INTRODUCTION
The zoonotic coronaviruses such as MERS-CoV, SARS-CoV, and SARS-CoV-2 have evolved into pathogens of global concerns causing episodic pandemics that resulted in alarming social, economic, and health issues only during the last two decades. Despite their overarching detrimental effects on human welfare, not a single pharmaceutical drug has been developed for their management. Repurposed medications, allegedly convenient and usable for the treatment of the symptoms, have not provided a solution or any practical clinical value (Martinez, 2022; Manóchio et al., 2023). These human viruses originated from animal coronaviruses with alarming successes in terms of virulence, infectivity, and mutational adaptations. As they rapidly diversify with new pathogenic adaptations in different hosts since they are first recorded (Lalchhandama, 2021), it is important to look for sources of therapeutic management. Attention has been drawn to natural products to identify antiviral compounds, which possibly offer insights into potential medications (Giordano et al., 2023; Saifulazmi et al., 2022).
Reflecting on the pandemic events due to viruses, it becomes apparent that global catastrophic infections as such are bound to appear from time to time. However, the imminence of such outbreaks is unpredictable. Technology helped tremendously in the fight against COVID-19 with rapid responses from pharmaceutical industries to produce vaccines. However, this measure does not offer the ultimate prevention or treatment. Several plants have been reported concerning their activities against SARS-CoV-2 with promising results, but without practical outcomes (Ebenezer et al., 2022). Among them, Gliricidia sepium, Piper tuberculatum, and Artemisia annua reportedly reduced the production of SARS-CoV-2 viral particles (Flórez-Álvarez et al., 2022; Nie et al., 2021). However, specific compounds are yet to be identified from these plants.
Lupeol (also fagarsterol) is a dietary triterpenoid found in edible fruits like fig, mango, strawberry; vegetables like cabbage, pepper, tomato; and in plants like aloe, Asiatic ginseng, black tea, ivy gourd, mulberry, pea, and soybean. The compound and its derivatives gained considerable attention as they show a wide range of pharmacological properties including anticancer, anti-inflammatory, and antiprotozoal activities (Osafo et al., 2023; Narendar et al., 2023; Sohag et al., 2022). They are further identified as efficient modulators of immune signaling molecules such as interleukin-2, interleukin-4, interleukin-5, interleukin-β, α-glucosidase, proteases, Bcl-2, cFLIP, and NFκB (Siddique et al., 2011). The medical application is predicted to be much more diverse than the experimental cases (Sohag et al., 2022). It reportedly inhibits the replication of herpes simplex virus 1 (Ye et al., 2021), dengue virus (Monsalve-Escudero et al., 2021), and Zika virus (Santos Pereira et al., 2021). However, its mode of antiviral interaction is not known. The established chemical and biological properties suggest that it exerts antipathogenic activity by specifically interfering with the key viral proteins. Based on this premise, the binding affinity of lupeol on different proteins of SARS-CoV-2 was analyzed, complemented with simulations of the biological properties using absorption, distribution, excretion, metabolism, and toxicity (ADMET) analyses, molecular interactions, and target predictions.
MATERIALS AND METHODS
Ligand retrieval and processing
The three-dimensional structure of lupeol (C30H50O, PubChem compound CID: 259846) was retrieved from the chemical database, PubChem (National Centre for Biotechnology Information, United States National Institutes of Health), in an SDF format as shown in Figure 1. Structure optimization and cumulative potential energy minimization were done in Chem Bio 3D Ultra using the force field MMFF94. The ligand was then saved in Protein Data Bank (PDB) format.
Protein retrieval and processing
SARS-CoV-2 proteins of well-established structures based on X-ray crystal structures were used for molecular binding predictions. The target proteins including main protease (Mpro) with PDB code 6Y2E (Zhang et al., 2020), spike glycoprotein with PDB code 6VXX (Walls et al., 2020), RNA-dependent RNA polymerase (RdRp) with PDB code 6M71 (Gao et al., 2020), papain-like protease (PLpro) with PDB code 6W9C (Gao et al., 2021), and nucleocapsid phosphoprotein with PDB code of 6VYO (El-Demerdash et al., 2021) were retrieved from the Research Collaboratory for Structural Bioinformatics PDB website (www.rcsb.org). Co-factors, water molecules, and unique ligands attached to each protein were removed with Molegro Molecular Viewer 2.5 software (Agrwal et al., 2022) to produce a clear and uninterrupted molecular binding.
Molecular docking
Molecular docking of lupeol to each of the recovered proteins was carried out on the AutoDock Vina platform (Trott and Olson, 2010). AutoDock Vina is considered the most powerful docking tool in predicting and reconstructing drug–molecule interactions (Tang et al., 2022). Polar hydrogens and Kollman charges were added to all the proteins in AutoDockTool-1.5.6 before saving the data in PDB partial charge (Q) and atom type (T) (PDBQT) format. Additionally, lupeol was saved in PDBQT format. Flexible docking was performed on all the proteins. For Mpro, the grid box was created with a coordinate of X dimension = 40 Å, Y dimension = 68 Å, Z dimension = 66 Å, X coordinate of the center = −16.524 Å, Y coordinate of the center = −26.137 Å, Z coordinate of the center = 17.525 Å. For nucleoprotein, grid box with a coordinate of X dimension = 44 Å, Y dimension = 40 Å, Z dimension = 54 Å and X coordinate of the center = −13.723 Å, Y coordinate of the center = −2.763 Å, Z coordinate of the center = −1.831 Å was created. For PLpro, RdRp, and spike proteins, the coordinates for grid boxes were created such as X dimension = 78 Å, Y dimension = 52 Å, Z dimension = 72 Å, X coordinate of the center = −32.692 Å, Y coordinate of the center = 34.009 Å, Z coordinate of the center = 25.535 Å, X dimension = 84 Å, Y dimension = 92 Å, Z dimension = 98 Å, X coordinate of the center = 119.712 Å, Y coordinate of the center = 123.621 Å, Z coordinate of the center = 120.3 Å, and X dimension = 80 Å, Y dimension = 100 Å, Z dimension = 126 Å, X coordinate of the center = 197.815 Å, Y coordinate of the center = 223.402 Å, Z coordinate of the center = 207.349 Å, respectively, to cover all the possible protein binding sites. With an exhaustiveness of 8, lupeol was docked to all the proteins and the outputs were saved for later visual analyses.
