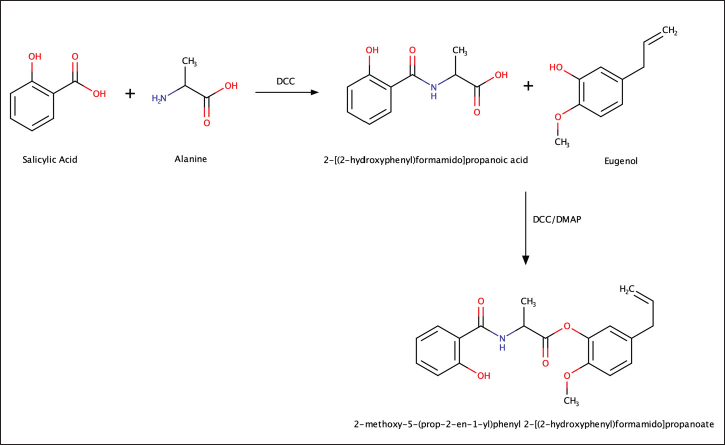 | Figure 2. Synthesis of eugenol and salicylic acid combination compounds with amino linker. [Click here to view] |
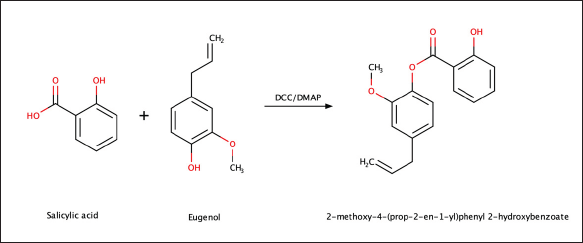 | Figure 3. Synthesis of eugenol and salicylic acid combination compounds with ester linker (Eugenyl Salicylate). [Click here to view] |
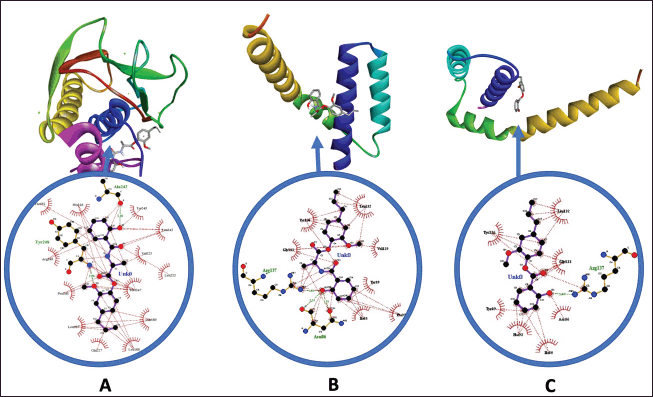 | Figure 4. Pose and ligan-receptors interaction: (A) Ligand complex 2-methoxy-4-(prop-2-en-1-yl) phenyl 2-[(2-hydroxyphenyl)formamido] propanoate with MMP9 (PDB ID: 4H1Q); (B) Ligand complex 2-methoxy-4-(prop-2-en-1-yl)phenyl 2-[(2-hydroxyphenyl)formamido] propanoate with BAK (PDB ID: 6UXM) ; (C) Eugenyl Salicylate complex witnh BAK. (PDB ID: 6UXM). [Click here to view] |
Synthesis of eugenol and salicylic acid combination compounds with ester linker (Eugenyl Salicylate)
The reaction equation for the synthesis of the combination compound of Eugenol and Salicylic Acid with an ester linker (Eugenyl Salicylate) can be seen in Figure 3. Salicylic acid (414 mg, 3 mmol) was reacted with DCC catalyst (118 mg, 3 mmol) and DMAP catalyst (50 mg, 0.4 mmol) in a solvent-free reaction. Eugenol (985 mg, 6 mmol) was added dropwise, and the reaction was carried out for 1–3 hours with monitoring using TLC. After the reaction, the mixture underwent filtration, and the liquid phase was sequentially washed with 5% HCl (2 × 5 ml), 5% NaHCO3 (3 × 5 ml), and H2O (3 × 5 ml). AnhydrousNa2SO4 was used for drying the organic phase before evaporating itwith nitrogen gas.
