INTRODUCTION
As of 2020, leukemia has resulted in not only over 474,000 cases but also more than 311,000 deaths [1,2]. The majority of the incident cases came from children between 0 and 5 years of age, whereas the highest death rates were recorded from patients above 60 years of age. In addition to the alarmingly high statistics, both rates are projected to increase continuously through 2030 [2].
Currently, chemotherapeutic drugs remain the main treatment method for leukemia, whereby regimens are given based on the patient’s subtype [3]. However, adverse effects have been reported from patients upon starting treatment [4–6]. Furthermore, the aforementioned higher death rates in patients above 60 years of age were majorly attributed to factors such as intensive chemotherapy [2], as this frequently results in increased toxicity towards older patients [7,8]. Therefore, it is of upmost importance to discover novel treatments that are both safe and have lesser adverse effects, to treat leukemia.
Melatonin, or N-acetyl-5-methoxytryptamine, although frequently known for its hormonal activities, has been reported to exert anticancer effects in different types of cancer [9]. The aforementioned properties have also been explored as an option to treat leukemia [10]. For instance, melatonin was reported to decrease cell proliferation in mixed-lineage leukemia-rearranged (MLL-r) leukemia cells by reducing NF-κB DNA binding [11], as well as causing cell arrest at the G1 phase in different lymphoid cell lines (Ramos, SU-DHL-4, and DoHH2) [12]. There also have been observations of melatonin-induced apoptosis in cancer cells via different pathways, such as the caspase-dependent [13], MAPK, and mTOR pathways [14]. It is worth noting that while melatonin does exert such effects on cancerous cells, it does not affect normal cells, but instead provides cytoprotective effects [15]. With these properties, melatonin is an excellent candidate for cancer therapeutics.
Many cellular pathways have been explored as potential pharmacological targets in order to find novel anticancer therapies. One such pathway, mitophagy, has been a conversational piece over the recent years. Mitophagy has been well-established as one of the quality control mechanisms for mitochondria, as it selectively removes highly dysfunctional mitochondria via autophagy to maintain homeostasis [16]. Depending on the tumor type and molecular context, mitophagy may either promote or inhibit tumorigenesis in cancer cells [17]. In terms of leukemia, the usage of dexamethasone in T-acute lymphoblastic leukemia (T-ALL) cancer cell lines have reported that while induction of mitophagy caused cell death in CCRF-CEM cells, it served as a survival mechanism for JURKAT cells [18]. A deeper insight into the effect of different drugs on modulating mitophagy in leukemic cell lines is required to not only discover potential anticancer compounds, but also understand cancer drug resistance.
Thus, to support our previous work [14,19], as well as the usage of melatonin as an anticancer therapeutic agent, we investigated whether melatonin could induce apoptosis via mitophagy-mediated pathways in JURKAT cells.
MATERIALS AND METHODS
Materials
The JURKAT cell line is a cell line originating from T-cell leukemia and has been established as a cancer cell model for T-ALL [20,21]. The JURKAT cell line and Chang Liver cell line were purchased from the American type culture collection. Dimethyl sulfoxide (DMSO) and bovine serum albumin (BSA) were purchased from Nacalai Tesque (Kyoto, Japan). Dulbecco’s modified Eagle medium (DMEM), fetal bovine serum (FBS), MTT, melatonin, and acridine orange (AO) were purchased from Sigma Aldrich (Saint Louis, MO, USA). TUDCA was purchased from Merck Millipore (Burlington, MA, USA). The primary antibodies β-actin (13E5) Rabbit mAb, BNIP3 (D7U1T) Rabbit mAb 44060, BNIP3L/Nix (D4R4B) Rabbit mAb 12396, Optineurin/OPTINEURIN (D2L8S) Rabbit mAb 58981, NDP52 (D1E4A) Rabbit mAb 60732, SQSTM1/p62 (D5E2) Rabbit mAb 8025, LC3B (D11) XP® Rabbit mAb 3868, and horseradish peroxidase-conjugated secondary antibodies (Anti-rabbit IgG, HRP-linked Antibody 7074, Anti-mouse IgG, HRP-linked Antibody 7076) were purchased from Cell Signalling Technology (Danvers, MA, USA). SuperSignalTM West Femto Maximum Sensitivity Substrate was purchased from Thermo Fisher Scientific (Waltham, MA, USA). The stock solution of melatonin (100 mg/ml) was dissolved in absolute ethanol. Preparations of different concentrations in the culture medium should have a final ethanol concentration of not more than 0.5%.
Cell culture
JURKAT cells and Chang liver cells were cultured in T-25 flasks with DMEM, supplemented with 10% FBS in an incubator with 5% CO2 at 37°C. Once 80% confluency is reached, the cells were collected and harvested by centrifugation (1,500 rpm, 5 minutes). The cells were then sub-cultured into new T-25 flasks or plated for assays [14].
Cell viability assay
JURKAT cells and Chang Liver cells were seeded at 5,000 cells (in 100 µl culture medium) per well in 96-well plates, followed by incubation for 24 hours. JURKAT cells were then treated with different concentrations of melatonin (0.1, 0.2, 0.3, 0.4, 0.5 mg/ml) and incubated for 72 hours (37°C, 5% CO2). Chang Liver cells were incubated for 72 hours with 0.5 mg/ml of melatonin. 20 µl of MTT solution was added into each well, followed by an additional 3 hour-incubation. Following centrifugation (2,000 rpm, 5 minutes), the supernatants were removed and 100 µl of DMSO was added to dissolve the formazan crystals. The assay was then read at 570 nm, with 630 nm as the reference wavelength, using a spectrophotometer. The procedures for the assay were repeated at least three times [14,19].