Visualization and analyses of interaction
BIOVIA Discovery Studio Visualizer 2016 v16.1.0.15350 was used for visualization and analytics. The software is a comprehensive tool suitable for micro- to macro-molecules for all types of molecular interactions related to biological and antipathogenic activities (Jejurikar and Rohane, 2021). The ligand output and protein PDBQT formats were opened and defined. Non-bond interactions and ligand interactions were selected and labels were added to each of the residues. For each protein, only the interaction with the lowest binding energy and root-mean-square deviation value was saved and converted to image files.
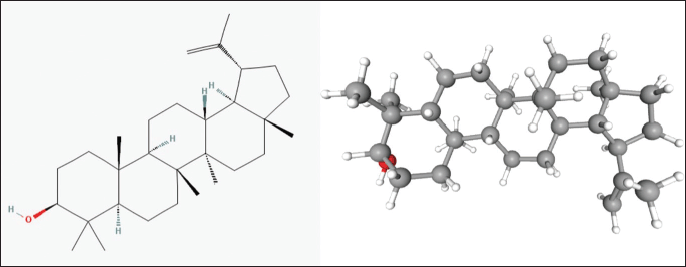 | Figure 1. Molecular structure of lupeol. [Click here to view] |
Prediction of activity spectra for substances (PASS)
PASS is an online tool designed to evaluate and predict the biological potential of molecules based on their structural resemblance to other known molecules (Rudik et al., 2019). For this prediction, analysis was done in Way2Drug (http://www.way2drug.com/passonline/predict.php). The SMILES data of lupeol retrieved from PubChem was uploaded for evaluation.
Target prediction
Analysis of molecular targets is critical to determining the probable side effects and bioactivities of drug candidates (Gfeller et al., 2014). For this, Swiss Target Prediction (http://www.swisstargetprediction.ch/) website was used where the SMILE data of lupeol retrieved from PubChem was uploaded for analysis.
ADMET prediction
Analyzing a compound for its cellular activities based on ADMET is an important step for screening compounds with pharmaceutical potential. For lupeol, an online database, pkCSM (https://biosig.lab.uq.edu.au/pkcsm/), was used where pharmacodynamics of the compound was predicted using SMILES data from PubChem. ADMET analysis was performed according to standard protocol (Pires et al., 2015). The SMILES data retrieved from PubChem were submitted and analyzed in terms of absorption (including Caco-2 cell permeability, intestinal absorption in human, skin permeability, P-glycoprotein substrate, P-glycoprotein I inhibitor, P-glycoprotein II inhibitor, water solubility), distribution (steady-state volume of distribution of human, fraction unbound of human, blood-brain barrier permeability, and central nervous system permeability), metabolism (cytochrome P450 substrates and inhibitors such as CYP1A2 inhibitor, CYP2C9 inhibitor, CYP2C19 inhibitor, CYP2D6 inhibitor, CYP3A3 inhibitor, CYP2D6 substrate, CYP3A4 substrate,), excretion (total clearance and renal organic cation transporter 2 substrate), and toxicity (Ames toxicity, maximum tolerated dose in human, human ether-à-go-go-related gene I inhibitor, hERG II inhibitor, oral rat acute toxicity (LD50), oral rat chronic toxicity with lowest observed adverse effect level (LOAEL), hepatotoxicity, skin sensitization, Tetrahymena pyriformis toxicity).
RESULTS AND DISCUSSION
Molecular docking
The molecular docking models showed that lupeol can interact with the vital proteins of SARS-CoV-2 with high binding affinities. The highest binding affinities were seen on lupeol-Mpro, lupeol-spike protein, and lupeol-PLpro, all of which showed binding affinities of −8.2 kcal/mol. Lupeol-RdRp and lupeol-nucleoprotein showed slightly less affinities of −7.5 and −7.0 kcal/mol, respectively (Table 1). In all the interactions analyzed, all complexes of lupeol with the different proteins showed negative binding energy. Lupeol interacts with Mpro through five amino acid residues namely valine (A:202), histidine (A:246), isoleucine (A:249), proline (A:293 and phenylalanine (A:294) (Figs. 2 and 3). It interacts with the spike protein at tryptophan (A:104), isoleucine (A:119), valine (A:126), phenylalanine (A:192), isoleucine (A:203), histidine (A:207), leucine (A:226), and valine (A:227) (Figs. 4 and 5). With RdRp, it binds at leucine (A:371), alanine (A:375), tryptophan (A:509), leucine (A:514), and tyrosine (A:515) (Figs. 6 and 7). With PLpro, it interacts with isoleucine (A:14), asparagine (A:15), tyrosine (A:71), and proline (A:130) (Figs. 8 and 9). It interacts with nucleoprotein at glutamine (A:70), valine (A:72), isoleucine (A:74), and proline (A:162) (Figs. 10 and 11).