RESULTS AND DISCUSSION
Anticancer potential through computational analysis: ADMET, lipinski rules, and in-silico Docking
Drug similarity analysis was conducted on two hybrid molecules with or without an alanine linker, using the five-parameter Lipinski rule approach. Table 2 presents the results of the drug similarity analysis for the two ester molecules. Both 2-methoxy-4-(prop-2-en-1-yl)phenyl 2-[(2-hydroxyphenyl) formamido]propanoate and eugenyl salicylate comply with Lipinski’s rule, exhibiting a molecular weight less than 500 Da, fewer than 5 hydrogen bond donors, fewer than 10 hydrogen bond acceptors, and a log p value of less than 5. Drugs with high lipid solubility, indicative of low hydrogen bonding and a larger polar surface area, can efficiently penetrate hydrophobic phospholipid bilayers to access organs in the body. Increasing a drug’s lipophilicity, especially for those with poor permeability, is thought to enhance cell penetration. Therefore, optimizing the physicochemical properties of drugs is crucial in designing central nervous system-active drugs for passive cell permeation. Values exceeding Lipinski’s rule may impact the interaction between drug molecules and lipophilic cell membranes. For hydrophilic central nervous system-active drugs, enhancing lipidization through the drug’s polar function can facilitate more efficient delivery into the central nervous system [17,18].
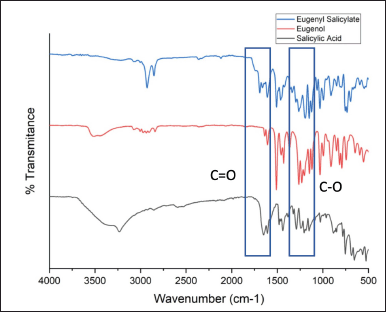 | Figure 5. FTIR of nonlinker hybrid molecules combination. [Click here to view] |
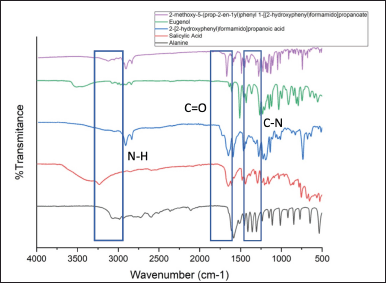 | Figure 6. FTIR of linker hybrid molecules combination. [Click here to view] |
The ADME profile summarizing the pharmacokinetic properties of the test ligands is presented in Table 3. These parameters cover various tests, including the Ames mutagenesis test (Ames), acute oral toxicity (AO), Caco-2 permeability (Caco2), P-glycoprotein substrate status (P-gp), P-glycoprotein inhibitor status (P-gpi), CYP450 substrate and inhibitor status (CYP2C9, CYP2D6, CYP3A4, CYP 1A2, CYP2C19, and CYP3A4), human intestinal absorption (HIA+), CYP enzyme activity inhibition (CYPPRO), carcinogenicity (CARC), hERG-related gene inhibition (human ether-a-go-go), and acute oral toxicity. Among these parameters, the first considers the oral absorption of drugs using the Caco-2 cell model, predicting permeability across the blood–brain barrier (BBB). For the compound 2-methoxy-4-(prop-2-en-1-yl)phenyl 2-[(2-hydroxyphenyl)formamido]propanoate, the BBB value is negative, indicating its inability to cross the BBB and reach the brain. In contrast, eugenyl salicylate shows a positive BBB penetration value, ranging, with a range of values from 0.7 to 0.8, suggesting its potential to pass through the BBB. Although the BBB value for this compound slightly exceeds the control [19].
