INTRODUCTION
Nile tilapia (Latin name Oreochromis niloticus) is the third most commonly cultured fish in the world according to the Food and Agriculture Organization statistics, with production which reached around 4.5 million metric tons in 2018. However, O. niloticus is normally exported in the form of fish fillet, which contributes only about 34%–37% of the total mass of the fish. In other words, about 2.7 million metric tons of the produced biomass will be discarded as waste (Choonpicharn et al., 2015; Sary et al., 2017).
Amongst the tilapia waste is its skin which still contains high-quality proteins. The skin has empirically been proven to aid in wound healing (Costa et al., 2019; Júnior et al., 2019). Some research revealed that the skin proteins impart this effect along with other biological activities. This essentially means that the wound healing capacity might be improved even further through hydrolysis of the skin proteins into peptides (Choonpicharn et al., 2015; Robert et al., 2015). Hydrolyzed proteins are generally better as they are easier to absorb due to their lower molecular weight (León-López et al., 2019; Lin et al., 2021). This is of utmost importance as wound healing compounds are normally applied topically, so they have to follow the 500 Da rule to allow better penetration across the skin barriers (Bos and Meinardi, 2000).
Several studies have characterized the wound healing potency of O. niloticus skin hydrolysates, such as the works by Hu et al. (2017) and Elbialy et al. (2020). Hu et al. (2017) characterized the Nile tilapia skin protein hydrolysates obtained through enzymatic hydrolysis with protease and reported its therapeutic effects on burns care. In vitro and in vivo tests done in this study supported the notion that the skin protein hydrolysates were potent for such an objective. Meanwhile, Elbialy et al. (2020) established a new understanding of the mechanism behind the acceleration of the wound healing process by tilapia skin protein hydrolysates. The hydrolysates were found to speed up the process through the upregulation of genes encoding growth factors such as transforming growth factor beta-1, basic fibroblast growth factor, and alpha-smooth muscle actin. The expression of genes for these growth factors speeds up the fibroblast and myofibroblast proliferation and, eventually, wound healing.
Proliferation, however, is only a part of the complicated wound healing mechanism. The process actually consists of four interlapping phases—hemostasis, inflammation, proliferation, and remodeling. In the first stage, platelets in the blood will form clots on the wound to prevent excessive blood loss. Next, white blood cells in the blood will clean up the wounded area during the inflammation period to prevent infection. Afterward, the proliferative growth factors are recruited to the site to induce proliferation of the fibroblast, allowing reepithelialization of the wound area. Last, the newly formed tissue is progressively digested and rebuilt with stronger material in the remodeling phase (Caley et al., 2015).
The wound healing process is tightly regulated by several factors, with the matrix metalloproteinase (MMPs) enzyme family being one of the most prominent regulators. MMPs play important roles in all stages of wound healing, and their activities are kept in check by their inhibitors (Caley et al., 2015; de Oliveira Gonzalez et al., 2016). Matrix metalloproteinase-2 (MMP-2) is amongst the MMPs involved in early wound healing (i.e., inflammation period). MMP-2 assists in platelet adhesion and aggregation, degrades potential microbial biofilm present, and triggers keratinocyte proliferation and migration (Caley et al., 2015). MMP-2 concentration will then decrease after progression into the proliferation phase. In fact, prolonged elevation of MMP-2 concentration is one of the indicators of a chronic or unhealed wound, and the use of its inhibitors improves the outcome of the wound (Sabino and auf dem Keller, 2015).
As in vivo trials with tilapia skin protein extract so far have never reported the incidence of chronic wounds, the skin extract and hydrolysate might have an additional inhibitory effect against MMP-2. In addition, it has previously been demonstrated with gelatin reverse zymography that a peptide from a tilapia skin hydrolysate exerted inhibitory activity against matrix metalloproteinase-2 (MMP-9), a homolog of MMP-2. This study revealed that the activity was maintained when tested in vivo, where the peptide was able to protect human keratinocytes from UV-triggered, MMP-9-mediated damage (Xiao et al., 2019). Ma et al. (2018) also observed similar results while testing the photoprotective capacity of another peptide of tilapia skin hydrolysate for mouse embryonic fibroblasts irradiated with UVB.
Hence, this work aimed to analyze the activity of hydrolyzed proteins from tilapia skin—termed DLBS3D33—as an MMP-2 inhibitor. A modified reverse gelatin zymography method was developed to fulfill this purpose. DLBS3D33 was obtained by hydrolysis of the tilapia skin with a protease from the pineapple stem (i.e., bromelain). DLBS3D33 was then characterized with size exclusion chromatography (SEC) and Fourier transform infrared (FTIR) spectroscopy to analyze the molecular weight distribution and the secondary structure of its peptides, respectively. In addition, peptides in DLBS3D33 were screened with thin-layer chromatography (TLC) and 2D electrophoresis.
MATERIALS AND METHODS
This research was divided into five major stages: protein extraction by enzymatic hydrolysis, quantitative analysis, protein characterization, screening and isolation of peptides, and in vitro study. The methods used are explained below, and the overall process flow is summarized in Figure 1.
Materials
The tilapia skin (as substrate) and the pineapple stem (as source of natural protease) were obtained from local suppliers. The skin was washed with running tap water to remove the leftover meat and other materials. The skin was then drained and stored in a −20°C freezer. The pineapple stem was washed prior to preparation work. After washing, the outer skin of the pineapple stem was peeled, and the peeled stem was cut and ground into pieces approximately 1 cm in length. The protease activity of the pineapple stem was quantified prior to use in hydrolysis following the methods of Rahayu et al. (2017). All reagents used were analytical-grade chemicals sourced from Merck (Darmstadt, Germany) and Sigma-Aldrich (St. Louis, MO, USA). The human MMP-2 protein used in the in vitro gelatin reverse zymography assay was purchased from Acro Biosystems (Newark, DE, USA).