Molecular modelings have indicated some natural products as having potential antiviral activities. Andrographolite from Andrographis paniculate inhibited Mpro with a high binding affinity (Enmozhi et al., 2021). Peonidin 3-O-glucoside, kaempferol 3-O-rutinoside, and 4-(3,4-dihydroxyphenyl)-7-methoxy-5-[(6-O-β-D-xylopyranosyl-β-D-glucopyranosyl) oxy]-2H-1-benzopyran-2-one, quercetin-3-D-xyloside, and quercetin 3-O-L-arabinopyranoside also showed good binding affinity (Majumder and Mandal, 2022). Kadsurenin L and methysticin from Piper nigrum were shown to have Mpro binding energies of −8.43 and −8.20 kcal/mol, respectively (Davella et al., 2022). Calendulaglycoside A showed a high binding affinity with a binding energy of −9.90 kcal/mol (Zaki et al., 2022).
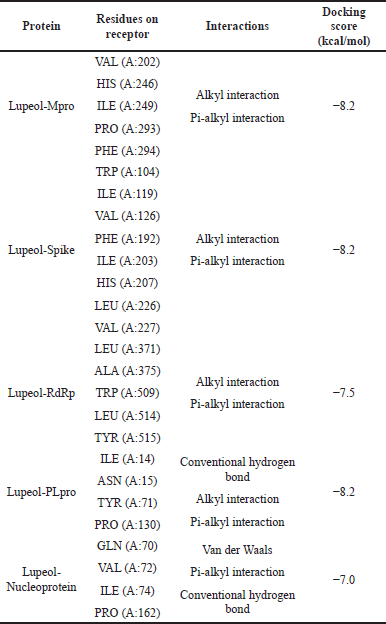 | Table 1. Predicted interactions of lupeol to different proteins of SARS-CoV-2. [Click here to view] |
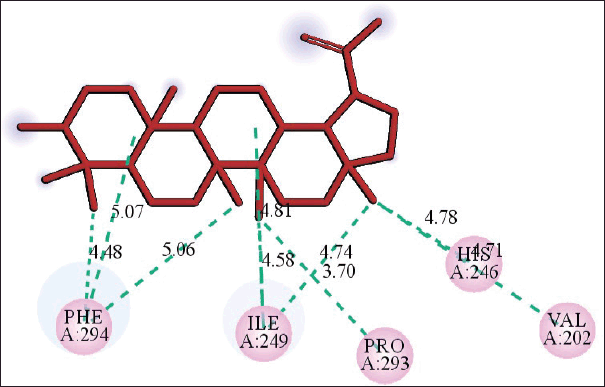 | Figure 2. 2D diagram of the interaction of lupeol and SARS-CoV-2 showing amino acid residues of Mpro involved and distance of hydrogen bonds (Å). [Click here to view] |
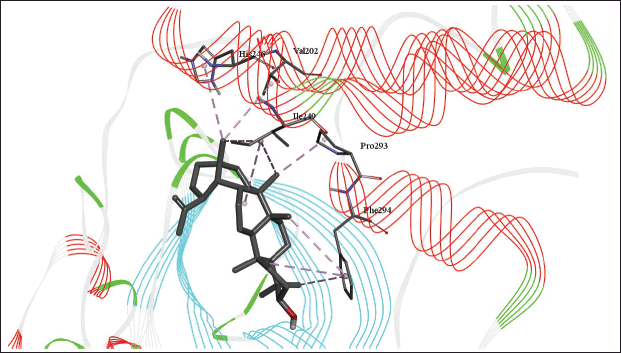 | Figure 3. 3D diagram of the site of lupeol binding with SARS-CoV-2 Mpro with interacting amino acid residues and hydrogen bonds. [Click here to view] |
Betulinic acid, azadirachtin A, ginsenoside Rg2, somniferine, soyasapogenol C, and saikosaponin A are also reported to have high binding affinities (Rudrapal et al., 2022). Anolignans, chebulic acid, mimusopic acid, mulberrosides, and punigluconin were shown to have similar binding affinity on Mpro as the standard drugs such as Remdesivir (RdRp inhibitor) and Darunavir (Mpro inhibitor) (Sharma et al., 2022). Prescription antibiotics, lefamulin, and bedaquiline showed dock scores of −9.153 and −8.989 kcal/mole, respectively on Mpro (Nandi et al., 2022). It has been reported that lupeol exhibited antiviral activity against Herpes simplex virus type 1 by inhibiting the viral genes (Ye et al., 2021), and the present study extends the potential antiviral activity against the coronavirus. Lupeol showed binding affinities with Mpro, RdRp, and Plpro of SARS-CoV-2 with affinities of −8.2, −7.5, and −8.2 kcal/ml, respectively, which indicate that the compound has a potential to be investigated as a lead molecule for the development of an antiviral drug against coronaviruses.