The equation used to calculate the cell viability is indicated below:
The half-maximal inhibitory concentration (1C50) value was obtained from a dose-response curve between the concentrations of melatonin and the percentage of cell viability. The IC50 value is defined as the concentration of melatonin that resulted in 50% cell viability [14].
Fluorescence staining
A 24-well plate was used to culture the JURKAT cells with the concentration of 2.4 × 105 cells per well for 24 hours (37°C, 5% CO2). The cells were then treated with melatonin at different concentrations (0.1, 0.2, 0.3, 0.4, 0.5 mg/ml). Following a 72-hour incubation, the cells were washed with 200 µl of PBS and stained with AO (5 µg/ml) for 15 to 20 minutes. Observations of the cells were done under an inverted fluorescence microscope Nikon Eclipse Ti (Tokyo, Japan). Photomicrographs of autophagic vacuoles were taken using an attached camera. ImageJ, a semi-automated software, was used for the merging of green and red images to facilitate the average calculation of green to red fluorescence ratio. The procedures for the assay were repeated at least three times [19].
The equation used to calculate the percentage of autophagy is indicated below:
Western blot analysis
JURKAT cells (1.6 × 105 cells per well) were cultured for 24 hours (37°C, 5% CO2) in a 60 mm2 culture dish. Various concentrations of melatonin were used to treat the cells (0.1, 0.2, 0.3, 0.4, 0.5 mg/ml). Following a 72-hour incubation, the cells were harvested and washed with PBS. Cell pellets were lysed in 200 µl of ice-cold DTT lysis buffer (62.5 mM Tris, 2% w/v SDS, 10% glycerol, pH 6.8, 100 mM DTT). Following centrifugation (10,000 rpm, 5 minutes, 4ºC), the supernatant was obtained and stored at –20°C. Protein quantification was done with Quick Start™ bradford protein assay (Bio-Rad, Hercules, CA, USA). Heat denaturation was applied to the lysates prior to loading into the wells. 50 µg of lysate protein was mixed with bromophenol blue (0.1%) and 10% SDS-PAGE gels were used to separate the proteins. Protein separation was done using the Mini-PROTEAN Tetra Vertical Electrophoresis Cell (Bio-Rad, Hercules, CA, USA) with a voltage of 120V for 1 hour. Electro-transferring of proteins was done on ice onto the PVDF membrane using the Mini Trans-Blot® Electrophoretic Transfer Cell (Bio-Rad, Hercules, CA, USA) with a voltage of 100V for 1 hour. Afterward, blocking for 2 hours with 3% BSA was done at room temperature. The membrane was then incubated with primary antibodies (1:1,000) at 4°C overnight. Later, diluted HRP-conjugated secondary antibodies (1:10,000) were incubated with the membrane at room temperature for 2 hours. After washing with TBST and TBS, protein expressions were visualized using SuperSignalTM West Femto Maximum Sensitivity Substrate. The bands’ intensity was then measured using ChemiDoc XRS+ with ImageLab software (Bio-Rad, Hercules, CA, USA). The procedures were then repeated at least three times [14,19].
Statistical analysis
All experiments were performed in biological triplicates; therefore, the given values are representative of at least three independent experiments. Each result was expressed as mean ± S.E.M. One-Way ANOVA (GraphPad Prism 10.2.0) was used as the statistical analysis method and Tukey’s Multiple Comparison test was selected as the preferred post-hoc test. Values < 0.05 were considered as statistically significant.
RESULTS AND DISCUSSION
Melatonin induces cytotoxicity in JURKAT ells
Büyükavci et al. [22] observed that melatonin causes cytotoxicity in leukemic cells, with moderate effects seen at 10–3 M. Furthermore, based on our previous work [14], it has been established that melatonin reduced the cell viability of JURKAT cells after 72 hours of treatment. Thus, the MTT assay was utilised to quantify the cell viability of JURKAT cells following the 72-hour incubation period.
Based on Figure 1, it was observed that melatonin treatment reduced the viability of JURKAT cells by 99.39%, 88.69%, 76.40%, 71.59%, and 61.55% at 0.1, 0.2, 0.3, 0.4, and 0.5 mg/ml respectively. The cell viability of the vehicle group was determined to be 96.67% when 0.5% ethanol was used (graph not provided). Results indicate that the cell viability decreased according to a dose-dependent manner; however, no statistical significance was found between dosage groups. The only statistically significant reduction (p < 0.05) was observed in the highest concentration (0.5 mg/ml) when compared with the control group (0 mg/ml) and 0.1 mg/ml of melatonin. This observation corresponds to our previous work by Kasi et al. [14] as they reported a decrease in JURKAT cell viability in a dose-dependent manner after treatment with melatonin for 72 hours, with 74% cell viability corresponding to 0.5 mg/ml [14]. The IC50 value was estimated to be higher than 0.74 mg/ml. Kasi et al. [14] also reported an IC50 value higher than 0.60 mg/ml for JURKAT cells [14]. Additionally, the cell viability of Chang liver cells following a 72-hour melatonin treatment was studied as well, in order to prove its selective cytotoxicity. As previously mentioned, melatonin was observed to have cytoprotective effects on normal cells [15]. This observation is concordant to our results, as the viability of Chang liver cells at the highest concentration (0.5 mg/ml) was determined to be 101% (graph not provided). Thus, this concludes that melatonin does exert selective antiproliferative properties on JURKAT cells in concentrations of 0.1–0.5 mg/ml.