Enzymatic extraction and hydrolysis of tilapia skin and posthydrolysis processing
The hydrolysis was done with optimized conditions (see Appendix A): reaction temperature and time of 60°C for 60 minutes (Fig. A1), protease solution (i.e., pineapple stem bromelain) with an activity of 60 U/ml (Fig. A2), and solid-to-liquid (STR) ratio of 1:5 (Fig. A3). The solvent used in the hydrolysis was purified water. The preoptimization was done in a duplicate manner, and both sets of results were used to obtain the average value shown in the graph.
After the hydrolysis, the hydrolysate was filtered twice with a 200-mesh nylon filter and 0.1 μm microfilter to remove larger and smaller insoluble particles. The filtrate was then heated at 90°C for 30 minutes. The resulting solution was regarded as DLBS3D33. Next, DLBS3D33 was evaporated to 10% of its initial volume with a rotary evaporator (Buchi Rotavapor R215, Flawil, Switzerland) at 40°C to obtain a DLBS3D33 concentrate. The DLBS3D33 solution was then powdered using a freeze dryer to yield DLBS3D33 powder (Martin Christ Alpha 1-2 LDplus, Osterode am Harz, Germany) at −50°C.
Quantitative analysis of DLBS3D33 protein and collagen
Protein and collagen quantification were used as determining factors in hydrolysis optimization and also for comparing DLBS3D33 quality throughout processing. Both values were measured using spectrophotometric-based methods (Agilent Cary 60 UV-Vis, Santa Clara, CA, USA). Quantification of protein in DLBS3D33 was done with the Lowry method (Lowry et al., 1951), whereas collagen was quantified indirectly based on Hydroxyproline (HYP) concentration following the method described by Cissell et al. (2017). HYP concentration was converted into collagen following the formula below:
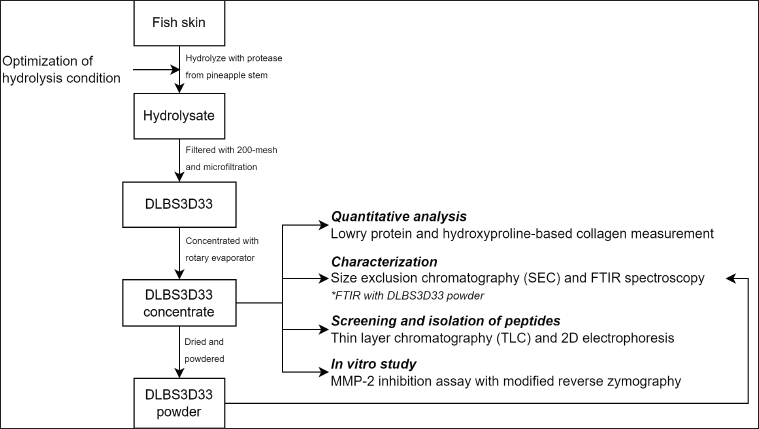 | Figure 1. Outline of the research. The processing done to the substrate (i.e., fish skin) to produce DLBS3D33 and the assays done to characterize and assess DLBS3D33 activity were detailed. [Click here to view] |
DLBS3D33 protein characterization
SEC was employed to analyze the molecular weight distribution of DLBS3D33. Superdex® 30 pg in HiLoad® 16/600 column (Cytiva Life Sciences, Marlborough, MA) was used as the stationary phase. The mobile phase employed was phosphate buffer pH 7.4 (contains 0.5 mM K2HPO4, 0.5 mM KH2PO4, and 10 mM NaCl) (López-Morales et al., 2019). 1 ml of DLBS3D33 was injected into the column with a flow rate of 0.5 ml/minutes, and the output was read at 280 nm. Vitamin B2 (375.36 Da), deoxidized glutathione (612 Da), and vitamin B12 (1355 Da) were employed as standard molecules and were injected in the same manner. Readouts from these standards molecules were used as comparators to map the molecular weight distribution of DLBS3D33 peptides.
The secondary structure of peptides in DLBS3D33 was observed indirectly through their infrared absorption spectra. FTIR spectroscopy (FTIR; PerkinElmer UATR Two, Waltham, MA) was utilized with the spectra produced in the wavenumber range of 4,000–700 cm−1.
Screening and isolation of DLBS3D33 peptides
TLC was used for the screening of peptides in DLBS3D33. Ten μl of DLBS3D33 was applied to the bottom part of the TLC plate (Merck) with an automatic sampler (CAMAG Automatic TLC Sampler 4, Muttenz, Switzerland). The eluent used was 0.4% ninhydrin dissolved in n-butanol, acetic acid, and water (3:1:1 ratio). The bromelain solution used in the hydrolysis process was also resolved along with a commercially available hydrolyzed tilapia skin collagen for use as a control and comparator, respectively.
The peptides in DLBS3D33 were resolved with 2D electrophoresis (2D-E) according to the method by Rahayu et al. (2016) with slight modification to include the tricine-SDS-PAGE system instead of glycine-SDS-PAGE. DLBS3D33 was mixed with a 70% trichloroacetic acid solution in a 9:1 ratio and then incubated in an ice bath for 15 minutes. Afterward, the mixture was centrifuged at 11,500 rpm for 30 minutes at 4°C. The pellet was dried and reconstituted with a rehydration buffer to the initial volume. Then, the first step of 2D-E, isoelectric focusing, was carried out by rehydrating an immobilized pH 3–10 gradient strip (IPG strip; Bio-Rad Ready Strip 7 cm, Hercules, CA) with 125 μl of the reconstituted pellet solution. The condition for isoelectric focusing was rapid ramping to 250 V for 15 minutes followed by gradual ramping to 4,000 V for 1 hour (Bio-Rad Protean i12 IEF Cell). The voltage was then kept at 4,000 V for 15,000 V-Hr.
For the second dimension, a tricine-SDS-PAGE gel was prepared. Two layers of resolving layer were made with polyacrylamide concentrations of 10% and 16%. Then, the IPG strip from the previous dimension was placed atop the 10% layer and secured with 0.5% agarose gel. After the agarose solidified, the gel was run at 120 V for 100 minutes (Bio-Rad PowerPac Basic & Mini-PROTEAN Tetra Cell) alongside an ultra-low molecular weight marker with the size range of 1.02–26.60 kDa. After the process, the Coomassie Brilliant Blue R-250 solution was used as the staining solution. The gel was soaked in the staining solution for 2 hours and then in the destaining solution for another 2 hours.