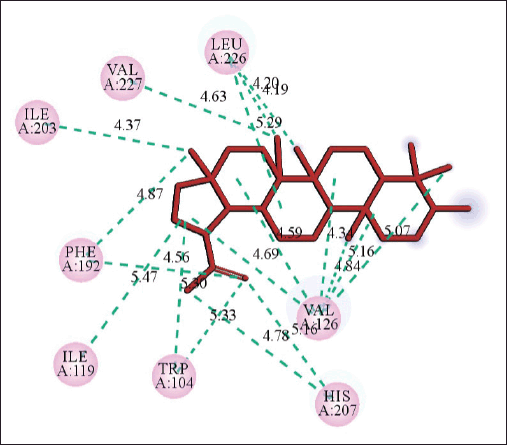 | Figure 4. 2D diagram of the interaction of lupeol and spike protein showing amino acid residues involved and distance of hydrogen bonds (Å). [Click here to view] |
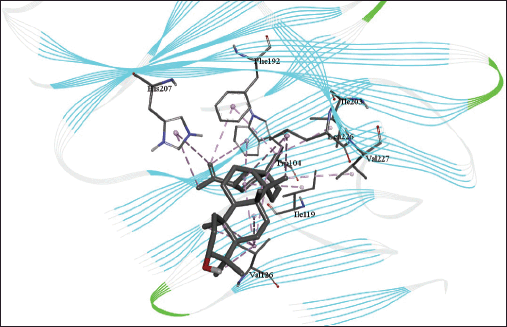 | Figure 5. 3D diagram of the site of lupeol binding with spike protein with interacting amino acid residues and hydrogen bonds. [Click here to view] |
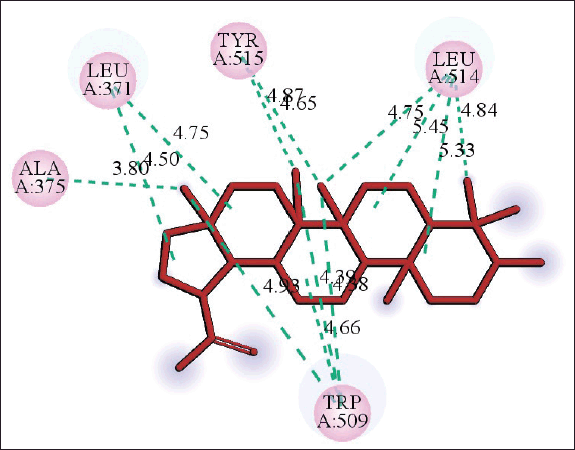 | Figure 6. 2D diagram of the interaction of lupeol and RdRp showing amino acid residues involved and distance of hydrogen bonds (Å). [Click here to view] |
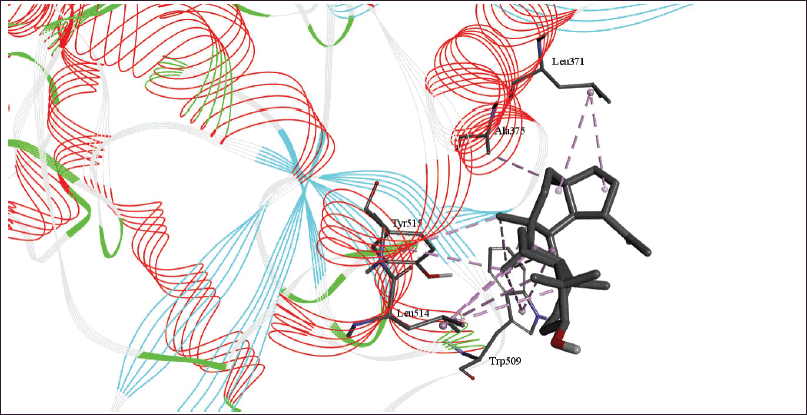 | Figure 7. 3D diagram of the site of lupeol binding with RdRp with interacting amino acid residues and hydrogen bonds. [Click here to view] |
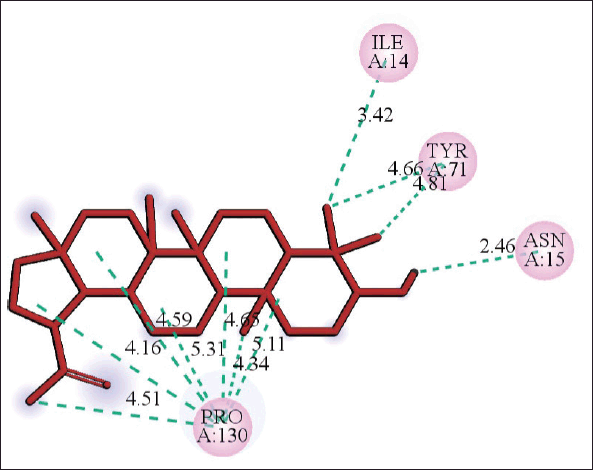 | Figure 8. 2D diagram of the interaction of lupeol and PLpro showing receptor amino acid residues involved and distance of hydrogen bonds (Å). [Click here to view] |
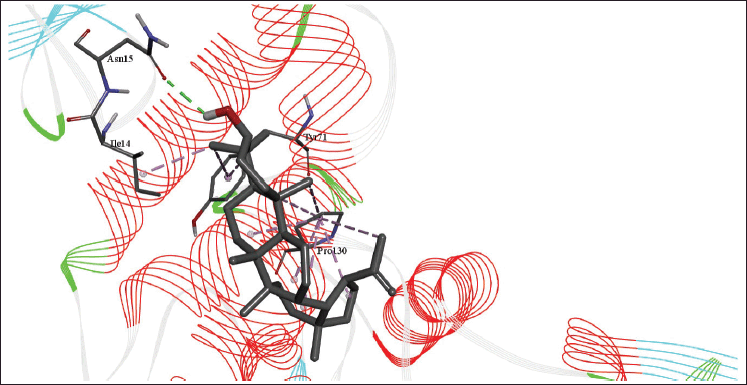 | Figure 9. 3D diagram of the site of lupeol binding with PLpro with interacting amino acid residues and hydrogen bonds. [Click here to view] |
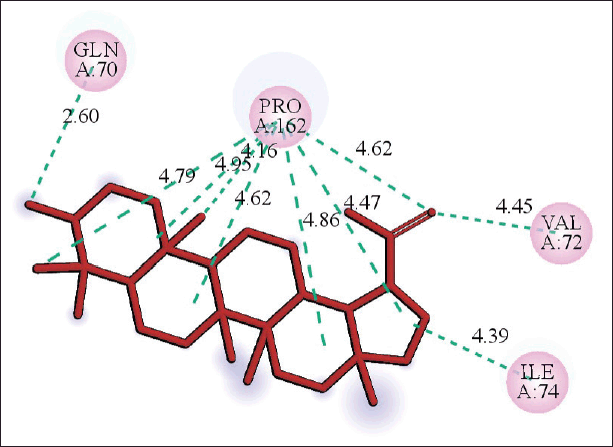 | Figure 10. 2D diagram of the interaction of lupeol and nucleoprotein showing receptor amino acid residues involved and distance of hydrogen bonds (Å). [Click here to view] |
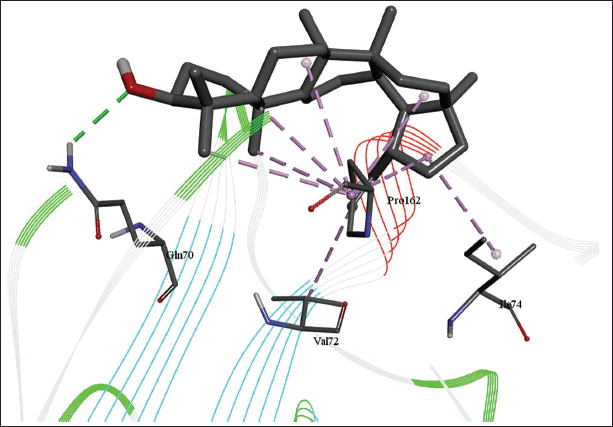 | Figure 11. 3D diagram of the site of lupeol binding with nucleoprotein with interacting amino acid residues and hydrogen bonds. [Click here to view] |
Less is known about the binding potential of probable antiviral compounds on the spike protein, a key component of coronaviruses that contributes to pathogenicity and evolution of viral strains (Lalchhandama, 2021). Oleanolic acid, tryptanthrin, chrysophanol, and rhein were predicted to block the S protein (Yu and Li, 2022). Our simulation indicates that lupeol binds with S protein with a relatively high binding affinity (−8.2 kcal/mol), which further suggests its possible inhibition of the interaction with the S protein with the ACE2 receptor, a critical event in coronavirus infection. Additionally, our data also indicate that lupeol can also interact with nucleoprotein, thereby intercepting viral replication.