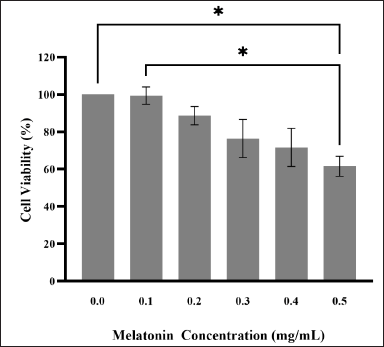 | Figure 1. MTT cell viability assay. JURKAT cells were treated with various concentrations of melatonin. The bar graph shows the means ± standard error of mean (SEM) from three separate experiments. One-Way ANOVA was performed to compare between concentration groups, followed by Tukey’s Multiple Comparison test as post-hoc testing. * indicates p < 0.05. [Click here to view] |
Melatonin induces autophagy in JURKAT cells
Autophagy, the process of degrading and eliminating damaged organelles and misfolded proteins, has been a controversial topic in cancer therapeutics as it exhibits dual roles in tumorigenesis by either inducing cell death or promote tumor survival [23]. Few studies have pointed out that the inhibition of autophagy would enhance apoptosis, leading to cell death [23]. However, there have been reported observations that autophagy may precede apoptosis or work in parallel with apoptosis [24,25]. Hence, we investigated the presence of autophagy in melatonin-treated JURKAT cells.
AO forms aggregates in acidic vesicles, as exemplified by autophagic vacuoles, emitting bright red fluorescence in the process. In contrast, green fluorescence is emitted from the cells’ nucleus and cytoplasm [26]. To investigate if autophagy is involved in the cytotoxicity of melatonin in JURKAT cells, fluorescent staining with AO was implemented after 72 hours of incubation. The calculations for the percent ratio of bright red fluorescent cells to green fluorescent cells were done.
Figure 2 A–F shows the obtained photomicrographs, depicting the formation of autophagic vacuoles in JURKAT cells after the 72-hour treatment period with various concentrations of melatonin (0.1, 0.2, 0.3, 0.4, 0.5 mg/ml). Results indicate that the percentage of red fluorescence in JURKAT cells increased as the concentration of melatonin increased. This qualitative trend was proven to be statistically significant, as according to Figure 3, the proportions of acidic vesicular formation after melatonin treatment (0.1–0.5 mg/ml) were significantly higher (p < 0.05, p < 0.01, p < 0.001) as compared to the control (34%, 35%, 44%, 47%, and 52%, respectively). Furthermore, this qualitative trend suggests a dose-dependent response; however, no statistical significance was found between dosages. This observation is concordant to our previous work by Chok et al. [19], who reported a significant difference in the percentage autophagic vacuole-positive HT-29 cells compared to the control, following treatment with melatonin. Similar observations were also reported in melatonin-cisplatin treated HepG2 cells [27].
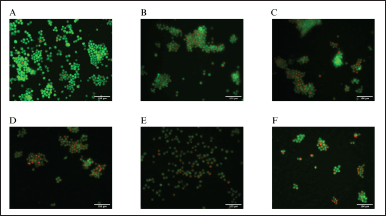 | Figure 2. Acridine orange fluorescence staining. JURKAT cells were treated with various concentrations of melatonin for 72 hours: (A) control, (B) 0.1 mg/ml, (C) 0.2 mg/ml, (D) 0.3 mg/ml, (E) 0.4 mg/ml, and (F) 0.5 mg/ml. Cells were stained with 5 µg/ml of the acridine orange dye for 15–20 minutes, followed by visualization under an inverted fluorescence microscope. [Click here to view] |
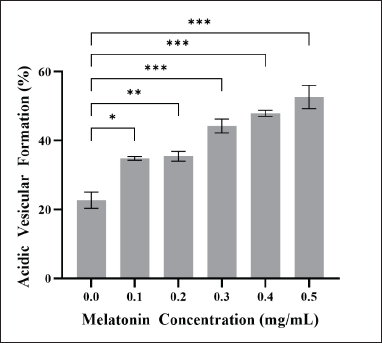 | Figure 3. % Autophagy (% Acidic Vesicular Formation). The bar graph shows the means ± standard error of mean (SEM) from three separate experiments. One-Way ANOVA was performed to compare between concentration groups, followed by Tukey’s Multiple Comparison test as post-hoc testing. * indicates p < 0.05; ** indicates p < 0.01; *** indicates p < 0.001. [Click here to view] |
Melatonin activates autophagy proteins
For cells to identify which cargo to be selectively degraded, autophagy receptors, such as SQSTM1/p62, NDP52, and optineurin proteins are required. These receptors will first recognize the ubiquitinated cargo, and then bind to the LC3 protein on the phagophore membrane [28]. The levels of the mentioned proteins were evaluated after exposing different concentrations of melatonin to the JURKAT cells. Based on Figure 4 and Figure 5, it was discovered that p62/SQSTM1, Optineurin, and NDP52 proteins expressions were generally increased as compared with control and decreased at higher concentrations of melatonin. However, no statistical significance between different concentrations in each investigated protein was discovered. Moreover, there is a decrease of LC3-II/LC3-I ratio if compared with the control (Fig. 5D).