In Vitro MMP-2 inhibitory study
The activity of DLBS3D33 as an MMP-2 inhibitor was assessed with cell-free reverse gelatin zymography. The technique was adapted from Ren et al. (2017) with several modifications, as detailed below.
The MMP-2 solution with a concentration of 5 μg/ml was prepared and activated by incubation at 37°C for 30 minutes. The incubated MMP-2 solution was mixed with samples in a 1:2 ratio and then reincubated at 37°C for 1.5 hours. The MMP-2/sample mix was run in the gelatin polyacrylamide gel at 100 V for 100 minutes. The gelatin reverse zymography gel was prepared following the standard formulation of glycine-SDS-PAGE gel with a polyacrylamide concentration of 12% and 0.1% gelatin as the substrate.
After electrophoresis, the gel was soaked and agitated in a 2.5% Triton X-100 solution for 30 minutes or until the blue stain disappeared. The gel was then incubated in a 50 mM Tris-HCl buffer solution pH 7.4 containing 5 mM calcium chloride overnight. The gel was stained in the Coomassie Brilliant Blue R-250 solution for 2 hours following the end of incubation and rinsed with the destaining solution for 30 minutes. The area and intensity of the resulting clear bands in the gel were analyzed with the ImageJ version 1.44 and GelAnalyzer version 19.1 software and compared to control—MMP-2 mixed with purified water in a 1:2 ratio.
RESULTS AND DISCUSSION
DLBS3D33 protein and collagen content
DLBS3D33 was obtained by hydrolysis of Nile tilapia skin with a natural protease from pineapple stem as the stem contains the highest level of protease activity (Rahayu et al., 2017). The hydrolysate was then processed with two-step filtration to remove the leftover solid material and evaporation to concentrate the solution. To determine the effectivity of the hydrolysis system and ensure retention of peptides, protein and collagen contents of DLBS3D33 throughout the processing were quantified (Table 1).
The hydrolysis of 144 g of tilapia skin resulted in 20,000 mg of protein in DLBS3D33 after filtration, which means the protein and collagen yields were estimated to be around 13.71% and 6.43%, respectively. The level of collagen obtained was more or less in agreement with previous publications on tilapia skin hydrolysis. Song et al. (2021) obtained collagen yields of as much as 7.60%–8.14% with pepsin hydrolysis and 4.27%–4.76% with acid hydrolysis. The commercial tilapia skin protein hydrolysate tested also gave a readout in a similar range (Table 1).
On the other hand, the protein yield was lower than that reported by Giraldo-Rios et al. (2020), which was around 15%–27.2% using alkaline. This slight discrepancy can be attributed to the different extraction methods and/or the difference in composition of tilapia fish based on their habitat. Nonetheless, the protein content of DLBS3D33 powder is still higher than the commercial product tested in this study (Table 1).
Previous studies revealed that the protein content of Nile tilapia skin (wet-mass basis) ranged from 21.30% to 29.68% (Alfaro et al., 2013; Giraldo-Rios et al., 2020). In relation to data presented in Table 1, the protein recovery in this study is 46.21%–64.40%. This data reciprocates that of Tian et al. (2014), who utilized a more complicated extraction method, ultrasound-assisted alkaline extraction, and achieved protein recovery of 62.6% from tilapia fillets.
Determination of molecular weight distribution of DLBS3D33 peptides
The molecular weight distribution of the peptides in DLBS3D33 was mapped by running the sample through SEC. Several compounds with known sizes—riboflavin (376 Da), oxidized glutathione (612 Da), and cobalamin (1355 Da)—were also eluted with the same setup to be used as comparison points (Fig. 2).
The area of each peak was measured in relation to the baseline from the mobile phase and then summed according to the molecular weight range (Table 2). From the data, it is clear that the hydrolysis process successfully digested Nile tilapia skin proteins into small peptides. This is in agreement with the observation by Sierra-Lopera and Zapata-Montoya (2021), who compared the protein content and degree of hydrolysis of Nile tilapia skeletal protein with endo- and exopeptidase. Their result suggests that the Nile tilapia skeleton hydrolysate obtained with an alkaline endopeptidase, similar to stem bromelain used in this study, has a higher protein content and degree of hydrolysis.
In this study, more than 50% of the DLBS3D33 peptides are below 500 Da while the portion of larger peptides (>1,355 Da) is only 26%. This data reciprocates those of Hu et al. (2017), whose hydrolysis of Nile tilapia skin resulted in 75% peptides under 1 kDa. DLBS3D33 theoretically has the ability to pass through the epidermis and enter the dermis as its peptides are predominantly below 500 Da. Hence, these peptides have the potential to exert better bioactivity. In addition, it has been suggested that bioactive peptides are mostly below 4 kDa (Sierra-Lopera and Zapata-Montoya, 2021).
Analysis of secondary structure of DLBS3D33 peptides
The FTIR spectrum of DLBS3D33 exhibited typical protein-specific peaks, such as amides A and B as well as amides I–III (Fig. 3). Amongst these peaks, amides I (around 1,700–1,600 cm−1) and II (around 1,570–1,540 cm−1) are the most sensitive to secondary structure changes. This is due to the IR corresponding to the C = O stretch and N-H bending, two bonds that are directly involved in the formation of protein secondary structure (Tatulian, 2019; Yang et al., 2015). Amide I and II peaks in DLBS3D33 were observed at 1,634 and 1,532 cm−1, respectively, indicating that the majority of the DLBS3D33 peptides are in α-helix and coiled structures (Tatulian, 2019; Yang et al., 2016).