PASS prediction
To fully understand the pharmacological potentials of any drug candidate, it is important to identify its probable target molecules for tracking its mechanism of action. Predictions of the potential biological properties showed that lupeol may have important therapeutic applications in various immune disorders, including cancer and parasitic diseases (Liu et al., 2021; Sohag et al., 2022). All the tested target molecules in the present study indicate probable activity (Pa) above the threshold, i.e., Pa > Pi (Table 2). The highest predicted activities, having Pa to Pi ratio more than 200:1, were on caspase 3 stimulant (244:1), transcription factor NF kappa B stimulant (947:1), transcription factor stimulant (947:1), antineoplastic (237:1), hepatoprotectant (453:1), antileishmanial (297:1), caspase 8 stimulant (865:1), melanoma (336:1), lung cancer (212:1), and colorectal cancer (209:1). Lupeol has been experimentally shown to have anticancer activities against different tumor cell lines (Bhatt et al., 2021; Maurya et al., 2020), and as a chemotherapy adjuvant (Khan et al., 2023), thereby corroborating with the predicted functional activities.
Target prediction
Lupeol was found to target a wide array of proteins from enzymes, membrane transporters, and ion channels to receptors (Table 3). The major target molecules, indicated in Figure 12, consisted of general enzymes (24%), nuclear receptor (12%), cytochrome P450 (12%), phosphatase (12%), hydrolase (8%), family A G protein-coupled receptor (8%), unclassified protein (8%), oxidoreductase (4%), electrochemical transporter (4%), lyase (4%), and voltage-gated ion channel (4%). These are the cellular molecules recognized for their roles in several pharmacological functions (Cao et al., 2012; Ju?ica et al., 2015).
Some of the predicted targets for lupeol include 11-beta-hydroxysteroid dehydrogenase 1, UDP-glycuronosyltransferase, cyclooxygenase-1, acetylcholinesterase, dual specificity phosphatase Cdc25A and Cdc25B, SUMO-activating enzymes, DNA polymerase beta, prostaglandin E synthase, aldo-keto reductase family 1 member B10, androgen receptor, LXR-alpha, nuclear receptor subfamily 1 group I member 3, and cytochrome P450 2C19, 19A1, 17A1.
