INTRODUCTION
Parkinson’s disease (PD) is a neurodegenerative disorder characterized by losing midbrain dopaminergic neurons in the substantia nigra [1]. It is estimated that PD affects 1%–2% of the geriatric population. The threat of the economic burden of PD in the national healthcare system continues to rise [2]. However, treatment for Parkinson’s is available, but it provides only symptomatic relief and does not stop progression because the exact mechanism is complex and unknown. Drugs were used to slow down the passage of the degeneration of neurons [3]. Understanding the mechanism of disease progression can help us target and get the right treatment choice. Many recent pieces of evidence indicate that mechanistic target of rapamycin (mTOR) has a critical role in PD pathogenesis.
The mammalian/mTOR is a serine–threonine kinase. It controls many important functions of mammalian cells, such as cell survival and protein synthesis [4]. During the early 2000s, neuroscientists began their interest in mTOR targets. 4E-binding proteins and p70 ribosomal S6 protein kinase 1 are initially studied [5]. Soon after identifying mTOR’s role in neuronal morphogenesis, survival, and differentiation, the target became popular, and many scientists observed its role in different diseases such as PD and Alzheimer’s disease (AD). The list of physiological conditions and neuropathologies associated with mTOR rapidly increased, but a thorough knowledge of mTOR modulation and its cellular effectors in neurons remained elusive. Autophagy, translation, cell signaling, transcription, and cytoskeleton dynamics are all influenced by changes in the mTOR activity [6]. According to new research, the overexpression of mTOR is linked to the pathogenesis of PD [7,8]. As a result, mTOR might be one of the possible treatment targets for PD [9]. mTOR activity is quite controversial. It has different actions in different pathological conditions [10]. The mTOR signaling system, its positive and negative functions in pathological conditions, its regulation, and its participation in autophagy are all discussed in this article. We also discuss possible mTOR modulators that can be used in treating PD.
We conducted a review of the literature in PubMed. Only articles in the English language are selected. The papers are reviewed until December 2021. The following terms are used to search articles: PD and each of the following terms or a combination of the following terms: mTOR, PD, akt. α-synuclein accumulation, Unc-51-like kinase 1 (ULK1), energy deficit mTOR, TSC1/2, autophagy, protein synthesis, cell cycle, inflammation, neuroprotective, toxicities mTOR, and small molecules (Fig. 1).
Components and structure of mTOR
mTOR or mammalian/mTOR is a serine/threonine protein kinase. It involves various cell functions such as gene transcription, protein synthesis, cell metabolism, survival, and proliferation [11]. The multidomain protein mTOR is found in two multiprotein complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) [12]. mTORC1 controls protein synthesis, whereas mTORC2 controls cell survival and cytoskeleton organization [13].
Each complex contains mTOR protein, mammalian lethal with SEC13 protein 8 (mLST8), DEP domain-containing mTOR interacting protein (DEPTOR), Tel two-interacting protein1, and telomere maintenance 2 (Tel2) [14,15]. Two proteins are specific to mTORC1, the regulatory-associated protein of mTOR (Raptor) and proline-rich AKT1 substrate 40 kDa (PRAS40) [16,17]. In these complexes, DEPTOR and PRAS40 can negatively regulate mTORC1 activity [18]. mTORC1 plays a major role in regulating cell growth and proliferation and senses stimuli such as insulin level and amino acid-like Leucine [19,20]. In addition, it is involved in protein synthesis, lipid metabolism, and autophagy [21,22].
mTORC2 contains a Rapamycin-insensitive companion of mTOR, mammalian stress-activated protein kinase-interacting protein1 (mSIN1), and protein observed with Rictor [23], mLST8, and Ttil/Tel2. Akt is an important substrate of mTORC2. Phosphorylation of Akt induces phosphorylation of mSIN1, which increases mTORC2 activity [24]. mTORC2 controls the remodeling of cytoskeleton and electrolyte homeostasis [25].
mTOR signaling pathways
mTORC1 signaling pathway
Insulin-like growth factor 1 (IGF-1) activates mTOR. IGF starts the conversion of Ras guanosine 5’-triphosphate (GTP) to Ras guanosine diphosphate (GDP) This conversion leads to the cascade of small molecule activation, which includes Ras, Mek, Erk, and Rsk. Erk and Rsk inhibit the activation of TSC1/2. TSC1/2complex is an essential regulator in this process [26]. TSC1/2 activates the conversion of GTPase Rheb to GDPase Rheb, an inactive form. This GDP-bound state negatively regulates the mTOR pathway [27]. Other factors, such as inflammation and hypoxia, can also lead to mTOR activation. In response to the cellular energy deficiency, LKB1 is activated. This activated LKB1 increases the adenosine monophosphate (AMP)/adenosine triphosphate (ATP) ratio. This elevated level of AMP leads to phosphorylation of AMP-activated protein kinase (AMPK), which consecutively activates TSC1/2. The activation of TSC1/2 leads to an increase in the GAP activity of the TSC1/2 complex and inhibits mTORC1 activity [28]. It is further noted that hypoxia can also inhibit mTORC1 activity by the increase in regulated in development and DNA damage responses (REDD1) expression [29]. Inflammation can also lead to the inhibition of TSC1/2 by activating Ikkβ.
The activated mTORC1 plays its role in protein regulation by two possible mechanisms. One by phosphorylation of ribosomal protein S6 kinase beta-1 (S6K1) and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) [30]. S6K1 is phosphorylated and activated by mTORC1, which again phosphorylates and activates S6 (a component of the S40 ribosomal subunit). S6K1 also activates several small molecules such as CBP80, Tripartite motif-containing protein-24 (TIF1A), SKAR, and eIF4B, and inhibits eukaryotic elongation factor 2 kinase (eEF2K) and PDCD4. S6K1 inhibits eEF2K by phosphorylation, thus inhibiting the translation and elongation of proteins. Phosphorylation of PDCD4 leads to the inhibition of eukaryotic initiation factor-4A, which is responsible for translation initiation [31]. S6K1 phosphorylates and activates SKAR and CBP80. This activation leads to mRNA biogenesis [32]. S6K1 also activates TIF1A, elevating rRNA expression through interaction through POL1.
 | Figure 1. Methodology. [Click here to view] |
Another pathway by which mTOR regulates protein synthesis is by 4E-BP1. By blocking the link between 4E-BP1 and eukaryotic translation initiation factor 4E, active mTORC1 phosphorylates and inactivates 4E-BP1, a regulator of mRNA translation (eIF-4E). These signaling pathways lead to protein and lipid synthesis and inhibit autophagy and lysosome biogenesis [33]. Autophagy is induced by the ULK1 complex when mTOR is inhibited. Hence, mTOR negatively regulates autophagy [34].