Based on the ADMET properties predicted using admetSAR, the acute oral toxicity value in animals categorizes this compound as belonging to Category III, both compounds exhibit HIA+, suggesting improved absorption from the digestive tract into the bloodstream after oral administration [20]. The next parameter considered is the potassium ion channel known as the human ether-a-go-go-related gene (hERG), which regulates cardiac action potential. Inhibition of hERG can lead to disturbances in ventricular repolarization, potentially increasing the risk of cardiac arrhythmias, including torsades de pointes. Both compounds demonstrate a lower ability to inhibit the hERG potassium ion channel with pIC50 values ≤6.0 mol/l, indicating a lower potential to cause cardiac arrhythmia issues [21]. Therefore, they could be predicted as safe for human use [22]. Based on further ADMET properties predicted by admetSAR, both compounds are non-AMES toxic, meaning they do not induce genetic changes according to the AMES test method. Eugenyl salicylate is classified as an inhibitor/nonsubstrate, due to distribution by P-glycoprotein (P-gp), while, 2-methoxy-4-(prop-2-en-1-yl)phenyl 2-[(2-hydroxyphenyl)formamido] propanoate is categorized as a noninhibitor/nonsubstrate. Additionally, both compounds are nonmutagenic, nontoxic, and noncarcinogenic. Eugenyl salicylate has a Log S value of −3.3913, and 2-methoxy-4-(prop-2-en-1-yl)phenyl 2-[(2-hydroxyphenyl)formamido] propanoate has a LogS value of −3.4397, indicating lower solubility in water for both compounds [23].
The results of in-silico analysis indicate that both combinations of salicylic acid with eugenol, whether with or without an amino acid linker, exhibit significant anticancer potential against the positive control on MMP9, MMP2, CDK2, P53, BAK, EGFR, and ADP Ribosepolymerase receptors (Table 4). Based on these seven receptors, the compound 2-methoxy-4-(prop-2-en-1-yl)phenyl 2-[(2-hydroxyphenyl) formamido] propanoate, containing an amino acid linker, excels in inhibiting MMP9 with a binding affinity of −8.9 kcal/mol. It forms hydrogen interactions (Figure 4A) with Ala242, Tyr248, and hydrophobic interactions with His226, Thr251, Arg249, Pro246, Leu187, Qln227, Leu188, Ala189, Met247, Leu222, Val223, Leu243, and Tyr245. Furthermore, the compound with an amino acid linker also demonstrates good anticancer potential against BAK with a binding affinity of −6.7 kcal/mol, forming hydrogen interactions (Figure 4B) with Asn86 and Arg137, as well as hydrophobic interactions with Gly133, Tyr136, Leu132, Val129, Tyr89, Phe93, and Ile85. In contrast, Eugenyl salicylate excels in BAK with a binding affinity of −6.7 kcal/mol, forming hydrogen interactions (Figure 4C) with Arg137 and hydrophobic interactions with Asn86, Tyr89, Phe93, Ile85, Gly133, Val129, Leu132, and Tyr136.
Docking results against combinations of salicylic acid with eugenol, incorporating amino acid linkers, suggest that compounds with these linkers can inhibit tumor cell proliferation, particularly concerning the MMP9 receptor. This aligns with the inhibition mechanism of MMP9, involving the formation of a 92 kDa type IV collagenase complex, typically 92TCI(MMP9)-TIMP. This complex leads to dimerization and activation of stromelysin (mediator) into 92TC-CII, capable of producing an active complex against gelatin and fibrillar collagen. [24]. Additionally, the in-silico results of the combination of salicylic acid with eugenol, with or without amino acid linkers, indicate their potential effect on the BAK receptor, suggesting that both compounds can act pro-apoptotically against cancer cells. BAK is part of the BCL2 pro-apoptotic group. Cancer cells, through stabilization by overexpression of the anti-apoptotic BCL-2 protein, can evade apoptosis [25]. Loss of BAK can decrease the permeability and conductance of the outer mitochondrial membrane without altering the function of the inner membrane MPTP (Mitochondrial permeability transition pore) leading to resistance against excessive mitochondrial calcium and cell death [26]. Thus, overcoming cancer involves inducing or activating BAK as a pro-apoptotic factor. BAK functions after receiving a death signal, its oligomerization leads to the formation of mitochondrial pores, increasing mitochondrial membrane permeability and releasing cytochrome c (Cyt c) into the cytosol, triggering cell death. During the docking process, it is crucial to ensure that the ligand compound does not inhibit the BAK protein receptor (having a high Ki value).