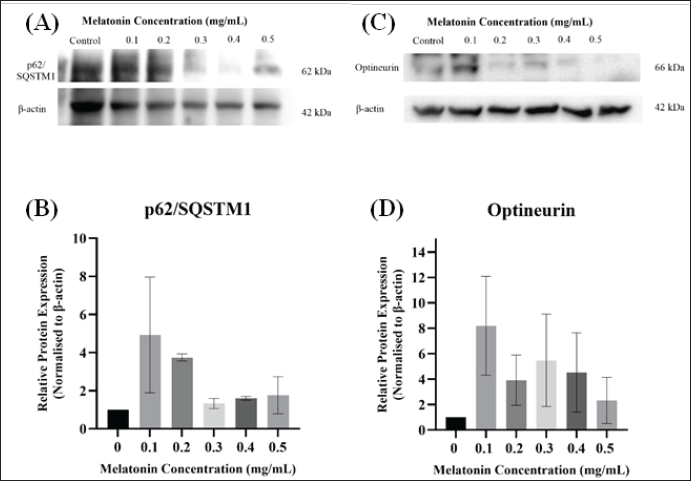 | Figure 4. Western blot analysis of the autophagy proteins, (A) p62/SQSTM1 (band), (B) p62/SQSTM1 (bar chart), (C) Optineurin (band), and (D) Optineurin (bar chart) in JURKAT cells after treatment with different concentrations of melatonin after 72 hours. Expression levels of autophagy proteins were measured by Western blot analysis. Beta-actin (β-actin) served as a loading control. The bar graph below shows the means ± standard error of mean (SEM) from three separate experiments. One-Way ANOVA was performed to compare between concentration groups, followed by Tukey’s Multiple Comparison test as post-hoc testing. [Click here to view] |
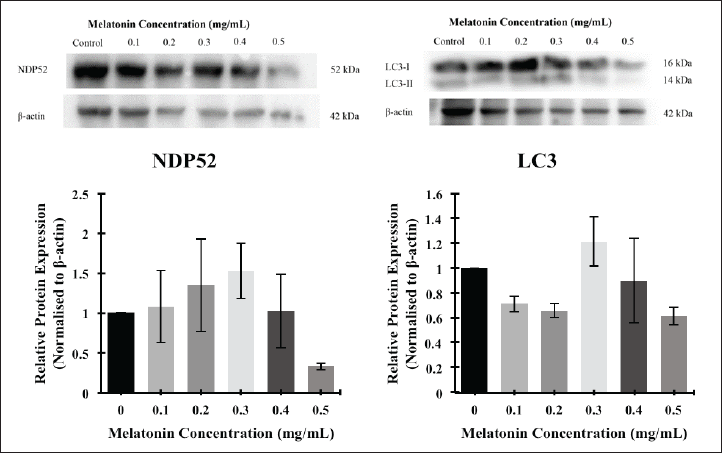 | Figure 5. Western blot analysis of the autophagy proteins, (A) NDP52 (band), (B) NDP52 (bar chart), (C) LC3 (band), and (D) LC3 (bar chart) in JURKAT cells after treatment with different concentrations of melatonin after 72 hours. Expression levels of autophagy proteins were measured by Western blot analysis. Beta-actin (β-actin) served as a loading control. The bar graph below shows the means ± standard error of mean (SEM) from three separate experiments. One-Way ANOVA was performed to compare between concentration groups, followed by Tukey’s Multiple Comparison test as post-hoc testing. [Click here to view] |
Both LC3 and p62/SQSTM1 are recognized as autophagy marker proteins in numerous literatures. The levels of p62/SQSTM1 are typically used as an indicator to study autophagic influx, as studies suggest that p62/SQSTM1 is a critical player in regulating autophagy and its interplay with cancer. Elevating p62/SQSTM1 levels can influence the autophagic process by acting as a selective autophagy receptor, facilitating the degradation of specific substrates and promoting cellular homeostasis [29,30]. In this study, the levels of p62/SQSTM1 were upregulated as the concentration of melatonin increased and downregulated at the highest concentration. This is due to p62/SQSTM1 being degraded during the autophagic process [29]. On the other hand, LC3 serves as an autophagy marker protein, as it is an ATG8 protein homologue associated with autophagosomal membranes. This protein can be cleaved at their carboxyl terminal to form LC3-I, and can be conjugated with phosphatidylethanolamine, forming LC3-II that firmly binds to autophagosomal membranes [31]. Our results showed that there is a decrease in of LC3-II/LC3-I ratio if compared with control (Fig. 5D). A decreased LC3-II/LC3-I ratio does not necessarily mean reduced autophagy. In some cases, a decreased ratio was associated with improved autophagic flux and increased autophagy efficiency. For example, in the study on exercise-induced cardioprotection, the LC3-II/LC3-I ratio decreased in the group with high-intensity exercise, but this was accompanied by increased levels of other autophagy markers like Beclin-1 and Cathepsin D. This indicates that the autophagic process was enhanced, despite the lowered LC3-II/LC3-I ratio [32].
The effects of melatonin have been documented in numerous signaling pathways used in cancer cells, such as the Notch signaling pathway, Wnt/ β-catenin pathway, and ERK/MAPK pathway, for numerous purposes including angiogenesis and cancer progression. The PI3K/Akt/mTOR pathway is an important pathway as it is involved in cell survival, apoptosis, and autophagy. Melatonin-induced inhibition of Akt and mTOR phosphorylation was observed to cause cell proliferation arrest in human neck squamous cell carcinoma (HNSCC) cells, as well as cell cycle arrest in glioma cells [33]. The inhibition of both Akt and mTOR is crucial as both proteins are antagonistic against the induction of autophagy [34]. In our previous work, the expression of mTOR was downregulated in melatonin-treated JURKAT cells [14]. Therefore, it could be speculated that melatonin induced autophagy through this method.