mTORC2 signaling pathway
mTORC2 also responds to growth factors like mTORC1, through the PI3K pathway. However, in the case of mTORC2, TSC1/2 activates mTORC2 rather than suppressing it like mTORC1. Some in-vitro studies reported that removal of TSC1/2 leads to loss of mTORC2 activity [35]. mTORC2 exerts its role by activating protein kinases A, G, and C which consist of Akt, serum/glucocorticoid regulated kinase 1 (SGK1), and protein kinase C alpha (PKCα) [36]. mTORC2 regulates cytoskeleton remodeling by phosphorylation of PKCα and ion transport by phosphorylation of SGK1 [37]. Akt paves the way for cell survival and metabolism [38]. Downstream of the Akt pathway includes activation of murine double minute 2 (MDM2) and inhibition of Forkhead box O (FOXO) and PRAS40. MDM2 is responsible for cytoskeleton organization, and FOXO inhibits proliferation and cell cycle arrest. PRAS40 regulates mTORC1 by directly inhibiting mTORC1 [39]. Figure 2 represents the signaling pathway of mTORC1 and mTORC2.
mTOR-double-edged sword
mTOR is vital in many neurodegenerative diseases such as AD and PD. In recent years, the role of mTOR in Parkinson’s has been studied widely. However, the role of mTOR in PD remains controversial [40]. Many studies have shown evidence of both neuroprotective and neurotoxic effects. For example, overexpression of mTOR in toxin-induced PD can partially prevent neuronal loss [41]. In contrast, another study indicates that PD toxins suppress mTOR signaling, which leads to a decrease in cell viability [42]. Here, we discuss the neuroprotective and neurotoxic effects of mTOR.
Neuroprotective effects of mTOR
The stimulation of mTOR in dopaminergic neurons may result in axon regrowth [43]. Deletion of phosphatase and tensin homolog deleted on Chromosome 10 activates mTOR and protects dopaminergic neurons by promoting Akt signaling [44]. REDD1 (regulated in development and DNA damage responses) inhibition causes mTOR activation and provides neuroprotection in cellular models [45]. Memory is similarly affected by mTOR. Inhibition of mTOR by rapamycin leads to memory deficits in PD models [46]. mTOR interacts with Raptor and produces signals for cell cycle regulation. The activation of mTOR significantly influences neurogenesis, particularly dendritic formation. Cell development needs average cell growth and advancement, governed by the mTOR pathway [47]. mTOR induces translation through direct or indirect phosphorylation in the presence of signals such as growth hormones and energy deprivation. mTOR also has other beneficial impacts, such as maintaining glucose homeostasis [48], regulating muscle mass [49], and increasing mitochondrial functions [50] (Fig. 3).
Neurotoxicity of mTOR
The neurotoxic effects of mTOR are mostly related to the overexpression of mTOR signaling. An elevated amount of mTOR is seen in the post-mortem brains of people with PD. Maneb and paraquat are the major environmental risk factors of PD. Among them, paraquat has been shown to drastically elevate mTOR levels in mice, resulting in defective axonal autophagy [51]. Inhibition of mTOR is crucial since it depends upon the availability of protein expression. Inhibition of mTOR may disrupt normal functions such as the production of proteins, cytoskeleton organization, and neurogenesis [52]. AMPK activation and suppression of Akt, led by mTOR inhibition through the S6K1 pathway, leads to neuronal cell death [53]. The neurotoxic effects of mTOR inhibitors are caused by side effects caused by the drugs themselves, not by mTOR. In Parkinson’s treatment, mTOR inhibitors are widely used. However, because of their side effects, mTOR inhibitors have numerous drawbacks over benefits. Drugs like Sirolimus exhibits hyperlipidemia, poor wound healing, and thrombocytopenia [54]. Tacrolimus, another mTOR inhibitor, shows nephrotoxicity and glucose intolerance [55]. Seizures are also a risk factor for these drugs. One study reported that using an mTOR inhibitor might decrease testosterone levels [56]. mTOR negatively regulates autophagy. mTOR signaling inhibition leads to the induction of autophagy [57]. Autophagy acts as a cell survival and death mechanism [58]. Autophagy causes cell death in the amoeba Dictyostelium discoideum when apoptosis is absent [59]. Autophagy is required for the apoptotic cell death of nurse cells in the Drosophila oocyte. Hence, autophagy is needed for both apoptotic and non-apoptotic cell death [60]. Levodopa is a standard medication used for PD, but it causes levodopa-induced dyskinesia in patients [61]. When it is given along with mTOR inhibitors, dyskinesia is significantly reduced [62]. The inability of mTOR inhibitors to block mTOR entirely is a significant disadvantage. In the short term, rapamycin partially suppresses mTORC1, and in the long term, it primarily inhibits mTORC2 and not mTORC1 [36]. Hence, treatment with rapamycin cannot suppress the activity of mTOR (Fig. 3).
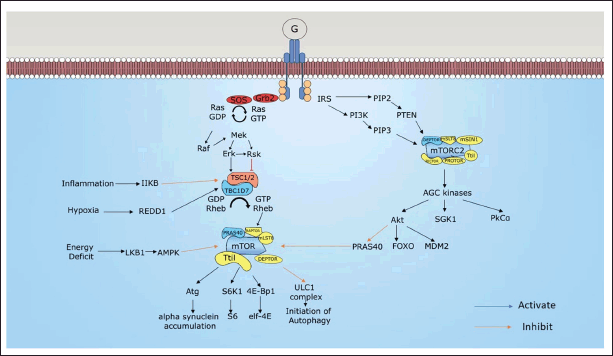 | Figure 2. Signaling pathway of mTORC1 and mTORC2. [Click here to view] |
Role of mTOR in neurodegenerative diseases
Neurodegenerative diseases such as AD, PD, and Huntington’s differ in their pathology. Among them, permanent loss of neurons, autophagy, genetic variability, and age are common. In AD, Amyloid-beta deposition is higher in patients, whereas alpha-synuclein deposition is seen in Parkinson’s disease. These are toxic or mutated protein aggregates [63]. The fact that these proteins cannot be cleared from the site due to their solubility and loss of autophagy worsens the condition. Autophagy is an essential process for a healthy system to function. This process clears the unwanted proteins and maintains homeostasis. Defective autophagy is commonly seen in neurodegenerative diseases [64]. mTOR is a critical protein that regulates autophagy [65]. It is also responsible for protein synthesis by the S6K1 pathway. In this context, the role of mTOR is studied in neurodegenerative diseases [25]. The various factors that modulate mTOR and autophagy which result in the onset of PD are aging, REDD1, PARKIN, PINK, UCHL1, and DJ1 (Fig. 4).