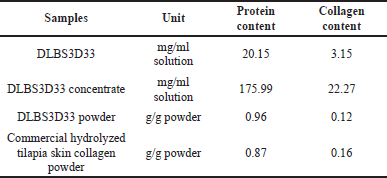 | Table 1. DLBS3D33 protein and collagen content throughout the processing. [Click here to view] |
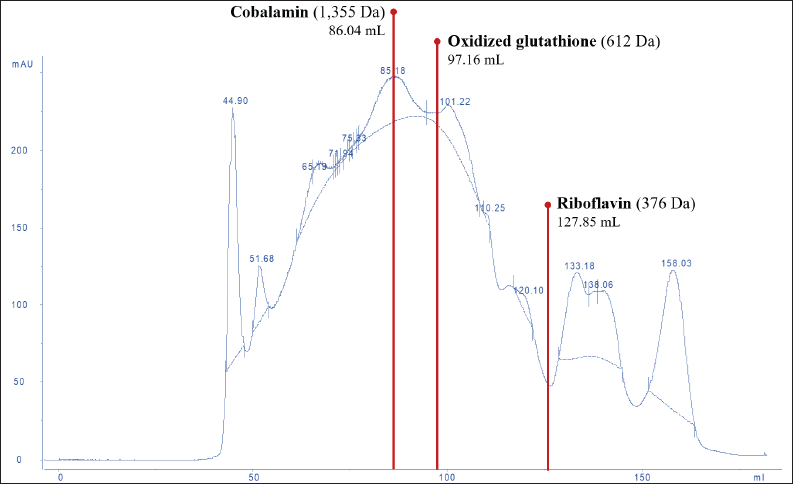 | Figure 2. SEC chromatogram of DLBS3D33. The chromatogram was analyzed and compared to the standard molecules—riboflavin, oxidized glutathione, and cobalamin—in order to reveal the molecular weight distribution of peptides in DLBS3D33. The solid line represented the readout of the sample whereas the dotted line was the baseline. [Click here to view] |
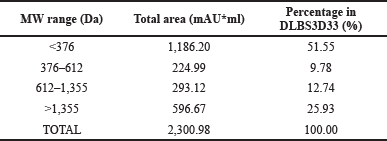 | Table 2. The molecular weight distribution of DLBS3D33 peptides. [Click here to view] |
Native collagen, the major protein in tilapia skin, has a special tertiary structure of a triple helix. The presence of this feature can be identified with FTIR analysis through the ratio of absorption at amide III and C-H stretch region. The closer the ratio to 1, the more the amount of the intact triple helix structure, indicating unsuccessful hydrolysis (Plepis et al., 1996; Zhang et al., 2019). The calculated amide III/C-H stretch ratio of DLBS3D33 is 0.78, identical to the ratio of a commercially available tilapia skin collagen hydrolysate. In addition, overlaid spectra of DLBS3D33 and the commercial sample (subset of Fig. 3) showed more or less similar peaks. This observation indicates that the current method of tilapia skin hydrolysis is successful in hydrolyzing the skin protein, at the very least the collagen part, into smaller peptides.
Screening, isolation, and identification of DLBS3D33 peptides
The peptides in DLBS3D33 were resolved with TLC together with the bromelain solution as a control (Fig. 4, lanes 1 and 2). The comparison is essential as TLC is a very sensitive method that could detect a small concentration of peptides potentially contributed by the bromelain solution. Corresponding to this fact, the bromelain solution produced seven bands with ninhydrin staining. These bands were all retained in DLBS3D33 even though bands 1, 2, 3, 6, and 7 were differently colored compared to the bromelain solution. As the color of the ninhydrin stain depends on the amino acid composition, the change in color indicates that these five bands might represent different peptides with similar Rf to the bromelain. Hence, DLBS3D33 potentially has six different peptides, including one additional band (band 8) found only in the DLBS3D33 lane.
The chromatogram of DLBS3D33 was compared to those of the commercial tilapia skin collagen hydrolysate (Fig. 4, lane 3). The collagen hydrolysate produced some bands with Rf and coloration similar to DLBS3D33—bands 1, 2, and 6. It is possible that these bands are indeed collagen peptides resulting from the hydrolysis of the skin collagen, though further research needs to be done to investigate the identity of the peptides definitively.
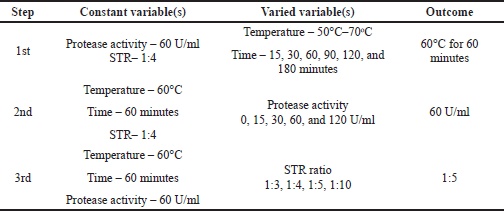 | Table 3. Depiction and outcome of step-by-step hydrolysis parameters optimization. [Click here to view] |
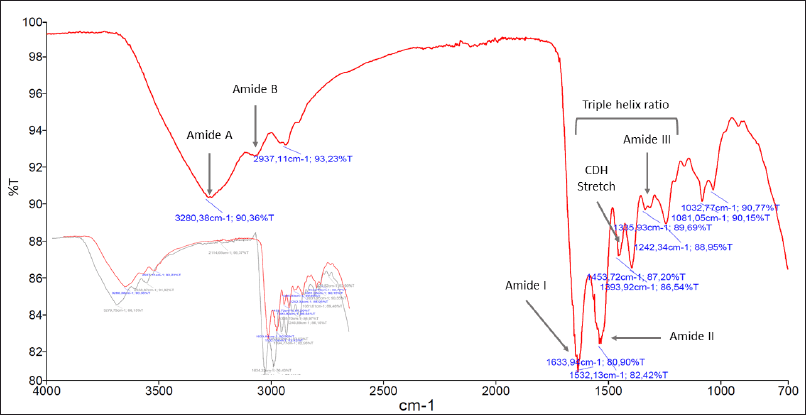 | Figure 3. FTIR spectrum of DLBS3D33. Crucial peaks attributed to the presence of proteins such as amides A and B along with amides I–III were present. In the subset, the reading of the DLBS3D33 spectrum (in red) was overlaid with the FTIR spectrum of commercial hydrolyzed tilapia skin collagen (in gray). Both readings showed comparable spectra, with the commercial product also producing five specific protein peaks in similar wavenumbers. [Click here to view] |
Meanwhile, the sequestration of the DLBS3D33 peptides with 2D electrophoresis (Fig. 5) resulted in five different spots, all sized above 6.5 kDa. Correlating it back to the molecular weight distribution of the DLBS3D33 peptides (Table 2), the peptides observed from the 2D-E separation account for only around 25% of the whole population. This is due to the fact that lower-sized peptides were underrepresented in the tricine-SDS-PAGE system as it can only detect peptides with a minimum size of 1 kDa (Jiang et al., 2016). The previous hydrolysis of fish skin protein with bromelain has yielded a hydrolysate with a higher peptide size range due to its high specificity (Cheung and Li-Chan, 2017), consistent with what was found in this study (Table 2). Hence, the combinatory approach of TLC and 2D-E implemented herein can provide a more complete insight into the peptides’ identity—2D-E focuses on larger-sized peptides and TLC on smaller-sized peptides.