ADMET prediction
Crucial information for any potential drug molecule is how it can affect physiology and cellular functions, and ADMET is a powerful tool for such preliminary screening (Pires et al., 2015). Lupeol is poorly soluble in water, having a logS = −5.861 log mol/l at 25°C, and only partially permeable as determined by Caco-2 permeability test (1.226 log Papp in 10−6 cm/s). 95.782% of lupeol is predicted to be absorbed via the intestine and has a skin permeability of −2.744 log Kp. While it does not act as a P-glycoprotein substrate, it acts as an inhibitor of both P-glycoprotein I and P-glycoprotein II. Steady-state volume of distribution and unbound fraction in blood plasma were predicted to be zero. It was predicted to be readily permeable across the blood-brain barrier (0.726 log BB) and central nervous system (−1.714 log PS). It showed no inhibition of CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4. It acts as a substrate of CYP3A4 but not of CYP2D6. It does not act as a substrate of organic cation transporter 2 and has a total clearance of 0.153 log ml/minute/kg.
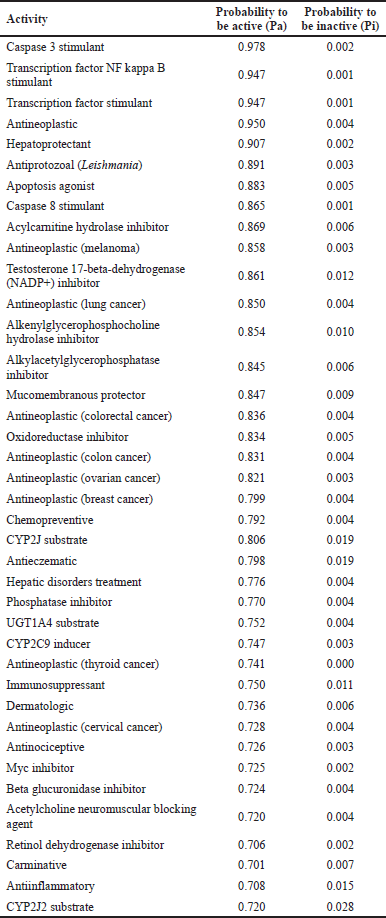 | Table 2. Prediction of probable biological activities of lupeol. [Click here to view] |
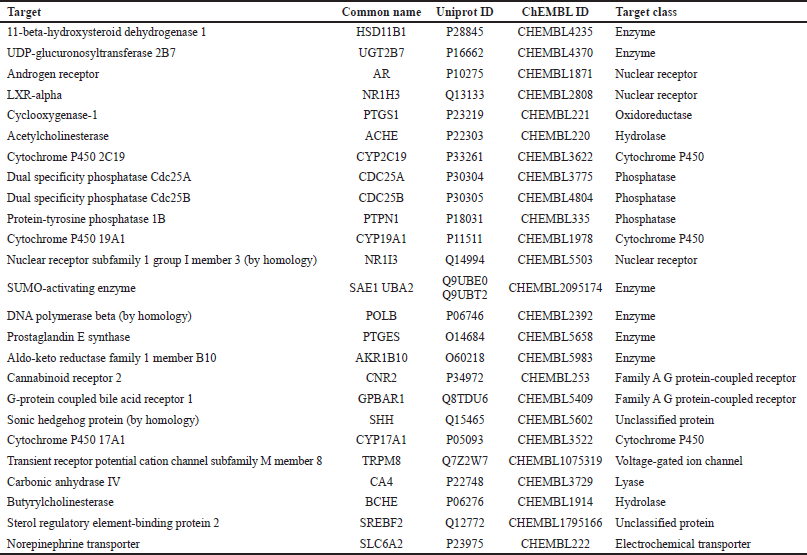 | Table 3. Probable targets of lupeol given by Swiss target prediction. [Click here to view] |
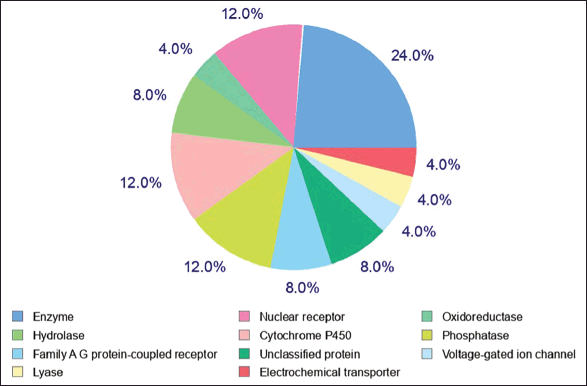 | Figure 12. Probable targets of lupeol as given by Swiss target prediction. [Click here to view] |
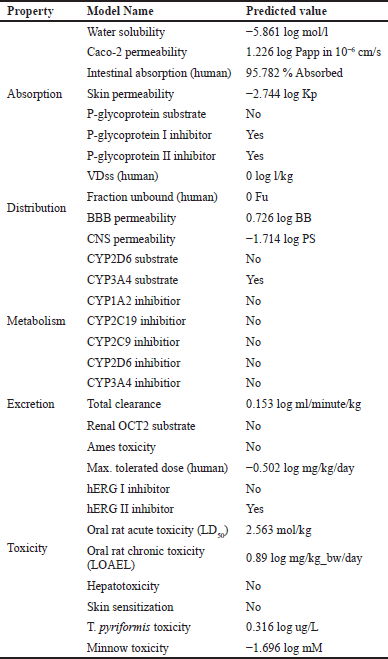 | Table 4. Prediction of ADMET of lupeol by pkCSM online platform. [Click here to view |
It showed no mutagenicity as given by the Ames test and is not toxic to the liver. Skin sensitization is predicted to be zero. It indicated the maximum tolerated dose as −0.502 log mg/kg/day. It does not act as an inhibitor of human hERG I, while it inhibits hERG II. The LD50 and LOAEL in rats are predicted to be 2.563 mol/kg and 0.89 log/mg bw/day, respectively. Minnow and T. pyriformis toxicity tests showed −1.696 log mM and 0.316 log ug/l, respectively (Table 4). The predicted physiological effects suggest that lupeol is highly favorable to cellular functions. It can be readily absorbed through the intestine and shows no predictable hepatotoxicity. It ostensibly acts as a protectant for the liver. These data strengthen the potential use of lupeol as medication based on the known biological effects (Bhatt et al., 2021; Maurya et al., 2020; Narendar et al., 2023; Osafo et al., 2023).