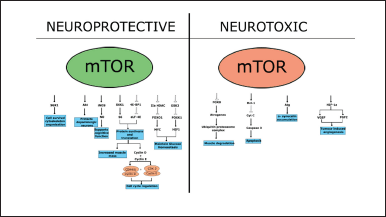 | Figure 3. Neuroprotective and neurotoxic effects of mTOR. [Click here to view] |
In a study, rapamycin which is a potent mTOR inhibitor, promotes autophagy in both in vivo and in vitro [66]. This exhibits the relationship between autophagy and mTOR inhibition. The inhibitors of mTOR are widely searched since they help in the inhibition of protein synthesis and induction of autophagy. Rapamycin (Sirolimus) is the first rapalog of this class. Everolimus and tacrolimus came into existence after that [67]. Despite blocking protein translation and autophagy induction, long-time exposure to these drugs causes tissue damage and impairment of metabolism. Small compounds that specifically alter the function of proteins governing autophagy downstream of mTORC1 signaling might be developed to trigger this process in a more targeted manner (Fig. 4).
Role of mTOR in autophagy
Autophagy is the highly conserved mechanism through which the misfolded proteins, and damaged organelles are removed from the body to maintain homeostasis [68]. Autophagy exerts its action through the lysosomal degradation pathway, thereby degrading the catabolic contents [69]. Autophagy is mediated through microautophagy, macroautophagy, and chaperone-mediated autophagy. In macroautophagy, the cytoplasmic portions are wrapped within the autophagosome and carried to lysosomes for bulk degradation [70]. Microautophagy includes autophagic tubes directly engulfing the cytoplasmic components [71]. Both micro and macroautophagy engulf cytoplasmic components through a selective and non-selective mechanism. In contrast, Chaperone-mediated autophagy exerts degradation in a non-specific manner, i.e., molecule-by-molecule fashion [72]. The major signaling pathway which controls autophagy is mTOR [73]. mTOR exerts its role in the various stages of autophagy, and its activity is tightly regulated by a complex interplay between stimulatory and inhibitory signals [74]. The various signaling pathways through which mTOR regulates autophagy are discussed below:
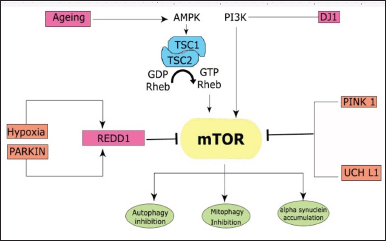 | Figure 4. The various factors that modulate mTOR signaling include aging, REDD1, PARKIN, PINK, UCHL1, and DJ1, which lead to the onset of PD. PINK and PARKIN cause mitophagy through mTOR. REDD1, UCHL1, and PINK inhibit mTOR, thereby reducing protein translation and leading to neuronal death. Aging inhibits AMPK, which activates mTOR and inhibits autophagy. DJ1 activates mTOR through the PI3K pathway resulting in alpha-synuclein accumulation and inhibition of autophagy. [Click here to view] |
ULK1 signaling pathway
The downstream of mTORC1 contains the ULK1 complex, which consists of ATG101, ATG13, and focal adhesion kinase family interacting protein of 200 kD. mTORC1 activates and phosphorylates ULK1 [75]. This phosphorylation leads to the inhibition of interaction with AMPK and inhibits autophagy. On cellular stress, ULK1 is phosphorylated by AMPK, which further leads to the induction of autophagy. Hence, mTORC1 negatively regulates autophagy by the ULK1 complex [76].
FEB signaling pathway
Transcription factor EB (TFEB) is a part of the bHLH leucine-zipper transcription factor family that regulates autophagy and lysosomal gene expression [77]. Normally inhibition of mTORC1 results in the release of TFEB, which releases transcriptional factors. These transcriptional factors cause autophagy and lysosome biogenesis [78].
Vacuolar protein sorting 34 (VPS34) complex signaling pathway
Vacuolar protein sorting 34 is another protein through which autophagy induction occurs. VPS34 consists of two complexes: Complex 1 and Complex 2. Complex 1 consists of VPS34, VPS15, beclin 1, and ATG14. Complex 2 consists of VPS34, VPS15, beclin 1, and UVRAG [79]. mTORC1 directly inhibits VPS34 by phosphorylating ATG14 at various sites. Autophagy might be improved by mutating these sites, which are resistant to mTOR inhibition [80].
Autophagic lysosome reformation (ALR)
Lysosomes are repurposed from autolysosomes at the end of autophagy by a process known as ALR, which comprises proto-lysosome tubule formation, elongation, and termination [81]. In short-term food deprivation, mTOR is inhibited, and in nutrient-rich conditions, mTOR is activated. Also, during prolonged starvation, mTOR is reactivated by a negative feedback loop of autophagy. For this reactivation, autophagic lysosomes have to be degraded. Spinster, a lysosomal efflux protein, is a regulator of ALR. Prolonged starvation leads to defective spinsters. mTOR reactivation failed due to the defective spinster [82]. The general amino acid control pathway is activated by starvation, which leads to the overexpression of amino acid transporters in the plasma membrane, increasing amino acid absorption and contributing to mTOR reactivation [83].
Lysosomal acidification and autophagy
Lysosomes are crucial organelles involved in the degradation of cellular waste. Lysosomal acidification is facilitated by proton pumps, predominantly the vacuolar H+-ATPase (V-ATPase), which actively pumps protons into the lysosomal lumen. The acidic pH inside lysosomes is crucial for the activation of hydrolytic enzymes, enabling efficient degradation of cellular components during autophagy [84].