Evaluation of bioactivity of DLBS3D33 peptides as MMP-2 inhibitors
The capacity of DLBS3D33 in inhibiting MMP-2 was analyzed with a modified gelatin reverse zymography method. In typical reverse zymography, the electrophoresis gel is impregnated with the substrate of the target enzyme and then used for the electrophoresis of a potential inhibitor. Afterward, the gel is incubated in the enzyme solution, causing the breakdown of the substrate in all parts of the gel beside the area where inhibitors are resolved. Subsequent staining of the gel will result in a deep blue band of inhibitors against a faint blue background.
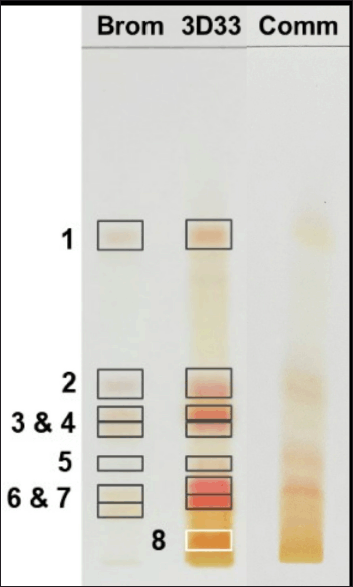 | Figure 4. TLC chromatogram of bromelain, DLBS3D33, and commercially available tilapia skin collagen hydrolysate. The separation resulted in seven and eight bands in bromelain protease (Brom; lane 1) and DLBS3D33 (3D33; lane 2), respectively. The chromatogram of DLBS3D33 was then compared to the chromatogram of commercial tilapia skin collagen hydrolysate (Comm; lane 3) to check for potential hydrolyzed collagen peptides in DLBS3D33. [Click here to view] |
In this study, the modification was done by mixing the enzyme (i.e., MMP-2) with the sample with potential inhibitory activity prior to loading into the gel. By doing so, the peptides in DLBS3D33 will bind to MMP-2 first and then travel through the gel together. Uninhibited MMP-2 resolved in the gel will digest gelatin around its bands and produce clear bands after the staining and destaining processes. On the other hand, if the DLBS3D33 peptides have inhibitory activity against MMP-2, the gelatin will not be broken; thus, no clear bands will be produced. After staining, the noninhibited MMP-2 will be depicted as clear zones whereas the inhibited MMP-2 will be of a dark blue color similar to the background. This modification reduced the amount of enzyme needed for the procedure and ensured the reaction between MMP-2 and the DLBS3D33 peptides. In this setup, pure MMP-2 needs to be run in the same gel as a reference. The difference between the typical and modified reverse zymography method and the result is portrayed in Figure 6.
DLBS3D33 was serially diluted for the concentration-dependent activity test. Initially, 10 μl of DLBS3D33 loaded into gels contained 2.83 mg of protein. DLBS3D33 was then diluted into four additional samples with protein contents of 1.415, 0.566, 0.378, and 0.283 mg. The lowest and highest concentrations of DLBS3D33 that were tested, without the addition of MMP-2, were used as negative controls to ensure the hydrolysate did not produce clear zones on its own (Fig. 7, lanes 1 and 2), whereas MMP-2 and MMP-2 diluted in purified water (in 1:2 ratio, similar to the MMP-2/sample ratio) were used as the positive controls (lanes 3 and 4). MMP-2 produced three distinguishable clear zones (lane 3), which were retained but reduced in their intensity after mixing with PW (lane 4). Since the addition of samples would cause the same dilution effect, the bands from MMP-2 diluted with PW were used as the reference.
DLBS3D33, containing 2.83 mg of protein, reduced the area and intensity of almost all MMP-2 bands, leaving only a faint band B (Fig. 7, lane 5). As the concentration of the DLBS3D33 protein was lowered, the MMP-2 clear zones reappeared and progressively became more similar to the positive control (lanes 6–9). To quantify these changes, the area and intensity of each band were measured, and both data were multiplied by each other to give the relative MMP-2 inhibition unit. The relative inhibition units were plotted and depicted in Figure 8 along with the total relative inhibition unit.