In contrast, the loss of Optineurin and NDP52 does not affect LC3 lipidation [35,36], and the presence of Optineurin, while required for mitophagy, is independent of p62-mediated pathways [35,37]. The role of Optineurin in cancer cells has been context-dependent, and there are limited studies based on its role in cancer [38]. While the knockdown of Optineurin was discovered to upregulate apoptosis in pancreatic ductal adenocarcinoma cells [39], Optineurin was involved in a crucial step in HACE-1-activated autophagy in facilitating tumor suppression in liver cancer [27]. Furthermore, the roles of Optineurin and NDP52 seem to be closely linked to the PINK1/Parkin pathway, not just because these receptors were best studied in this pathway, but also because only Optineurin and NDP52 were considered essential to this pathway in cancer cells [40].
Additionally, it was found that Optineurin and NDP52 were involved in several unconventional pathways. [41,42] For instance, the recruitment of NDP53 could be due to the activation of Rab35, which was implied to be involved in Parkin-mediated mitophagy [43]. TBK1, another kinase, has been implicated in several selective autophagic pathways, including the PINK1/Parkin pathway. An investigation on a similar matter also discovered that TBK1 was used in Optineurin and NDP52-mediated mitophagy [44]. These findings further prove that Optineurin and NDP-52 are intertwined mechanistically with PINK1 and Parkin mitophagy pathways. In our study, we discovered that although the expression levels of NDP52 were upregulated compared to the control, they decreased at the highest doses of melatonin. Nonetheless, there is a notable spike in the expression levels of NDP52 at low doses of melatonin, suggesting that there was activity from NDP52 but was inhibited by melatonin at higher concentrations.
Melatonin modulates mitophagy pathways
Mitophagy is often intertwined with mitochondrial dynamics, as exemplified by the need to isolate dysfunctional mitochondria to ensure efficient removal without affecting the healthy system. These intricacies involve two mechanisms; one, the receptor-mediated mitophagy, where activated membrane-bound receptors bind directly to LC3; and two, ubiquitin-mediated mitophagy, which in this case would be the PINK1/Parkin pathway [44]. A variety of factors can trigger mitophagy, including mitochondrial depolarization, hypoxic conditions, iron chelation, and others [22]. Mitophagy, similar to autophagy, plays a dual role in cancer cell biology. It can either promote cell survival or death, depending on the molecular context [45]. Furthermore, it should be mentioned that these pathways are not mutually exclusive, as there has been crosstalk between several pathways. For instance, certain components of the PINK1/Parkin pathway have been found to interact with proteins involved in receptor-mediated mitophagy pathways [46].
In the recent decade, melatonin has been discovered to have effects on mitophagy pathways in different contexts. For instance, melatonin was found to prevent PINK1/Parkin activation to block mitophagy-mediated cell death in cardiac microcirculation endothelial cells [47]. Regarding cancer cells, melatonin was mostly explored as an adjuvant and yielded promising results. Melatonin together with rapamycin induced Nix-mediated mitophagy in HNSCC cells [48,49] while inhibiting hypoxia-mediated mitophagy in HCC cells [50]. To confirm our preliminary observation that melatonin-induced cell death in JURKAT cells involves the modulation of mitophagy, the investigation into key mitophagy proteins in their respective pathways is warranted.
Melatonin activates receptor-mediated mitophagy
Mitophagy occurring independent of the PINK1/Parkin pathway is known as receptor-mediated mitophagy. These pathways have receptors that contain an LIR motif, enabling them to recruit LC3 and mitophagophore [51]. Under this family, BNIP3 and Nix (BNIP3L) were reported to be involved in eliminating mitochondria under both physiological and pathological conditions. BNIP3 and Nix have been demonstrated as tumor suppressors in different types of cancers, as BNIP3 deletion was associated with mammary tumor growth, while upregulation of Nix contributes to tumor cell apoptosis [17]. In this study, the protein levels of those involved in the BNIP3/Nix pathway were investigated after subjecting JURKAT cells to varying concentrations of melatonin. Based on Figure 6, results showed that BNIP3 and Nix /BNIP3L protein expression increased as compared to the control. Upon statistical analysis, no significances were found in the trend across differing concentration groups.
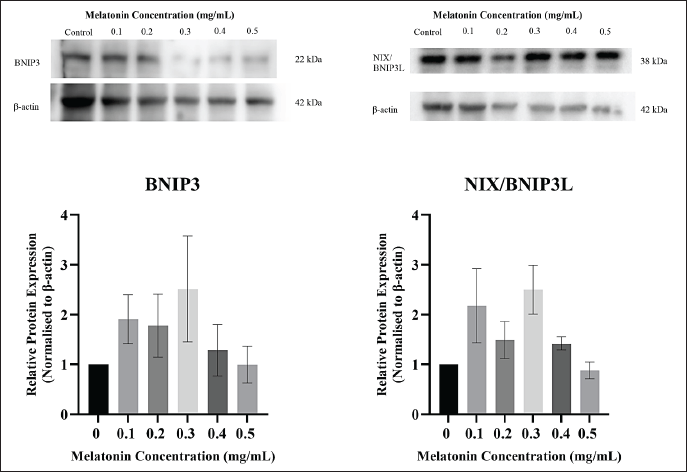 | Figure 6. Western blot analysis of BNIP3 and Nix/BNIP3L in JURKAT cells after treatment with different concentrations of melatonin after 72 hours. (A) BNIP3 (band), (B) BNIP3 (bar chart), (C) BNIP3L/Nix (band), and (D) BNIP3L/Nix (bar chart). Expression levels of the BNIP3 and Nix/BNIP3L were measured by Western blot analysis. Beta-actin (β-actin) served as a loading control. The bar graph below shows the means ± standard error of mean (SEM) from three separate experiments. One-Way ANOVA was performed to compare between concentration groups, followed by Tukey’s Multiple Comparison test as post-hoc testing. [Click here to view] |
Initial reports of BNIP3 and NIX/BNIP3L associated the function of both proteins with promotion of apoptosis and necrosis, but recent evidence has established their roles in mitophagy [16]. However, mechanistic information for both BNIP3 and Nix/BNIP3L-mediated mitophagy is not well-elucidated [52], therefore imposing the need to further investigate these pathways. The current consensus on mitophagy states that hypoxia is considered the most common factor that induces BNIP3/NIX-mediated mitophagy, as the stabilization of HIF-1α stimulates the production of both proteins [52]. The expression of BNIP3 would cause the formation of a complex comprising of Beclin-1 and PI3K-III, which can be regulated through the PI3K/Akt pathway [53]. Meanwhile, there have been multiple perspectives on how NIX modulates mitophagy, including the disruption of the BCL-BECN1 complex or the inhibition of MTORC1 to induce autophagy. However, the consensus on NIX-mediated mitophagy is that it occurs through interactions with Atg8 family proteins to recruit autophagosomes [54].