The interplay between mTOR and lysosomal acidification
An intriguing feedback loop exists between mTOR and lysosomal acidification. Amino acids, critical activators of mTORC1, are transported into lysosomes, where they play a pivotal role in recruiting and activating mTORC1 at the lysosomal surface. The V-ATPase, responsible for lysosomal acidification, is central to this process [85]. Understanding the interplay between mTOR and lysosomal acidification holds significant implications for cellular health and disease. Dysregulation of either process can lead to disruptions in autophagy, compromising cellular homeostasis and contributing to the pathogenesis of various diseases, such as cancer, neurodegenerative disorders, and metabolic conditions [86]. In PD, proper lysosomal acidification is necessary for the optimal function of lysosomal enzymes [87]. Genetic investigations have unveiled a significant association between lysosomal function and PD. Several autosomal dominant and recessive genes linked to PD, as well as various genetic risk factors, encode proteins involved in lysosomal, autophagic, and endosomal pathways. Mutations in these PD-associated genes can lead to lysosomal dysfunction, and considering that α-synuclein degradation is primarily reliant on lysosomal processes, this impairment can hinder α-synuclein turnover. Consequently, this disruption contributes to elevated intracellular levels of α-synuclein, facilitating its accumulation and subsequent aggregation, among other consequences.
Feedback loop-AMPK, ULK1, and mTOR
The main reasons for many neurodegenerative diseases are the overproduction of abnormal proteins and autophagy dysregulation [88]. Much evidence suggests that mTOR signaling is overactivated in PD patients. Overexpression of this mTOR will lead to autophagy inhibition [89]. The activity of mTOR is reduced in animals that cause autophagy induction. Hence, the direct correlation between mTOR and autophagy inducer ULK1 is studied. On the other hand, during energy deficit conditions, AMPK is activated, which inhibits mTORC1, thereby activating ULK1 [90]. This AMPK is further inactivated by mTOR. These shreds of evidence suggest that mTOR, ULK1, and AMPK have a direct relationship. A feedback loop is a mechanism that restores the body to its original state. All stable system has a feedback mechanism to retain control internally. Protein regulator mTOR and autophagy regulator ULK1 and AMPK have a direct feedback link. Two possible cycles exist between them. AMPK activates ULK1 as a process of autophagy induction. This activation leads to direct inhibition of AMPK by ULK1 as a feedback loop. This feedback loop leads to delayed inhibition of ULK1, which results in oscillation of autophagy induction [57]. The double negative-feedback loop in which AMPK directly phosphorylates the mTOR complex and inhibits it. This inhibition results in the inhibition of ULK1, followed by AMPK inhibition [91].
We know that mTOR is activated in nutrient-rich conditions and inhibits autophagy by phosphorylating ULK1. On starvation, phosphorylated AMPK inhibits mTOR, thereby activating autophagy. The factors that activate AMPK are LKB1, CaMKKβ, TAK1, DNA damage, and a decrease in ATP/AMP ratio [76]. On the other hand, ULK1 is activated by AMPK and inhibited by mTOR. ULK1 also counteracts and inhibits mTOR [92]. mTOR inhibition cannot be permanent. Prolonged inhibition of mTOR leads to reactivation [93]. A recent study suggests that AMPK alone cannot induce autophagy when both mTOR and ULK1 are inhibited [94]. AMPK remains dephosphorylated until mTOR is activated [95]. Therefore, the balance between autophagy and mTOR is challenging to achieve. Currently, mTOR, ULK1, and AMPK gained an attractive target for the treatment of PD. These feedback loops help maintain homeostasis and regulate the balance between protein synthesis and clearance. Hence, the complex interplay of the feedback loops has to be explored, and a better understanding of their autophagy modulation will help us treat PD.
Potential PD treatment by targeting mTOR and autophagy
Targeting mTOR and autophagy has emerged as a potential therapeutic strategy for PD due to their critical roles in cellular homeostasis and protein degradation. Dysregulation of mTOR and autophagy pathways has been implicated in the accumulation of toxic protein aggregates, like alpha-synuclein, and the degeneration of dopaminergic neurons, hallmark features of PD [96]. Here are some approaches for potential PD treatment by targeting mTOR and autophagy:
mTOR inhibitors
Rapamycin and its analogs (rapalog) are mTOR inhibitors that have shown promise in preclinical studies for their neuroprotective effects in PD models. These inhibitors promote autophagy and reduce the accumulation of toxic protein aggregates [97]. However, further research is needed to optimize their dosing and delivery to the brain while minimizing side effects. Small molecules that control autophagy and mTOR can be targeted, which assists in mTOR and autophagy balance (Table 1).
Activators of autophagy
Enhancing autophagy through pharmacological interventions has been explored as a therapeutic strategy for PD. Certain compounds, such as trehalose [98] and lithium [99,100], have been shown to induce autophagy and promote the clearance of toxic proteins. Clinical trials to evaluate their efficacy and safety in PD patients are ongoing.
Lysosomal enzyme modulators
Enhancing lysosomal function and promoting efficient degradation of toxic protein aggregates is another potential approach. Small molecules that modulate lysosomal enzymes, such as glucocerebrosidase (GBA), have shown promise in preclinical studies and may hold therapeutic potential for PD with GBA mutations [101].
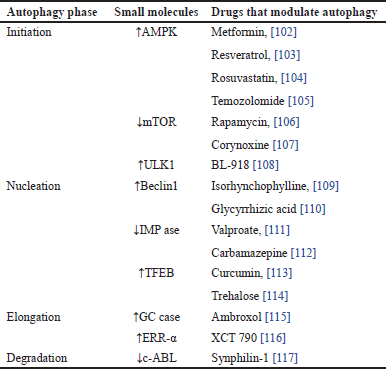 | Table 1. The action of small molecules and drugs on various phases of autophagy. [Click here to view] |
CONCLUSION
Due to its dual function of neuroprotective and neurotoxic effects, the role of mTOR in neurodegenerative diseases is still debated. Because mTOR is critical for protein synthesis and cell survival processes, it is worth noting that mTOR inhibition causes autophagy to activate, which may play a role in neuroprotection. Long-term inhibition, however, can trigger the feedback loop and mTORC1, preventing autophagy. As a result, mTOR is a double-edged sword. It can play both neuroprotective and neurotoxic. Hence, the signaling between mTOR and autophagy has to be balanced. If the balance between mTOR and autophagy can be achieved, mTOR might be a possible target for the therapy of PD.
ACKNOWLEDGMENTS
The authors thank JSS College of Pharmacy, Mysuru for providing the facilities during the manuscript preparation. The authors express their gratitude to Biorender.com (free trial version) for enabling the creation of all the figures.