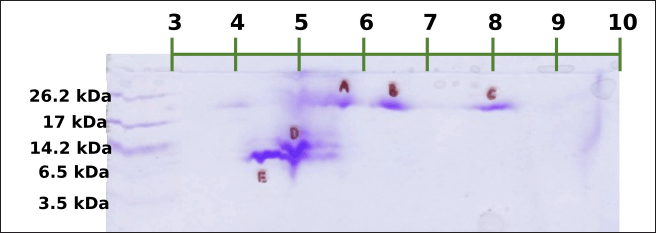 | Figure 5. 2D electrophoresis of DLBS3D33. Five spots were formed in total. The first three spots (A–C) were sized around 17–26.2 kDa with estimated pI of 5.7, 6.4, and 8.1, respectively. The fourth (D) and fifth (E) spots were sized around 6.5–14.2 kDa with estimated pI of 4.9 and 4.4, respectively. [Click here to view] |
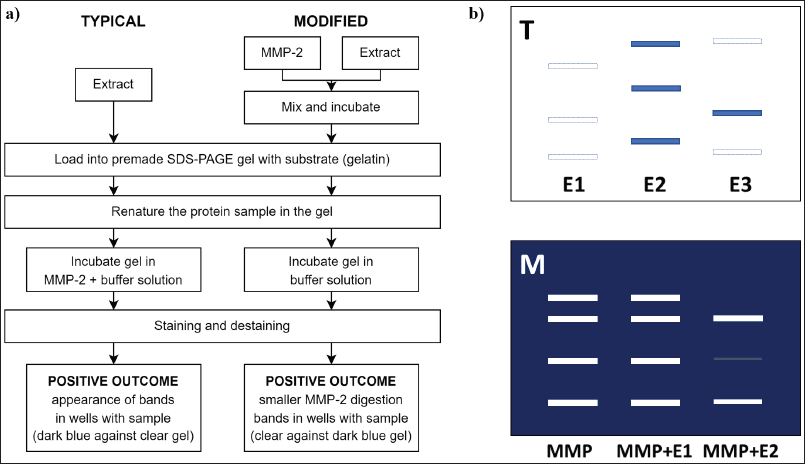 | Figure 6. Comparison of a typical and modified gelatin reverse zymography. (a) The flowchart of both methods reveals the main difference between them lies in the timing of MMP-2 and DLBS3D33 reaction. In the typical reverse zymography method, DLBS3D33 was exposed to MMP-2 after it was resolved in the gel. Meanwhile, the reaction between DLBS3D33 and MMP-2 was established prior to electrophoresis. (b) Illustration of results with typical (T) and modified (M) reverse zymography. In the presence of inhibitors, dark blue bands were formed with the typical reverse zymography method. On the other hand, clear zones were formed by MMP-2, and the reduction of these clear zones’ area and intensity were the signs of MMP-2 inhibitors. [Click here to view] |
 | Figure 7. Gelatin reverse zymography of DLBS3D33 in various concentrations. Each well was loaded with 10 μl of the designated sample. The amount of MMP-2 in lanes 4–8 was 0.017 μg. [Click here to view] |
Figure 8 confirms that DLBS3D33 inhibits the MMP-2 in a dose-dependent manner. As the amount of DLBS3D33 protein increased, the relative inhibition unit decreased for every MMP-2 band. Nonetheless, DLBS3D33 protein as little as 0.283 mg can already induce a reduction of the clear zones, especially in bands A and C. Based on the total relative inhibition unit, it is estimated that around 0.6–0.7 mg of the DLBS3D33 protein will be needed to give 50% inhibition of MMP-2 in this setup. As MMP-2 present in each sample was 0.017 μg, it can be deduced that, for every 1 μg of MMP-2, 35.3–41.2 mg of the DLBS3D33 protein is needed to inhibit 50% of the MMP-2 activity.
Generally, inhibition or downregulation of MMP-2 can be achieved through several methods (Mohan et al., 2016, Fields, 2019). First is the use of a chelating agent to bind the Zn2+-dependent active site in the catalytic region. This will directly prevent the binding of substrate and subsequently prevent its breakdown. However, this type of inhibitor is disadvantageous as the catalytic site sequence is generally conserved across all MMPs. The second method is by utilizing molecules that can bind to the hemopexin-like domain, whose sequence is more diverse, thus offering better specificity. The specificity can also be further escalated by the third method— targeting of the collagen-binding domain. This approach improves the specificity as said domain is found only in gelatinase-type MMPs (i.e., MMP-2 and MMP-9). Moreover, the hemopexin-like domain function is to aid in opening the triple helix and positioning the collagen molecules in regard to the catalytic site (Fields, 2019; Mohan et al., 2016; Ndinguri et al., 2012; Van Doren, 2015).
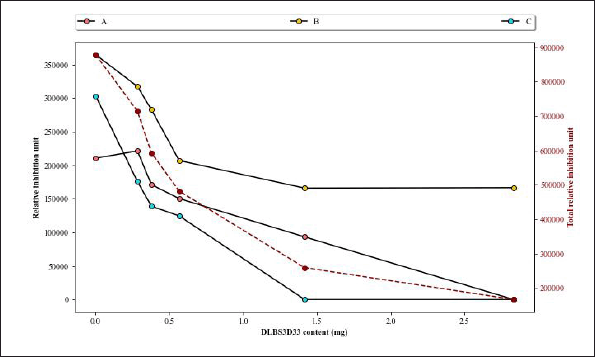 | Figure 8. Analysis of area and intensity of gelatin reverse zymography bands. Three digestion bands from MMP-2 as seen in Figure 7 were assigned as bands A–C. The intensity of bands produced by MMP-2 diluted with PW in a 1:2 ratio (Fig. 7, lane 4) was used as the comparison in this graph, its intensity reads included as “0 mg DLBS3D33.” The secondary axis represents the total relative inhibition unit (red dashed line), which is the sum of relative inhibition unit values from all three bands at a certain concentration. [Click here to view] |
It has been established that DLBS3D33 consists of smaller-sized peptides, some of which are collagen peptides. Small peptides have been shown to inhibit MMPs mostly through binding of the enzyme active site or the hemopexin-like domain (Das et al., 2021; Mohan et al., 2016). For example, Ma et al. (2018) revealed that a peptide derived from tilapia skin gelatin inhibited MMP-1 and MMP-9 through the binding of the Zn2+ catalytic site. Meanwhile, other studies have derived the sequence of substrates typically recognized by the enzyme as inhibitors (Fields, 2019; Mohan et al., 2016; Van Doren, 2015). This is most definitely the case for many collagen peptides, as established by Van Doren (2015). Thus, it is highly likely that DLBS3D33 might work in the same way, with the peptide preventing the breakdown of the substrate through binding of the hemopexin-like or collagen-binding domain. These kinds of inhibitions might allow not only specified targeting of the MMP but also specified effect on the MMP. In the case of MMP-2, the binding of inhibitors to the collagen-binding domain might prevent the gelatinase activity but not its other activities (Van Doren, 2015). This way, the enzyme is not totally incapacitated and hence prevents dysfunction in the body mechanism.