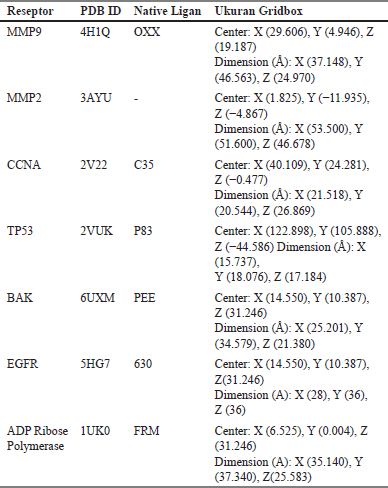 | Table 1. Docking condition. [Click here to view] |
Although both combinations of salicylic acid and eugenol, whether with or without an amino acid linker, do not exhibit optimal interactions with receptors such as MMP2, CDK, P53, EGFR, and ADP Ribose Polymerase, the presence of binding affinity values and some similarities in amino acid residues still indicates the potential of both compounds as cancer drugs. The hydrogen interaction between a substrate’s chemical structure and the receptor results in the synergistic action of volatile essential oil compounds as drugs [27]. Furthermore, their structures also influence the differences in the anticancer abilities of the two combinations of eugenol and salicylic acid, with or without an amino acid linker. Compounds containing an amino acid linker feature structures with a higher probability of hydrogen interactions compared to ester linkers. Consequently, the eugenol and salicylic acid compounds with an amino acid linker exhibit lower binding affinity but engage in more interactions with their receptors.
Synthesis of eugenol and salicylic acid combination compounds with ester linker (Eugenyl salicylate)
The synthesis was initiated by reacting salicylic acid, DCC, and DMAP in a reaction flask. As the reaction was carried out without a solvent, all reactants were combined, and eugenol was added dropwise. Upon the addition of eugenol, the reactants dissolved, resulting in the synthesis reaction. The resulting product, eugenyl salicylate, appeared as a brownish-yellow liquid. Monitoring using TLC (Silicagel 60 F254; n-hex:EtOAc(9:1)) revealed new spots (Rf 0.78) indicative of the synthesis of eugenyl salicylate. FTIR analysis (Figure 5) confirmed specific absorption bands, including C = O vibration (1,680–1,650 cm−1) and asymmetric C–O stretch (1,200–1,175 cm−1). Based on LCMS analysis, the yield of eugenyl salicylate was determined to be 59.85%.
 | Table 2. Drug likeness of hybrid molecules. [Click here to view] |