LIST OF ABBREVIATIONS
AD, Alzheimer’s disease; ALR, Autophagic lysosome reformation; AMPK, Adenosine monophosphate-activated protein kinase; DEPTOR, DEP domain-containing mTOR interacting protein; eEF2K, Eukaryotic elongation factor 2 kinase; IGF-1, Insulin-like growth factor 1; mSIN1, Mammalian stress-activated protein kinase-Interacting protein 1; mTOR, Mammalian/mTOR; mTORC1 mTOR complex 1; mTORC2, mTOR complex 2; PD, Parkinson’s disease; PKCα, Protein kinase C alpha; Raptor, Regulatory-associated protein of mTOR; REDD1, Regulated in development and DNA damage responses; SGK1, Serum/glucocorticoid regulated kinase 1; Tel2, Telomere maintenance 2; TIF1A, Tripartite motif-containing protein-24; ULK1, Unc-51-like kinase 1.
AUTHOR CONTRIBUTION
TLN, SKM, DT, AB, YMT, SK, and SNM made a significant contribution to the work reported, whether that is in the conception, execution, or the acquisition, analysis, or interpretation of data, or all the areas; took part in drafting, revising, or critically reviewing the article; and gave final approval of the version to be published. All have read and agreed to the published version of the manuscript.
FINANCIAL SUPPORT
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
CONSENT TO PARTICIPATE
It is a review article, thus it is not applicable
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
The data that support the findings of this study are available in standard research databases such as PubMed, Science Direct, or Google Scholar, and/or on public domains that can be searched with either key words or DOI numbers.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Ping LA, Chen J, Zhao Y, Chai Z, Hu Y. mTOR signaling in Parkinson’s disease. Neuromolecular Med. 2017;19(1):1–10. CrossRef
2. Yang W, Hamilton JL, Kopil C, Beck JC, Tanner CM, Albin RL, et al. Current and projected future economic burden of Parkinson’s disease in the U.S. NPJ Parkinsons Dis [Internet]. 2020 Dec 1 [cited 2023 Aug 21];6(1):15. CrossRef
3. Darden L. Mechanisms and models. Camb Companion Philos Biol. 2007;39:139–59. CrossRef
4. Laplante M, Sabatini DM. MTOR signaling in growth control and disease. Cell. 2012;149(2):274–93. CrossRef
5. Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95(4):1432–7. CrossRef
6. Deleyto-Seldas N, Efeyan A. The mTOR–autophagy axis and the control of metabolism. Front Cell Dev Biol. 2021 Jul 1;9:655731. CrossRef
7. Crews L, Spencer B, Desplats P, Patrick C, Paulino A, Rockenstein E, et al. Selective molecular alterations in the autophagy pathway in patients with lewy body disease and in models of α-synucleinopathy. PLoS One. 2010;5(2):e9313. CrossRef
8. Tan C, Ai J, Zhu Y. mTORC1-dependent protein and Parkinson’s disease: a mendelian randomization study. Brain Sci [Internet]. 2023 Apr 1 [cited 2023 Aug 21];13(4):536. Available from: /pmc/articles/PMC10137243/ CrossRef
9. Zhou Q, Liu C, Liu W, Zhang H, Zhang R, Liu J, et al. Rotenone induction of hydrogen peroxide inhibits mTOR-mediated S6K1 and 4E-BP1/eIF4E pathways, leading to neuronal apoptosis. Toxicol Sci. 2015;143(1):81–96. CrossRef
10. Tuo Y, Xiang M. mTOR: a double-edged sword for diabetes. J Leukoc Biol. 2019;106(2):385–95. CrossRef
11. Yang M, Lu Y, Piao W, Jin H. The translational regulation in mTOR pathway. Biomolecules [Internet]. 2022 Jun 1 [cited 2023 Aug 2];12(6):802. CrossRef
12. Yu L, Wei J, Liu P. Attacking the PI3K/Akt/mTOR signaling pathway for targeted therapeutic treatment in human cancer. Semin Cancer Biol [Internet]. 2022 Oct 1 [cited 2023 Aug 21];85:69–94. Available from: https://pubmed.ncbi.nlm.nih.gov/34175443/ CrossRef
13. Sun Y, Wang H, Qu T, Luo J, An P, Ren F, et al. mTORC2: a multifaceted regulator of autophagy. Cell Commun Signal [Internet]. 2023 Dec 1 [cited 2023 Aug 21];21(1):4. CrossRef
14. Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, Takehana K, et al. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem. 2010;285(26):20109–16. CrossRef
15. Feng SW, Wu ZS, Chiu YL, Huang SM. Exploring the functional roles of telomere maintenance 2 in the tumorigenesis of glioblastoma multiforme and drug responsiveness to temozolomide. Int J Mol Sci. 2023;24:9256. CrossRef
16. Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25(6):903–15. CrossRef
17. Eum WS, Kim DW, Yeo EJ, Yeo HJ, Choi YJ, Cha HJ, et al. Transduced Tat-PRAS40 prevents dopaminergic neuronal cell death through ROS inhibition and interaction with 14-3-3σ protein. Free Radic Biol Med [Internet]. 2021 Aug 20 [cited 2023 Aug 21];172:418–29. CrossRef
18. de la Cruz López KG, Toledo Guzmán ME, Sánchez EO, García Carrancá A. mTORC1 as a regulator of mitochondrial functions and a therapeutic target in cancer. Front Oncol. 2019 Dec 13;9:492202. CrossRef
19. Di Malta C, Siciliano D, Calcagni A, Monfregola J, Punzi S, Pastore N, et al. Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science (1979). 2017;356(6343):1188–93. CrossRef
20. Beirowski B, Wong KM, Babetto E, Milbrandt J. MTORC1 promotes proliferation of immature Schwann cells and myelin growth of differentiated Schwann cells. Proc Natl Acad Sci U S A. 2017;114(21):E4261–70. CrossRef
21. Chiarini F, Evangelisti C, McCubrey JA, Martelli AM. Current treatment strategies for inhibiting mTOR in cancer. Trends Pharmacol Sci. 2015;36(2):124–35. CrossRef
22. Caron A, Richard D, Laplante M. The roles of mTOR complexes in lipid metabolism. Annu Rev Nutr. 2015;35(1):321–48. CrossRef