It has been discussed that melatonin can either directly or indirectly inhibit hypoxia-inducible factor 1 (HIF-1) by destabilizing HIF-1α. The destabilization effect would then potentially lead to reactive oxygen species (ROS) production and cell death [55]. Although melatonin is known for its antioxidant properties, it has been observed that pro-oxidant actions could be exerted on cancer cells to a certain extent [55], as exemplified by its ability to induce ROS production in HNSCC cells [48], HT-29 cells [19], and JURKAT cells [14,22].
As previously mentioned, both BNIP3 and NIX-mediated mitophagy are regulated through HIF-1α. Our findings indicated that both BNIP3 and NIX were generally upregulated after treatment with melatonin. A study found that mitochondrial depolarization has been proposed as a method for BNIP3 to induce mitophagy under hypoxic conditions. Furthermore, accumulation of BNIP3 may also occur due to the inhibition of AMPK-mediated phosphorylation on ULK1 or ULK2 [56]. Similarly, the expression of NIX has been correlated to numerous triggers related to other cellular stress not limited to hypoxia [57], such as the presence of cytotoxic drugs [58]. This is further supported by reports that melatonin alone or paired with rapamycin has induced an accumulation of NIX protein in HNSCC cells, resulting in NIX-mediated mitophagy [46,47]. In both studies, the activation of mitophagy was attributed to the increase in oxidative stress caused by melatonin.
CONCLUSION
Based on the obtained data, it can be concluded that melatonin does induce cytotoxicity in a dose-dependent manner upon exposing JURKAT cells to concentrations of 0.1–0.5 mg/ml. Furthermore, melatonin also induced autophagy in JURKAT cells, as proven by the AO fluorescence microscopy. Finally, melatonin was proven to be able to modulate mitophagy-mediated pathways to induce autophagy. It should be remembered that this study serves as a surface analysis on how melatonin modulates mitophagy pathways in JURKAT cells, therefore it has its fair share of limitations regarding the depth of investigation. Nevertheless, it is hoped that this study could provide some insights into how melatonin causes autophagy in JURKAT cells via mitophagy.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
The research funding was supported by International Medical University; the Grant Number is BMSc I-2022 (03).
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021;149(04):778–89. CrossRef
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of incidence and mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. CrossRef
3. Davis AS, Viera AJ, Mead MD. Leukemia: an overview for primary care. Am Fam Physician. 2014 May 1;89(9):731–8.
4. Ness KK, Armenian SH, Kadan-Lottick N, Gurney JG. Adverse effects of treatment in childhood acute lymphoblastic leukemia: general overview and implications for long-term cardiac health. Expert Rev Hematol. 2011;4(2):185–97. CrossRef
5. Crossnohere NL, Richardson DR, Reinhart C, O’Donoghue B, Love SM, Smith BD, et al. Side effects from acute myeloid leukemia treatment: results from a national survey. Curr Med Res Opin. 2019;35(11):1965–70. CrossRef
6. Mughal TI, Schrieber A. Principal long-term adverse effects of imatinib in patients with chronic myeloid leukemia in chronic phase. Biologics. 2010;4:315–23. CrossRef
7. Ross K, Gillespie-Twardy AL, Agha M, Raptis A, Hou JZ, Farah R, et al. Intensive chemotherapy in patients aged 70 years or older newly diagnosed with acute myeloid leukemia. Oncol Res. 2015;22(2):085–92. CrossRef
8. Aldoss I, Forman SJ, Pullarkat V. Acute lymphoblastic leukemia in the older adult. J Oncol Pract. 2019;15(2):067–75. CrossRef
9. Talib WH, Alsayed AR, Abuawad A, Daoud S, Mahmod AI. Melatonin in cancer treatment: current knowledge and future opportunities. Molecules. 2021;26(9):2506. CrossRef