CONCLUSION
Molecular predictions indicate that lupeol can be a lead compound as an antiviral drug for its ability to interact with the vital cell surface proteins of coronavirus. It showed high binding affinities with Mpro, RdRp, Plpro, and S protein with affinities of −8.2, −7.5, −8.2, and −8.2 kcal/ml, respectively. These are the critical proteins of SARS-CoV-2 for infectivity, virulence, and adaptation. Lupeol also has potential additional pharmacological properties as it can interact with signaling proteins that are fundamental to normal cellular functions. Our data further add to the repertoire of lupeol as an anticancer agent as it is predicted to interact with a cohort of neoplastic conditions including breast cancer, cervical cancer, colorectal cancer, colon cancer, lung carcinoma, melanoma, ovarian carcinoma, and thyroid cancer. Therefore, our findings warrant experimental investigations to understand lupeol for pharmaceutical applications as antiviral and other medicinal activities.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
The study was supported by DBT-BUILDER (BT/INF/22/SP41398/2021) of the Department of Biotechnology, Government of India. PBL and CLR are research staff under the scheme.
CONFLICT OF INTERESTS
All authors declare no conflict of interests.
ETHICS APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Agrwal A, Juneja S, Dwivedi S, Kasana V. Molecular docking and antimicrobial analyses of synthesized imidazole derivatives in solvent less condition, adjacent to human pathogenic bacterial strains. Mater Today Proc, 2022; 57:2250–4.
Bhatt M, Patel M, Adnan M, Reddy MN. Anti-metastatic effects of lupeol via the inhibition of MAPK/ERK pathway in lung cancer. Anticancer Agents Med Chem, 2021; 21:201–6.
Cao DS, Liu S, Xu QS, Lu HM, Huang JH, Hu QN, Liang YZ. Large-scale prediction of drug-target interactions using protein sequences and drug topological structures. Anal Chim Acta, 2012; 752:1–10.
Davella R, Gurrapu S, Mamidala E. Phenolic compounds as promising drug candidates against COVID-19—an integrated molecular docking and dynamics simulation study. Mater Today Proc, 2022; 51:522–7.
Ebenezer O, Bodede O, Awolade P, Jordaan MA, Ogunsakin RE, Shapi M. Medicinal plants with anti-SARS-CoV activity repurposing for treatment of COVID-19 infection: a systematic review and meta-analysis. Acta Pharm, 2022; 72(2):199–224.
El-Demerdash A, Hassan A, Abd El-Aziz TM, Stockand JD, Arafa RK. Marine brominated tyrosine alkaloids as promising inhibitors of SARS-CoV-2. Molecules, 2021; 26:6171.
Enmozhi SK, Raja K, Sebastine I, Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J Biomol Struct Dyn, 2021; 39:3092–8.
Flórez-Álvarez L, Martínez-Moreno J, Zapata-Cardona MI, Galeano E, Alzate-Guarin F, Zapata W. In vitro antiviral activity against SARS-CoV-2 of plant extracts used in Colombian traditional medicine. Vitae, 2022; 29:347854.
Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science, 2020; 368:779–82.
Gao X, Qin B, Chen P, Zhu K, Hou P, Wojdyla JA, Wang M, Cui S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm Sinica B, 2021; 11:237–45.
Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res, 2014; 42:32–8.
Giordano D, Facchiano A, Carbone V. Food plant secondary metabolites antiviral activity and their possible roles in SARS-CoV-2 treatment: an overview. Molecules, 2023; 28(6):2470.
Jejurikar BL, Rohane SH. Drug designing in discovery studio. Asian J Rer Chem, 2021; 14(2):135–8.
Ju?ica J, Dovrt?lová G, Nosková K, Zendulka O. Bile acids, nuclear receptors and cytochrome P450. Physiol Res, 2016; 65(4):S427–40.
Khan MA, Singh D, Fatma H, Akhtar K, Arjmand F, Maurya S, Siddique HR. Antiandrogen enzalutamide induced genetic, cellular, and hepatic damages: amelioration by triterpene lupeol. Drug Chem Toxicol, 2023; 46(2):380–91.
Lalchhandama K. A history of coronaviruses. WikiJ Med, 2021; 9:5(1–19).
Liu K, Zhang X, Xie L, Deng M, Chen H, Song J, Long J, Li X, Luo J. Lupeol and its derivatives as anticancer and anti-inflammatory agents: molecular mechanisms and therapeutic efficacy. Pharmacol Res, 2021; 164:105373.
Majumder R, Mandal M. Screening of plant-based natural compounds as a potential COVID-19 main protease inhibitor: an in silico docking and molecular dynamics simulation approach. J Biomol Struct Dyn, 2022; 40:696–711.