23. Wang B, Jie Z, Joo D, Ordureau A, Liu P, Gan W, et al. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature. 2017;545(7654):365–9. CrossRef
24. Zhang Z, Sun X, Wang K, Yu Y, Zhang L, Zhang K, et al. Hydrogen-saturated saline mediated neuroprotection through autophagy via PI3K/AKT/mTOR pathway in early and medium stages of rotenone-induced Parkinson’s disease rats. Brain Res Bull. 2021;172(January):1–13. CrossRef
25. Querfurth H, Lee HK. Mammalian/mechanistic target of rapamycin (mTOR) complexes in neurodegeneration. Mol Neurodegener. 2021;16(1):1–25. CrossRef
26. Fitzian K, Brückner A, Brohée L, Zech R, Antoni C, Kiontke S, et al. TSC1 binding to lysosomal PIPs is required for TSC complex translocation and mTORC1 regulation. Mol Cell. 2021 Jul 1;81(13):2705–21.e8. CrossRef
27. Zhong Y, Zhou X, Guan KL, Zhang J. Rheb regulates nuclear mTORC1 activity independent of farnesylation. Cell Chem Biol. 2022 Jun 16;29(6):1037–45.e4. CrossRef
28. Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1(1):15–25. CrossRef
29. Murakami T. Isometric contraction induces transient increase of REDD1 expression in non-contracted muscles partly through glucocorticoids. Physiol Rep [Internet]. 2023 Jun 1 [cited 2023 Aug 22];11(11):e15745. CrossRef
30. Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev. 2013;23(1):53–62. CrossRef
31. Andreou AZ, Klostermeier D. The DEAD-box helicase eIF4A. RNA Biol. 2013;10(1):19–32. CrossRef
32. Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol. 2008;49(11):1634–44. CrossRef
33. Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10(5):307–18. CrossRef
34. Maiti P, Manna J, Dunbar GL, Maiti P, Dunbar GL. Current understanding of the molecular mechanisms in Parkinson’s disease: targets for potential treatments. Transl Neurodegener. 2017;6(1):1–35. CrossRef
35. Huang J, Wu S, Wu CL, Manning BD. Signaling events downstream of mammalian target of rapamycin complex 2 are attenuated in cells and tumors deficient for the tuberous sclerosis complex tumor suppressors. Cancer Res. 2009;69(15):6107–14. CrossRef
36. Rosner M, Hengstschläger M. Cytoplasmic and nuclear distribution of the protein complexes mTORC1 and mTORC2: rapamycin triggers dephosphorylation and delocalization of the mTORC2 components rictor and sin1. Hum Mol Genet. 2008;17(19):2934–48. CrossRef
37. Fu W, Wu G. Targeting mTOR for anti-aging and anti-cancer therapy. Molecules. 2023;28:3157. CrossRef
38. Tsai PJ, Lai YH, Manne RK, Tsai YS, Sarbassov D, Lin HK. Akt: a key transducer in cancer. J Biomed Sci. 2022;29:1–17. CrossRef
39. Knight J, Caseldine C, Boykoff MT. Forum review. Geogr J. 2010;176(3):267–9. CrossRef
40. Zhu Z, Yang C, Iyaswamy A, Krishnamoorthi S, Sreenivasmurthy SG, Liu J, et al. Balancing mTOR signaling and autophagy in the treatment of Parkinson’s disease. Int J Mol Sci. 2019;20(3):1–15. CrossRef
41. Malagelada C, Jin ZH, Jackson-Lewis V, Przedborski S, Greene LA. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson’s disease. J Neurosci. 2010;30(3):1166–75. CrossRef
42. Chen L, Xu B, Liu L, Luo Y, Yin J, Zhou H, et al. Hydrogen peroxide inhibits mTOR signaling by activation of AMPKα leading to apoptosis of neuronal cells. Lab Invest. 2010;90(5):762–73. CrossRef
43. Kim SR, Chen X, Oo TF, Kareva T, Yarygina O, Wang C, et al. Dopaminergic pathway reconstruction by Akt/Rheb-induced axon regeneration. Ann Neurol. 2011;70(1):110–20. CrossRef
44. Domanskyi A, Geiβler C, Vinnikov IA, Alter H, Schober A, Vogt MA, et al. Pten ablation in adult dopaminergic neurons is neuroprotective in Parkinson’s disease models. FASEB J. 2011;25(9):2898–910. CrossRef
45. Deyoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22(2):239–51. CrossRef
46. Ainslie GR, Gibson KM, Vogel KR. mTOR, autophagy, aminoacidopathies, and human genetic disorders. Molecules to medicine with mTOR: translating critical pathways into novel therapeutic strategies. Amsterdam, The Netherlands: Elsevier Inc.; 2016. 143–66 pp. CrossRef
47. Licausi F, Hartman NW. Role of mTOR complexes in neurogenesis. Int J Mol Sci. 2018;19(5):1544. CrossRef
48. Mao Z, Zhang W. Role of mTOR in glucose and lipid metabolism. Int J Mol Sci. 2018;19(7):1–14. CrossRef
49. Yoon MS. mTOR as a key regulator in maintaining skeletal muscle mass. Front Physiol. 2017;8(OCT):1–9. CrossRef
50. Morita M, Gravel SP, Hulea L, Larsson O, Pollak M, St-Pierre J, et al. MTOR coordinates protein synthesis, mitochondrial activity. Cell Cycle. 2015;14(4):473–80. CrossRef
51. Wills J, Credle J, Oaks AW, Duka V, Lee JH, Jones J, et al. Paraquat, but not maneb, induces synucleinopathy and tauopathy in striata of mice through inhibition of proteasomal and autophagic pathways. PLoS One. 2012;7(1):1–12. CrossRef
52. Mao B, Zhang Q, Ma L, Zhao DS, Zhao P, Yan P, et al. Overview of research into mTOR inhibitors. Molecules. 2022;27:5295. CrossRef
53. Xu Y, Liu C, Chen S, Ye Y, Guo M, Ren Q, et al. Activation of AMPK and inactivation of Akt result in suppression of mTOR-mediated S6K1 and 4E-BP1 pathways leading to neuronal cell death in in vitro models of Parkinson’s disease. Cell Signal. 2014;26(8):1680–9. CrossRef
54. Watson CJE, Clatworthy MR. mTOR inhibitors: sirolimus and everolimus. Kidney transplantation—principles and practice. 8th ed. Amsterdam, The Netherlands: Elsevier; 2019. pp 261–82. CrossRef
55. Van Duijnhoven EM, Boots JMM, Christiaans MHL, Wolffenbuttel BHR, Van Hooff JP. Influence of tacrolimus on glucose metabolism before and after renal transplantation: a prospective study. J Am Soc Nephrol. 2001;12(3):583–8. CrossRef