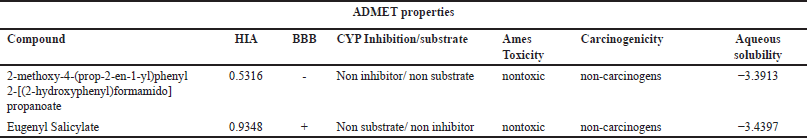 | Table 3. ADMET properties hybrid molecules. [Click here to view] |
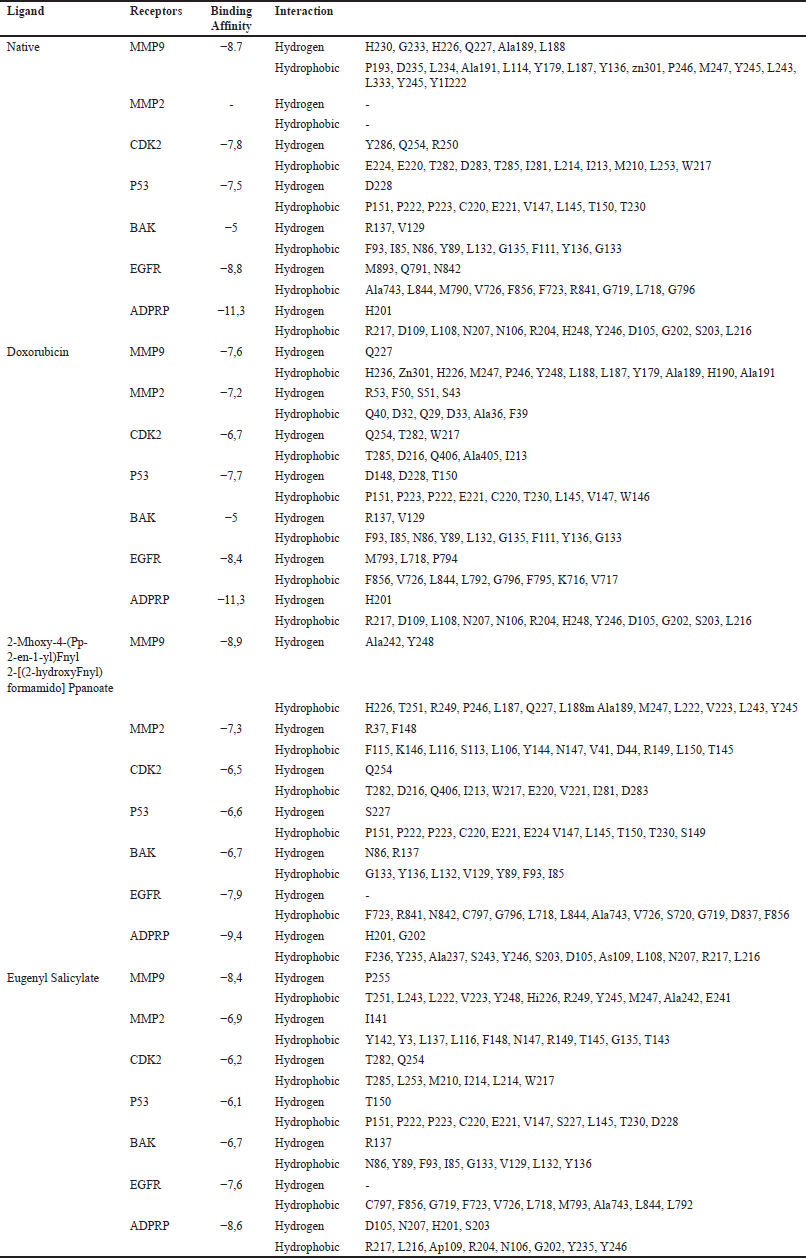 | Table 4. Docking result against 7 cancer receptors. [Click here to view] |
Synthesis of eugenol and salicylic acid combination compounds with amino linker
The reaction of 2-[(2-hydroxyphenyl)formamido] propanoic acid produced a colorless solution, monitored through qualitative TLC using n-hexane: ethyl acetate eluent (9:1), generating new spots with an Rf value of 0.58. Subsequent FTIR analysis of the resulting compound, 2-[(2 hydroxyphenyl)formamido]propanoic acid (Figure 6), indicating the formation of an amide group, evidenced by absorption bands at 1246 cm−1 (C–N), 3200 cm−1 (N–H), and 1,600 cm−1 (C = O) LCMS analysis further confirmed the presence of the product compound, 2-[(2-hydroxyphenyl)formamido] propanoic acid, with a yield of 57.79%. Previously, esterification reactions were conducted by Silva et al. [28] who reacted eugenol with benzoic acid. The reaction involvedmixing eugenol, DMAP, DCC, and benzoic acid in dichloromethane solvent. The mixture was stirred at room temperature for 24 hours, yielding a 70% product yield. Similarly, Intan Nurjaya adopted a comparable method to formester compounds by reacting salicylic acid and quinidine, resulting in a 97% yield [29].
The reaction of 2-methoxy-4-(prop-2-en-1-yl)phenyl 2-[(2-hydroxyphenyl)formamido] propanoate produced a yellow solution, observed using qualitative TLC with n-hexane: ethyl acetate eluent (9:1), resulting in new spots with an Rf value of 0.71. Subsequent FTIR analysis of the product compound, 2-methoxy-4-(prop-2-en-1-yl)phenyl 2-[(2-hydroxyphenyl)formamido] propanoate, revealed the presence of the C = O ester group at 1,220 cm–1 (C-O) and 1,687 cm–1. Additionally, LCMS analysis confirmed the presence of the product compound, 2-methoxy-4-(prop-2-en-1-yl)phenyl 2-[(2-hydroxyphenyl)formamido] propanoate, with a yield of 93.89%.