Manóchio C, Torres-Loureiro S, Scudeler MM, Miwa B, Souza-Santos FC, Rodrigues-Soares F. Theranostics for COVID-19 antiviral drugs: prospects and challenges for worldwide precision/personalized medicine. OMICS J Integr Biol, 2023; 27:0151.
Martinez MA. Efficacy of repurposed antiviral drugs: lessons from COVID-19. Drug Discov Today, 2022; 27:1954–60.
Maurya SK, Shadab GG, Siddique HR. Chemosensitization of therapy resistant tumors: targeting multiple cell signaling pathways by lupeol, a pentacyclic triterpene. Curr Pharm Des, 2020; 26:455–65.
Monsalve-Escudero LM, Loaiza-Cano V, Zapata-Cardona MI, Quintero-Gil DC, Hernández-Mira E, Pájaro-González Y, Oliveros-Díaz AF, Diaz-Castillo F, Quiñones W, Robledo S, Martinez-Gutierrez M. The antiviral and virucidal activities of voacangine and structural analogs extracted from Tabernaemontana cymosa depend on the dengue virus strain. Plants, 2021; 10(7):1280.
Nandi S, Kumar M, Saxena AK. Repurposing of drugs and HTS to combat SARS-CoV-2 main protease utilizing structure-based molecular docking. Lett Drug Des Discov, 2022; 19:413–27.
Narendar K, Rao BS, Tirunavalli S, Jadav SS, Andugulapati SB, Ramalingam V, Babu KS. Synthesis of novel thiazoles bearing lupeol derivatives as potent anticancer and anti-inflammatory agents. Nat Prod Res, 2023; 5:1–8.
Nie C, Trimpert J, Moon S, Haag R, Gilmore K, Kaufer BB, Seeberger PH. In vitro efficacy of Artemisia extracts against SARS-CoV-2. Virol J, 2021; 18:1–7.
Osafo T, Philips TJ, Adomako AK, Borquaye LS, Ekuadzi E, Appiah-Opong R, Dickson RA. In vitro antileishmanial activity and molecular docking studies of lupeol and monostearin, isolated from Parkia biglobosa. Sci Afr, 2023; 19:e01464.
Pires DE, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem, 2015; 58:4066–72.
Rudik A, Bezhentsev V, Dmitriev A, Lagunin A, Filimonov D, Poroikov V. Metatox-Web application for generation of metabolic pathways and toxicity estimation. J Bioinform Comput Biol, 2019; 17:1940001.
Rudrapal M, Gogoi N, Chetia D, Khan J, Banwas S, Alshehri B, Alaidarous MA, Laddha UD, Khairnar SJ, Walode SG. Repurposing of phytomedicine-derived bioactive compounds with promising anti-SARS-CoV-2 potential: molecular docking, MD simulation and drug-likeness/ADMET studies. Saudi J Biol Sci, 2022; 29:2432–46.
Saifulazmi NF, Rohani ER, Harun S, Bunawan H, Hamezah HS, Nor Muhammad NA, Azizan KA, Ahmed QU, Fakurazi S, Mediani A, Sarian MN. A review with updated perspectives on the antiviral potentials of traditional medicinal plants and their prospects in antiviral therapy. Life, 2022; 22(12):1287.
Santos Pereira R, Vasconcelos Costa V, Luiz Menezes Gomes G, Rodrigues Valadares Campana P, Maia de Pádua R, Barbosa M, Oki Y, Heiden G, Fernandes GW, Menezes de Oliveira D, Souza DG. Anti-Zika virus activity of plant extracts containing polyphenols and triterpenes on Vero CCL-81 and human neuroblastoma SH-SY5Y cells. Chem Biodivers, 2022; 19(4):e202100842.
Sharma A, Vora J, Patel D, Sinha S, Jha PC, Shrivastava N. Identification of natural inhibitors against prime targets of SARS-CoV-2 using molecular docking, molecular dynamics simulation and MM-PBSA approaches. J Biomol Struct Dyn, 2022; 40:3296–311.
Siddique HR, Saleem M. Beneficial health effects of lupeol triterpene: a review of preclinical studies. Life Sci, 2011; 88:285–93.
Sohag AA, Hossain T, Rahaman A, Rahman P, Hasan MS, Das RC, Khan MK, Sikder MH, Alam M, Uddin J, Rahman H. Molecular pharmacology and therapeutic advances of the pentacyclic triterpene lupeol. Phytomedicine, 2022; 99:154012.
Tang S, Chen R, Lin M, Lin Q, Zhu Y, Ding J, Hu H, Ling M, Wu J. Accelerating autodock Vina with GPUS. Molecules, 2022; 27(9):3041.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem, 2010; 31:455–61.
Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 2020; 181:281–92.
Ye J, Wang Z, Jia J, Li F, Wang Y, Jiang Y, Ren Z, Pu H. Lupeol impairs herpes simplex virus type 1 replication by inhibiting the promoter activity of the viral immediate early gene α0. Acta Virol, 2021; 65:254–63.
Yu R, Li P. Screening of potential spike glycoprotein/ACE2 dual antagonists against COVID-19 in silico molecular docking. J Virol Methods, 2022; 301:114424.
Zaki AA, Ashour A, Elhady SS, Darwish KM, Al-Karmalawy AA. Calendulaglycoside A showing potential activity against SARS-CoV-2 main protease: molecular docking, molecular dynamics, and SAR studies. J Trad Complement Med, 2022; 12:16–34.
Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-Cov-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science, 2020; 368:409–12.