CONCLUSIONS
Hydrolysis of Nile tilapia skin with bromelain successfully produced hydrolysate DLBS3D33, which contained a high level of protein and collagen. The peptides in DLBS3D33 were found to be sized predominantly below 600 Da and were in an α-helix and coiled-coil conformation. It was estimated that DLBS3D33 consisted of six different species of peptides, five of which were isolated and identified. In vitro analysis with a modified gelatin reverse zymography method demonstrated the MMP-2 inhibitory activity of DLBS3D33. Quantification of the area and intensity of the clear zones revealed that 50% inhibition of 1 μg MMP-2 was achieved with around 35.3–41.2 mg of the DLBS3D33 protein. Further study needs to be done to unravel the specific molecular mechanisms underlying the MMP-2 inhibitory activity of DLBS3D33.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the facilities and materials provided by Dexa Laboratories of Biomolecular Sciences. They also would like to thank Affina Musliha and Ireno Mega Putera (DLBS scientists) for their excellent assistance in the data collection process.
AUTHORS’ CONTRIBUTIONS
RRT and PR conceptualized the experiments; PR and SPW developed the methodology; SPW performed the experiment and curated the data; SPW and PR analyzed the data; SPW prepared the original draft; MTS and RRT reviewed the manuscript and supervised the project. All authors have read and agreed to the published version of the manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
FUNDING
This research and the APC were funded by internal funding from PT Dexa Medica.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Alfaro ADT, Fonseca GG, Balbinot E, Machado A, Prentice C. Physical and chemical properties of wami tilapia skin gelatin. Food Sci Technol, 2013; 33:592–5; doi:org/10.1590/s0101-20612013005000069 CrossRef
Arshad ZIM, Amid A, Yusof F, Jaswir I, Ahmad K, Loke SP. Bromelain: an overview of industrial application and purification strategies. Appl Microbiol Biotechnol, 2014; 98:7283–97; doi:org/10.1007/s00253-014-5889-y CrossRef
Bala M, Ismail NA, Mel M, Saedi M, Salleh H, Salleh A, Amid A. Bromelain production: current trends and perspective. Arch Des Sci, 2012; 65:1464–661.
Bos JD, Meinardi MMHM. The 500 Dalton rule for the skin penetration of chemical compounds and drugs. Exp Dermatol, 2000; 9:165–9; doi:org/10.1034/j.1600-0625.2000.009003165.x CrossRef
Caley MP, Martins VLC, OToole EA. Metalloproteinases and wound healing. Adv Wound Care, 2015; 4:225–34; doi:org/10.1089/wound.2014.0581. CrossRef
Cheung IWY, Li-Chan ECY. Enzymatic production of protein hydrolysates from steelhead (Oncorhynchus mykiss) skin gelatin as inhibitors of dipeptidyl-peptidase IV and angiotensin-I converting enzyme. J Funct Foods, 2017; 28:254–64; doi:org/10.1016/j.jff.2016.10.030 CrossRef
Choonpicharn S, Tateing S, Jaturasitha S, Rakariyatham N, Suree N, Niamsup H. Identification of bioactive peptide from Oreochromis niloticus skin gelatin. J Food Sci Technol, 2015; 53:1222–9; doi:org/10.1007/s13197-015-2091-x CrossRef
Cissell DD, Link JM, Hu JC, Athanasiou KA. A modified hydroxyproline assay based on hydrochloric acid in ehrlichs solution accurately measures tissue collagen content. Tissue Eng Part C: Methods, 2017;23:243–50; doi:org/10.1089/ten.tec.2017.0018 CrossRef
Costa BA, Júnior EML, de Moraes Filho MO, Fechine FV, de Moraes MEA, Júnior FRS, do Nascimento Soares MFA, Rocha MBS. Use of tilapia skin as a xenograft for pediatric burn treatment: a case report. J Burn Care Res, 2019; 40:714–7; doi:org/10.1093/jbcr/irz085 CrossRef
Das S, Amin SA, Jha T. Inhibitors of gelatinases (MMP-2 and MMP-9) for the management of hematological malignancies. Eur J Med Chem, 2021; 223:113623; doi: 10.1016/j.ejmech.2021.113623 CrossRef
Das S, Amin SA, Jha T. Inhibitors of gelatinases (MMP-2 and MMP-9) for the management of hematological malignancies. Eur J Med Chem, 2021; 223:113623; doi: 10.1016/j.ejmech.2021.113623 CrossRef
de Oliveira Gonzalez AC, Costa TF, de Araújo Andrade Z, Medrado ARAP. Wound healing - a literature review. An Bras Dermatol, 2016; 91:614–20; doi:org/10.1590/abd1806-4841.20164741 CrossRef
Elbialy ZI, Atiba A, Abdelnaby A, Al-Hawary II, Elsheshtawy A, El-Serehy HA, Abdel-Daim MA, Fadl SE, Assar DH. Collagen extract obtained from Nile tilapia (Oreochromis niloticus L.) skin accelerates wound healing in rat model via up regulating VEGF, bFGF, and α-SMA genes expression. BMC Vet Res, 2020; 16:352; doi:org/10.1186/s12917-020-02566-2. CrossRef
Fields GB. Mechanisms of Action of Novel Drugs Targeting Angiogenesis-Promoting Matrix Metalloproteinases. Frontiers in Immunology, 2019; doi: 10.3389/fimmu.2019.0127 CrossRef
Fields GB. Mechanisms of action of novel drugs targeting angiogenesis-promoting matrix metalloproteinases. Front Immunol, 2019; doi: 10.3389/fimmu.2019.0127 CrossRef
Giraldo-Rios DE, Rios LA, Zapata-Montoya JE. Kinetic modeling of the alkaline deproteinization of Nile tilapia skin for the production of collagen. Heliyon, 2020; 6:e03854; doi:org/10.1016/j.heliyon.2020.e03854 CrossRef
Hu Z, Yang P, Zhou C, Li S, Hong P. Marine collagen peptides from the skin of Nile tilapia (Oreochromis niloticus): characterization and wound healing evaluation. Mar Drugs, 2017; 15:102; doi:org/10.3390/md15040102 CrossRef
Jafari H, Lista A, Siekapen MM, Ghaffari-Bohlouli P, Nie L, Alimoradi H, Shavandi A. Fish collagen: extraction, characterization, and applications for biomaterials engineering. Polymers (Basel), 2020; 12:2230; doi:org/10.3390/polym12102230. CrossRef
Jiang S, Liu S, Zhao C, Wu C. Developing protocols of tricine-SDS-PAGE for separation of polypeptides in the mass range 1–30 kDa with minigel electrophoresis system. Int J Electrochem Sci, 2016; 11:640–9.