10. Ng MG, Ng KY, Koh RY, Chye SM. Potential role of melatonin in prevention and treatment of leukaemia. Horm Mol Biol Clin Investig. 2021;42(4):445–61. CrossRef
11. Tang YL, Sun X, Huang LB, Liu XJ, Qin G, Wang LN, et al. Melatonin inhibits MLL-rearranged leukemia via RBFOX3/hTERT and NF-κB/COX-2 signaling pathways. Cancer Lett. 2019;443:167–78. CrossRef
12. Sánchez Hidalgo M, Lee M, de la Lastra CA, Guerrero JM, Packham G. Melatonin inhibits cell proliferation and induces caspase activation and apoptosis in human malignant lymphoid cell lines. J Pineal Res. 2012;53(4):366–73. CrossRef
13. Perdomo J, Cabrera J, Estévez F, Loro J, Reiter RJ, Quintana J. Melatonin induces apoptosis through a caspase-dependent but reactive oxygen species-independent mechanism in human leukemia Molt-3 cells. J Pineal Res. 2013;55(2):195–206. CrossRef
14. Kasi R, Yeo PL, Yen NK, Koh RY, Ponnudurai G, Tiong YL, et al. Melatonin induces apoptosis and inhibits the proliferation of cancer cells via reactive oxygen species-mediated MAPK and mTOR pathways. Clin Cancer Drugs. 2019;7(1):044–56. CrossRef
15. Sainz RM, Mayo JC, Rodriguez C, Tan DX, Lopez-Burillo S, Reiter RJ. Melatonin and cell death: differential actions on apoptosis in normal and cancer cells. Cell Mol Life Sci. 2003;60(7):1407–26. CrossRef
16. Iorio R, Celenza G, Petricca S. Mitophagy: molecular mechanisms, new concepts on parkin activation and the emerging role of AMPK/ULK1axis. Cells. 2021;11(1):030. CrossRef
17. Denisenko TV, Gogvadze V, Zhivotovsky B. Mitophagy in carcinogenesis and cancer treatment. Discov Oncol. 2021;12(1):058. CrossRef
18. PMI Olivas Aguirre M, Pérez Chávez J, Torres López L, Hernández Cruz A, Pottosin I, Dobrovinskaya O. Dexamethasone-induced fatty acid oxidation and autophagy/mitophagy are essential for T-ALL glucocorticoid resistance. Cancers (Basel). 2023;15(2):445. CrossRef
19. Chok KC, Koh RY, Ng MG, Ng PY, Chye SM. Melatonin induces autophagy via reactive oxygen species-mediated endoplasmic reticulum stress pathway in colorectal cancer cells. Molecules. 2021;26(16):5038. CrossRef
20. Arredondo Beltrán IG, Ramírez Sánchez DA, Zazueta García JR, Canizalez Roman A, Angulo Zamudio UA, Velazquez Roman JA, et al. Antitumor activity of bovine lactoferrin and its derived peptides against HepG2 liver cancer cells and Jurkat leukemia cells. Biometals. 2023;36(3):639–55. CrossRef
21. Rostami Yasuj S, Obeidi N, Khamisipou G, Gharehdaghi Z, Zangeneh Z. Overexpression of MiR-506 in Jurkat (Acute T Cell Leukemia) Cell Line. Iran J Pathol. 2020;15(4):282–91. CrossRef
22. Büyükavci M, Ozdemir O, Buck S, Stout M, Ravindranath Y, Sava?an S. Melatonin cytotoxicity in human leukemia cells: relation with its pro-oxidant effect. Fundam Clin Pharmacol. 2006;20(1):073–9. CrossRef
23. Yun CW, Lee SH. The roles of autophagy in cancer. Int J Mol Sci. 2018;19(11):3466. CrossRef
24. Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4(10):e838. CrossRef
25. Das S, Shukla N, Singh SS, Kushwaha S, Shrivastava R. Mechanism of interaction between autophagy and apoptosis in cancer. Apoptosis. 2021;26(9-10):512–33. CrossRef
26. Murugan S, Amaravadi RK. Methods for studying autophagy within the tumor microenvironment. Adv Exp Med Biol. 2016;899:145–66. CrossRef
27. Yu Z, Li Y, Han T, Liu Z. Demethylation of the HACE1 gene promoter inhibits the proliferation of human liver cancer cells. Oncol Lett. 2019;17(5):4361–8. CrossRef
28. Leyva Paredes K, Castrejón Jiménez NS, Arrieta Oliva HI, Baltierra Uribe SL, García Pérez BE. Choosing lunch: the role of selective autophagy adaptor proteins. In: Gorbunov N, Ed. Autophagy in current trends in cellular physiology and pathology. Intech, Rijeka: Croatia; 2016. Pp. 1–20. 29.Puissant A, Fenouille N, Auberger P. When autophagy meets cancer through p62/SQSTM1. Am J Cancer Res. 2012;2(4):397–413.