56. Guo Z, Chen X, Feng P, Yu Q. Short-term rapamycin administration elevated testosterone levels and exacerbated reproductive disorder in dehydroepiandrosterone-induced polycystic ovary syndrome mice. J Ovarian Res. 2021;14(1):1–10. CrossRef
57. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–41. CrossRef
58. Jung S, Jeong H, Yu SW. Autophagy as a decisive process for cell death. Exp Mol Med. 2020;52(6):921–30. CrossRef
59. Denton D, Kumar S. Autophagy-dependent cell death. Cell Death Differ. 2019;26(4):605–16. CrossRef
60. Yonekawa T, Thorburn A. Autophagy and cell death. Essays Biochem. 2013;55(1):105–17. CrossRef
61. Pandey S, Srivanitchapoom P. Levodopa-induced Dyskinesia: clinical features, pathophysiology, and medical management. Ann Indian Acad Neurol [Internet]. 2017 Jul 1 [cited 2023 Aug 2];20(3):190. Available from: /pmc/articles/PMC5586110/ CrossRef
62. Jankovic J. Motor fluctuations and dyskinesias in Parkinson’s disease: clinical manifestations. Mov Disord. 2005;20(SUPPL. 11):S11–6. CrossRef
63. Gorman AM. Neuronal cell death in neurodegenerative diseases: recurring themes around protein handling: apoptosis review series. J Cell Mol Med. 2008;12(6A):2263–80. CrossRef
64. Guo F, Liu X, Cai H, Le W. Autophagy in neurodegenerative diseases: pathogenesis and therapy. Brain Pathol. 2018;28(1):3–13. CrossRef
65. Mushtaq A, Ashraf NU, Altaf M. The mTORC1-G9a-H3K9me2 axis negatively regulates autophagy in fatty acid-induced hepatocellular lipotoxicity. J Biol Chem [Internet]. 2023 Mar 1 [cited 2023 Aug 22];299(3):102937. CrossRef
66. Tanemura M, Ohmura Y, Deguchi T, MacHida T, Tsukamoto R, Wada H, et al. Rapamycin causes upregulation of autophagy and impairs islets function both in vitro and in vivo. Am J Transplant. 2012;12(1):102–14. CrossRef
67. Meng LH, Zheng XF. Toward rapamycin analog (rapalog)-based precision cancer therapy. Acta Pharmacol Sin. 2015;36(10):1163–9. CrossRef
68. Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. 2009;335:3112–23.
69. Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta Mol Cell Res. 2009;1793(4):664–73. CrossRef
70. Levine B, Kroemer G. SnapShot: macroautophagy. Cell. 2008;132(1):162.e1–162.e3. CrossRef
71. Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69(7):1125–36. CrossRef
72. Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3(4):295–9. CrossRef
73. Dunlop EA, Tee AR. MTOR and autophagy: a dynamic relationship governed by nutrients and energy. Semin Cell Dev Biol. 2014;36:121–9. CrossRef
74. Kim YC, Guan KL. MTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125(1):25–32. CrossRef
75. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221(1):3–12. CrossRef
76. Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32(1):2–11. CrossRef
77. Settembre C, Di Malta C, Polito VA, Arencibia MG, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science (1979). 2011;332(6036):1429–33. CrossRef
78. Chen Y, Gucek M, Puertollano R, Martina JA. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8(6):877–6. CrossRef
79. Jaber N, Zong WX. Class III PI3K Vps34: essential roles in autophagy, endocytosis, and heart and liver function. Ann N Y Acad Sci. 2013;1280(1):48–51. CrossRef
80. Jaber N, Zong WX. Class III PI3K Vps34: essential roles in autophagy, endocytosis, and heart and liver function. Ann N Y Acad Sci. 2013 Mar;1280:48–51. CrossRef
81. Chen Y, Yu L. Autophagic lysosome reformation. Exp Cell Res. 2013;319(2):142–6. CrossRef
82. Rong Y, McPhee C, Denga S, Huanga L, Chen L, Liu M, et al. Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc Natl Acad Sci U S A. 2011;108(19):7826–31. CrossRef
83. Chen Y, Yu L. Development of research into autophagic lysosome reformation. Mol Cells. 2018;41(1):45–9.
84. Navarro-Romero A, Montpeyó M, Martinez-Vicente M. The emerging role of the lysosome in Parkinson’s disease. Cells [Internet]. 2020 Nov 2 [cited 2023 Aug 2];9(11):2399. Available from: /pmc/articles/PMC7692401/ CrossRef
85. Abu-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, et al. Lysosomal metabolomics reveals V-ATPase-and mTOR-dependent regulation of amino acid efflux from lysosomes. Science [Internet]. 2017 Nov 10 [cited 2023 Aug 2];358(6364):807–13. CrossRef
86. Arotcarena ML, Soria FN, Cunha A, Doudnikoff E, Prévot G, Daniel J, et al. Acidic nanoparticles protect against α-synuclein-induced neurodegeneration through the restoration of lysosomal function. Aging Cell [Internet]. 2022 Apr 1 [cited 2023 Aug 2];21(4):e13584. CrossRef
87. Fujii T, Nagamori S, Wiriyasermkul P, Zheng S, Yago A, Shimizu T, et al. Parkinson’s disease-associated ATP13A2/PARK9 functions as a lysosomal H+,K+-ATPase. Nat Commun. 2023;14:1–11. CrossRef
88. Deneubourg C, Ramm M, Smith LJ, Baron O, Singh K, Byrne SC, et al. The spectrum of neurodevelopmental, neuromuscular and neurodegenerative disorders due to defective autophagy. Autophagy [Internet]. 2022 [cited 2023 Aug 21];18(3):496–517. CrossRef
89. Wen Z, Zhang J, Tang P, Tu N, Wang K, Wu G. Overexpression of miR-185 inhibits autophagy and apoptosis of dopaminergic neurons by regulating the AMPK/mTOR signaling pathway in Parkinson’s disease. Mol Med Rep. 2018;17(1):131–7. CrossRef
90. Rey V, Tamargo-Gómez I. From kinases to diseases: investigating the role of AMPK in human pathologies. Kinases Phosphatases. 2023;1:181–205. Available from: https://www.mdpi.com/2813-3757/1/3/12/htm CrossRef