CONCLUSION
Based on the research findings, hybrid molecules of eugenol and salicylic acid, both without a linker and with a linker, exhibit properties that meet the criteria for drug candidates based on ADMET and Lipinski approaches. Furthermore, in silico analysis suggests that hybrid molecules without a linker have the potential as pro-apoptotic agents (BAK), while hybrid molecules with an alanine linker show promise as both pro-apoptotic agents (BAK) and inhibitors of enzymes responsible for cancer metastasis (MMP9). The synthesis results indicate that hybrid molecules without a linker yield 59.85%, while those with an alanine linker yield 93.89%. This research demonstrates the potential of hybridization approaches, particularly using natural compounds such as essential oil compounds. Further research on native tumor cells is needed for future investigations.
ACKNOWLEDGMENTS
We would like to thank Brawijaya University and the Faculty of Science for their support through the Excellence Professor Grant (HGB), Fiscal Year 2023, with contract number: 4158.24/UN10.F09/PN/2023.
AUTHOR CONTRIBUTION
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors do not have any conflict of interest against this research.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008 Nov;4(11):682–90.
2. Mohamed MFA, Abuo-Rahma GEDA. Molecular targets and anticancer activity of quinoline–chalcone hybrids: literature review. RSC Adv. 2020;10(52):31139–55.
3. Raghavendra NM, Pingili D, Kadasi S, Mettu A, Prasad SVUM. Dual or multi-targeting inhibitors: the next generation anticancer agents. Eur J Med Chem. 2018 Jan;143:1277–300.
4. Li X, Li X, Liu F, Li S, Shi D. Rational multitargeted drug design strategy from the perspective of a medicinal chemist. J Med Chem. 2021 Aug 12;64(15):10581–605.
5. Zhou J, Jiang X, He S, Jiang H, Feng F, Liu W, et al. Rational design of multitarget-directed ligands: strategies and emerging paradigms. J Med Chem. 2019 Oct 24;62(20):8881–914.
6. Tilekar K, Shelke O, Upadhyay N, Lavecchia A, Ramaa CS. Current status and future prospects of molecular hybrids with thiazolidinedione (TZD) scaffold in anticancer drug discovery. J Mol Struct. 2022 Feb;1250:131767.
7. Randjelovic P, Veljkovic S, Stojiljkovic N, Sokolovic D, Ilic I, Laketic D, et al. The beneficial biological properties of salicylic acid. Acta Fac Med Naiss. 2015 Dec 1;32(4):259–65.
8. Ulanowska M, Olas B. Biological properties and prospects for the application of eugenol—a review. IJMS. 2021 Apr 1;22(7):3671.
9. Das AJ, Das MK, Singh SP, Saikia PP, Singh N, Islam J, et al. Synthesis of salicylic acid phenylethyl ester (SAPE) and its implication in immunomodulatory and anticancer roles. Sci Rep. 2022 May 24;12(1):8735.
10. Ausina P, Branco JR, Demaria TM, Esteves AM, Leandro JGB, Ochioni AC, et al. Acetylsalicylic acid and salicylic acid present anticancer properties against melanoma by promoting nitric oxide-dependent endoplasmic reticulum stress and apoptosis. Sci Rep. 2020 Nov 12;10(1):19617.
11. Bashir A, Kankipati C, Jones S, Newman R, Safrany S, Perry C, et al. A novel mechanism for the anticancer activity of aspirin and salicylates. Int J Oncol [Internet]. 2019 Jan 29 [cited 2023 Apr 19]; Available from: http://www.spandidos-publications.com/10.3892/ijo.2019.4701
12. Kumar V, Mudgal MM, Rani N, Jha A, Jaggi M, Singh AT, et al. Synthesis of functionalized amino acid derivatives as new pharmacophores for designing anticancer agents. J Enzyme Inhib Med Chem. 2009 Jun 1;24(3):763–70.