30. Kumar AV, Mills J, Lapierre LR. Selective autophagy receptor p62/SQSTM1, a pivotal player in stress and aging. Front Cell Dev Biol. 2022;10:793328. CrossRef
31. Klionsky DJ, Abdel Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy. 2021;17(1):1–382. CrossRef
32. Yuan JQ, Yuan Y, Pan SS, Cai K. Altered expression levels of autophagy-associated proteins during exercise preconditioning indicate the involvement of autophagy in cardioprotection against exercise-induced myocardial injury. J Physiol Sci. 2020;70:10. CrossRef
33. Kma L, Baruah TJ. The interplay of ROS and the PI3K/Akt pathway in autophagy regulation. Biotechnol Appl Biochem. 2022;69(1):248–64. CrossRef
34. Ainaz Mihanfar, Yousefi B, Bita Azizzadeh, Majidinia M. Interactions of melatonin with various signaling pathways: implications for cancer therapy. Cancer Cell Int. 2022;22(1):420. CrossRef
35. Inokuchi S, Yoshizumi T, Toshima T, Itoh S, Yugawa K, Harada N, et al. Suppression of optineurin impairs the progression of hepatocellular carcinoma through regulating mitophagy. Cancer Med. 2021;10(5):1501–14. CrossRef
36. Petkova DS, Verlhac P, Rozières A, Baguet J, Claviere M, Kretz-Remy C, et al. Distinct contributions of autophagy receptors in measles virus replication. Viruses. 2017;9(5):123. CrossRef
37. Hou X, Watzlawik JO, Fiesel FC, Springer W. Autophagy in Parkinson’s Disease. J Mol Biol. 2020;432(8):2651–72. CrossRef
38. Guo Q, Wang J, Weng Q. The diverse role of optineurin in pathogenesis of disease. Biochem Pharmacol. 2020;180(3):114157. CrossRef
39. Ali DM, Ansari SS, Zepp M, Knapp-Mohammady M, Berger MR. Optineurin downregulation induces endoplasmic reticulum stress, chaperone-mediated autophagy, and apoptosis in pancreatic cancer cells. Cell Death Discov. 2019;5:128. CrossRef
40. Weil R, Laplantine E, Curic S, Génin P. Role of optineurin in the mitochondrial dysfunction: potential implications in neurodegenerative diseases and cancer. Front Immunol. 2018;9:1243. CrossRef
41. Viret C, Rozières A, Faure M. Novel insights into NDP52 autophagy receptor functioning. Trends Cell Biol. 2018;28(4):255–7. CrossRef
42. Minowa Nozawa A, Nozawa T, Okamoto Furuta K, Kohda H, Nakagawa I. Rab35 GTPase recruits NDP52 to autophagy targets. EMBO J. 2017;36(18):2790–807. CrossRef
43. Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTINEURIN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell. 2015;60(1):07–20. CrossRef
44. Nguyen TN, Sawa-Makarska J, Khuu G, Lam WK, Adriaenssens E, Fracchiolla D, et al. Unconventional initiation of PINK1/Parkin mitophagy by Optineurin. Mol Cell. 2023;83(10):1693–709.e9. CrossRef
45. Panigrahi DP, Praharaj PP, Bhol CS, Mahapatra KK, Patra S, Behera BP, et al. The emerging, multifaceted role of mitophagy in cancer and cancer therapeutics. Semin Cancer Biol. 2020;66:045–58. CrossRef
46. Zimmermann M, Reichert A. How to get rid of mitochondria: crosstalk and regulation of multiple mitophagy pathways. Biol Chem. 2018;399(1):029–45. CrossRef
47. Zhou H, Zhang Y, Hu S, Shi C, Zhu P, Ma Q, et al. Melatonin protects cardiac microvasculature against ischemia/reperfusion injury via suppression of mitochondrial fission-VDAC1-HK2-mPTP-mitophagy axis. J Pineal Res. 2017;63(1):e12413. CrossRef
48. Guerra Librero A, Fernandez Gil BI, Florido J, Martinez Ruiz L, Rodríguez Santana C, Shen YQ, et al. Melatonin targets metabolism in head and neck cancer cells by regulating mitochondrial structure and function. Antioxidants (Basel). 2021;10(4):603. CrossRef
49. Shen YQ, Guerra Librero A, Fernandez Gil BI, Florido J, García López S, Martinez Ruiz L, et al. Combination of melatonin and rapamycin for head and neck cancer therapy: suppression of AKT/mTOR pathway activation, and activation of mitophagy and apoptosis via mitochondrial function regulation. J Pineal Res. 2018;64(3):e12461.CrossRef
50. Prieto Domínguez N, Méndez Blanco C, Carbajo Pescador S, Fondevila F, García Palomo A, González Gallego J, Mauriz JL. Melatonin enhances sorafenib actions in human hepatocarcinoma cells by inhibiting mTORC1/p70S6K/HIF-1α and hypoxia-mediated mitophagy. Oncotarget. 2017;8(53):91402–14. CrossRef
51. Miyazaki N, Shiratori R, Oshima T, Zhang Z, Valencia R, Kranrod J, et al. PINK1-dependent and Parkin-independent mitophagy is involved in reprogramming of glycometabolism in pancreatic cancer cells. Biochem Biophys Res Commun. 2022;625:167–73. CrossRef
52. Li Y, Zheng W, Lu Y, Zheng Y, Pan L, Wu X, et al. BNIP3L/NIX-mediated mitophagy: molecular mechanisms and implications for human disease. Cell Death Dis. 2021;13(1):14. CrossRef
53. Zhou X, Zhao X, Zhou W, Qi H, Zhang H, Han TL, et al. Impaired placental mitophagy and oxidative stress are associated with dysregulated BNIP3 in preeclampsia. Sci Rep. 2021;11(1):20469. CrossRef
54. Rogov VV, Suzuki H, Marinkovi? M, Lang V, Kato R, Kawasaki M, et al. Phosphorylation of the mitochondrial autophagy receptor nix enhances its interaction with LC3 proteins. Sci Rep. 2017;7(1):1131. CrossRef
55. Lee YJ, Lee JH, Moon JH, Park SY. Overcoming hypoxic-resistance of tumor cells to TRAIL-induced apoptosis through melatonin. Int J Mol Sci. 2014;15(7):11941–56. CrossRef
56. Park CW, Hong SM, Kim ES, Kwon JH, Kim KT, Nam HG, et al. BNIP3 is degraded by ULK1-dependent autophagy via MTORC1 and AMPK. Autophagy. 2013 Mar;9(3):345–60. CrossRef
57. Chen L, Liu L, Li Y, Gao J. Melatonin increases human cervical cancer HeLa cells apoptosis induced by cisplatin via inhibition of JNK/Parkin/mitophagy axis. In Vitro Cell Dev Biol Anim. 2018;54(1):01–10. CrossRef
58. Zhang S, Wang Y, Cao Y, Wu J, Zhang Z, Ren H, et al. Inhibition of the PINK1-Parkin pathway enhances the lethality of sorafenib and regorafenib in hepatocellular carcinoma. Front Pharmacol. 2022;13:851832. CrossRef