91. Holczer M, Hajdú B, Lorincz T, Szarka A, Bánhegyi G, Kapuy O. Fine-tuning of AMPK–ULK1–mTORC1 regulatory triangle is crucial for autophagy oscillation. Sci Rep. 2020;10(1):1–12. CrossRef
92. Zou L, Liao M, Zhen Y, Zhu S, Chen X, Zhang J, et al. Autophagy and beyond: unraveling the complexity of UNC-51-like kinase 1 (ULK1) from biological functions to therapeutic implications. Acta Pharm Sin B. 2022 Oct 1;12(10):3743–82. CrossRef
93. Hua R, Han S, Zhang N, Dai Q, Liu T, Li J. CPKCγ-modulated sequential reactivation of mTOR inhibited autophagic flux in neurons exposed to oxygen glucose deprivation/reperfusion. Int J Mol Sci. 2018;19(5):1380. CrossRef
94. Huang WQ, Wen JL, Lin RQ, Wei P, Huang F. Effects of mTOR/NF-κB signaling pathway and high thoracic epidural anesthesia on myocardial ischemia-reperfusion injury via autophagy in rats. J Cell Physiol. 2018;233(9):6669–78. CrossRef
95. Holczer M, Hajdú B, Lorincz T, Szarka A, Bánhegyi G, Kapuy O. A double negative feedback loop between MTORC1 and AMPK kinases guarantees precise autophagy induction upon cellular stress. Int J Mol Sci. 2019;20(22):5543. CrossRef
96. Tewari D, Patni P, Bishayee A, Sah AN, Bishayee A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: a novel therapeutic strategy. Semin Cancer Biol [Internet]. 2022 May 1 [cited 2023 Aug 21];80:1–17. CrossRef
97. Wang D, Eisen HJ. Mechanistic target of rapamycin (mTOR) inhibitors. Handb Exp Pharmacol [Internet]. 2022 [cited 2023 Aug 2];272:53–72. CrossRef
98. Darabi S, Noori-Zadeh A, Abbaszadeh HA, Rajaei F, Bakhtiyari S. Trehalose neuroprotective effects on the substantia nigra dopaminergic cells by activating autophagy and non-canonical Nrf2 pathways. Iran J Pharm Res [Internet]. 2019 Jun 1 [cited 2023 Aug 2];18(3):1419. Available from: /pmc/articles/PMC6934986/
99. Lazzara CA, Kim YH. Potential application of lithium in Parkinson’s and other neurodegenerative diseases. Front Neurosci [Internet]. 2015 [cited 2023 Aug 2];9(OCT):403. Available from: /pmc/articles/PMC4621308/ CrossRef
100. Puglisi-Allegra S, Lazzeri G, Busceti CL, Giorgi FS, Biagioni F, Fornai F. Lithium engages autophagy for neuroprotection and neuroplasticity: translational evidence for therapy. Neurosci Biobehav Rev [Internet]. 2023 May 1 [cited 2023 Aug 21];148:105148. CrossRef
101. Schapira AHV. Glucocerebrosidase and Parkinson disease: recent advances. Mol Cell Neurosci [Internet]. 2015 May 1 [cited 2023 Aug 2];66(Pt A):37–42. CrossRef
102. Lu G, Wu Z, Shang J, Xie Z, Chen C, Zhang C. The effects of metformin on autophagy. Biomed Pharmacother. 2021;137(January):111286. CrossRef
103. Tian Y, Song W, Li D, Cai L, Zhao Y. Resveratrol as a natural regulator of autophagy for prevention and treatment of cancer. Onco Targets Ther. 2019;12:8601–9. CrossRef
104. Kang SY, Lee SB, Kim HJ, Kim HT, Yang HO, Jang W. Autophagic modulation by rosuvastatin prevents rotenone-induced neurotoxicity in an in vitro model of Parkinson’s disease. Neurosci Lett. 2017;642:20–6. CrossRef
105. Zou Y, Wang Q, Li B, Xie B, Wang W. Temozolomide induces autophagy via ATM-AMPK-ULK1 pathways in glioma. Mol Med Rep. 2014;10(1):411–6. CrossRef
106. Lamming DW. Inhibition of the mechanistic target of rapamycin (mTOR)–rapamycin and beyond. Cold Spring Harb Perspect Med. 2016;6(5):a025924. CrossRef
107. Chen LL, Song JX, Lu JH, Yuan ZW, Liu LF, Durairajan SSK, et al. Corynoxine, a natural autophagy enhancer, promotes the clearance of alpha-synuclein via Akt/mTOR pathway. J Neuroimmune Pharmacol. 2014;9(3):380–7. CrossRef
108. Ouyang L, Zhang L, Zhang S, Yao D, Zhao Y, Wang G, et al. Small-molecule activator of UNC-51-like kinase 1 (ULK1) that induces cytoprotective autophagy for Parkinson’s disease treatment. J Med Chem. 2018;61(7):2776–92. CrossRef
109. Lu JH, Tan JQ, Durairajan SS, Liu LF, Zhang ZH, Ma L, et al. Isorhynchophylline, a natural alkaloid, promotes the degradation of alpha-synuclein in neuronal cells via inducing autophagy. Autophagy. 2012 Jan;8(1):98–108. CrossRef
110. Laconi S, Madeddu MA, Pompei R. Autophagy activation and antiviral activity by a licorice triterpene. Phytother Res. 2014;28(12):1890–2. CrossRef
111. Salsaa M, Aziz K, Lazcano P, Schmidtke MW, Tarsio M, Hüttemann M, et al. Valproate activates the Snf1 kinase in Saccharomyces cerevisiae by decreasing the cytosolic pH. J Biol Chem. 2021;297(4):101110. CrossRef
112. Motoi Y, Shimada K, Ishiguro K, Hattori N. Lithium and autophagy. ACS Chem Neurosci. 2014;5(6):434–42. CrossRef
113. Zhang J, Wang J, Xu J, Lu Y, Jiang J, Wang L, et al. Curcumin targets the TFEB-lysosome pathway for induction of autophagy. Oncotarget. 2016;7(46):75659–71. CrossRef
114. Rusmini P, Cortese K, Crippa V, Cristofani R, Cicardi ME, Ferrari V, et al. Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy. 2019;15(4):631–51. CrossRef
115. Kopytova AE, Rychkov GN, Nikolaev MA, Baydakova GV, Cheblokov AA, Senkevich KA, et al. Ambroxol increases glucocerebrosidase (GCase) activity and restores GCase translocation in primary patient-derived macrophages in Gaucher disease and Parkinsonism. Parkinsonism Relat Disord. 2021;84(January):112–21. CrossRef
116. Suresh SN, Jayaprakash Rao M, Manjithaya R. XCT 790 is a pharmacological aggrephagy inducer in a yeast model of proteotoxicity. Cell Biol Int. 2021;45(3):654–61. CrossRef
117. Zhou ZH, Wu YF, Wang XM, Han YZ. The c-Abl inhibitor in Parkinson disease. Neurol Sci. 2017;38(4):547–52. CrossRef