13. Mahgoub RE, Atatreh N, Ghattas MA. Chapter Three - Using filters in virtual screening: a comprehensive guide to minimize errors and maximize efficiency. In: Caballero J, editor. Annual Reports in Medicinal Chemistry [Internet]. Academic Press; 2022. 59: pp 99–136. [cited 2023 Nov 16]. Available from: https://www.sciencedirect.com/science/article/pii/S0065774322000100
14. Coumar MS. Molecular docking for computer-aided drug design fundamentals, techniques, resources and applications [Internet]. 2021 [cited 2022 May 27]. Available from: http://www.vlebooks.com/vleweb/product/openreader?id=none&isbn=9780128223130
15. Chen J, Li W, Yao H, Xu J. Insights into drug discovery from natural products through structural modification. Fitoterapia [Internet]. 2015 Jun [cited 2022 May 27];103:231–41. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0367326X15001021
16. Bew SP. Carboxylic acid. In: Comprehensive organic functional group transformations II (COFGT-II). New York, NY: Elsevier; 2005. Vol. 5, no. 1, pp 19–125.
17. Turner JV, Agatonovic-Kustrin S. 5.29 In Silico Prediction of Oral Bioavailability. In: John B. Taylor and David J. Triggle, editors. Comprehensive medicinal chemistry II. Amsterdam, The Netherlands: Elseiver; 2007. pp. 699–724.
18. Li G, Shao K, Umeshappa CS. Recent progress in blood-brain barrier transportation research. In: Gao H, Gao X, editors. Brain targeted drug delivery system. Amsterdam, The Netherlands: Elsevier; 2019. pp. 33–51.
19. Vardhan S, Sahoo SK. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput Biol Med. 2020 Sep;124:103936.
20. Nisha CM, Kumar A, Nair P, Gupta N, Silakari C, Tripathi T, et al. Molecular Docking and In Silico ADMET study reveals acylguanidine 7a as a potential inhibitor of β -secretase. Adv Bioinform. 2016 Apr 10;2016:1–6.
21. Kravchenko AD, Pyatigorskaya NV, Brkich GE, Yevsieieva LV, Kyrychenko AV, Kovalenko SM. Synthesis, molecular docking, ADMET study and in vitro pharmacological research of 7-(2-chlorophenyl)-4-(4-methylthiazol-5-yl)-4,6,7,8-tetrahydroquinoline-2,5(1H,3H)-dione as a promising non-opioid analgesic drug. RRP. 2022 Mar 30;8(1):1–11.
22. Vimal A, Jha A, Kumar A. Eugenol derivatives prospectively inhibit l-asparaginase: a heady target protein of Salmonella typhimurium. Microb Pathog. 2018 Jan;114:8–16.
23. Pruthviraj K, Lohith TN, Yeshwanth M, Usha GH, Vanajakshi HV, Venugopal KBet al. Molecular iodine catalyzed solvent free one pot synthesis, characterization, insilico Adme, Herg analysis, Molecular Docking Studies and DFT Analysis of 2- substituted 4,5-diphenyl-1h-imidazole derivatives. JASR. 2021 Nov 30;12(04):211–29.
24. Strongin AY, Collier IE, Krasnov PA, Genrich LT, Marmer BL, Goldberg GI. Human 92kDa type IV collagenase: functional analysis of fibronectin and carboxyl-end domains. Kidney Int. 1993 Jan;43(1):158–62.
25. Matsuura K, Canfield K, Feng W, Kurokawa M. Metabolic regulation of apoptosis in cancer. Int Rev Cell Mol Biol [Internet]. 2016;327:43–87. [cited 2022 Nov 24]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1937644816300570
26. Karch J, Kwong JQ, Burr AR, Sargent MA, Elrod JW, Peixoto PM, et al. Bax and bak function as the outer membrane component of the mitochondrial permeability pore in regulating necrotic cell death in mice. Elife. 2013 Aug 27;2:e00772.
27. Ribeiro-Santos R, Andrade M, Sanches-Silva A, de Melo NR. Essential oils for food application: natural substances with established biological activities. Food Bioprocess Technol. 2018 Jan;11(1):43–71.
28. da Silva FFM, Monte FJQ, de Lemos TLG, do Nascimento PGG, de Medeiros Costa AK, de Paiva LMM. Eugenol derivatives: synthesis, characterization, and evaluation of antibacterial and antioxidant activities. Chem Central J. 2018 Dec;12(1):34.
29. Nurjaya I, Hanafi M, Lotulung PDN, Ernawati T, Mursiti S. The synthesis of quinidine salicylate ester compound. JKimTerapIndones. 2019 Jan 28;20(2):98–102.