Júnior EML, de Moraes Filho MO, Forte AJ, Costa BA, Fechine FV, Alves APNN, de Moraes MEA, Rocha MBS, Júnior FRS, do Nascimento Soares MFA, Bezerra AN, Martins CB, Mathor MB. Pediatric burn treatment using tilapia skin as a xenograft for superficial partial-thickness wounds: a pilot study. J Burn Care Res, 2019; doi:org/10.1093/jbcr/irz149
Jutamongkon R, Charoenrein S. Effect of temperature on the stability of fruit bromelain from smooth cayenne pineapple. Kasetsart J Nat Sci, 2010; 44:943–8.
León-López A, Morales-Peñaloza A, Mart\’{\i}nez-Juárez VM, Vargas-Torres A, Zeugolis DI, Aguirre-Álvarez G. Hydrolyzed collagen sources and applications. Molecules, 2019; 24:4031; doi:org/10.3390/molecules24224031 CrossRef
Lin H, Zheng Z, Yuan J, Zhang C, Cao W, Qin X. Collagen peptides derived from Sipunculus nudus accelerate wound healing. Molecules, 2021; 26:1385; doi:org/10.3390/molecules26051385. CrossRef
López-Morales CA, Vázquez-Leyva S, Vallejo-Castillo L, Carballo-Uicab G, Muñoz-Garc\’{\i}a L, Herbert-Pucheta JE, Zepeda-Vallejo LG, Velasco-Velázquez M, Pavón L, Pérez-Tapia SM, Medina-Rivero E. Determination of peptide profile consistency and safety of collagen hydrolysates as quality attributes. J Food Sci, 2019; 84:430–9; doi:org/10.1111/1750-3841.14466 CrossRef
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem, 1951; 193:265–75; doi:org/10.1016/s0021-9258(19)52451-6. CrossRef
Ma Q, Liu Q, Yuan L, Zhuang Y. Protective effects of LSGYGP from fish skin gelatin hydrolysates on UVB-induced MEFs by regulation of oxidative stress and matrix metalloproteinase activity. Nutrients, 2018; 10:420; doi:org/10.3390/nu10040420 CrossRef
Mohan V, Talmi-Frank D, Arkadash V, Papo N, Sagi I. Matrix metalloproteinase protein inhibitors: highlighting a new beginning for metalloproteinases in medicine. Metalloproteinases Med, 2016; 3:31–47; doi:org/10.2147/MNM.S65143 CrossRef
Ndinguri M, Bhowmick M, Tokmina-Roszyk D, Robichaud T, Fields G. Peptide-based selective inhibitors of matrix metalloproteinase-mediated activities. Molecules, 2012; 17(12):14230–48; doi:org/10.3390/molecules171214230 CrossRef
Plepis AMDG, Goissis G, Das-Gupta DK. Dielectric and pyroelectric characterization of anionic and native collagen. Polym Eng Sci, 1996; 36:2932–8; doi:org/10.1002/pen.10694 CrossRef
Rahayu P, Agustina L, Tjandrawinata RR. Tacorin, an extract from Ananas comosus stem, stimulates wound healing by modulating the expression of tumor necrosis factor α, transforming growth factor β and matrix metalloproteinase 2. FEBS Open Bio, 2017; 7:1017–25; doi:org/10.1002/2211-5463.12241 CrossRef
Rahayu P, Marcelline F, Sulistyaningrum E, Suhartono MT, Tjandrawinata RR. Potential effect of striatin (DLBS0333), a bioactive protein fraction isolated from Channa striata for wound treatment. Asian Pac J Trop Biomed, 2016; 6:1001–7; doi:org/10.1016/j.apjtb.2016.10.008 CrossRef
Ren Z, Chen J, Khalil RA. Zymography as a research tool in the study of matrix metalloproteinase inhibitors. Zymography. Springer, New York, NY, pp 79–102, 2017; doi:org/10.1007/978-1-4939-7111-4_8 CrossRef
Robert M, Zatylny-Gaudin C, Fournier V, Corre E, Le Corguillé G, Bernay B, Henry J. Molecular characterization of peptide fractions of a tilapia (Oreochromis niloticus) by-product hydrolysate and in vitro evaluation of antibacterial activity. Proc Biochem, 2015; 50:487–92; doi:org/10.1016/j.procbio.2014.12.022 CrossRef
Robinson PK. Enzymes: principles and biotechnological applications. Essays Biochem, 2015; 59:1–41; doi:org/10.1042/bse0590001 CrossRef
Sabino F, auf dem Keller U. Matrix metalloproteinases in impaired wound healing. Metalloproteinases Med, 2015; 1; doi:org/10.2147/mnm.s68420 CrossRef
Sary C, de Paris LD, Bernardi DM, Lewandowiski V, Signor A, Boscolo WR. Tilapia by-product hydrolysate powder in diets for Nile tilapia larvae. Acta Sci Anim Sci, 2017; 39:1; doi:org/10.4025/actascianimsci.v39i1.32805 CrossRef
Sierra-Lopera LM, Zapata-Montoya JE. Optimization of enzymatic hydrolysis of red tilapia scales (Oreochromis sp.) to obtain bioactive peptides. Biotechnol Rep, 2021; 30:e00611; doi:org/10.1016/j.btre.2021.e00611 CrossRef
Song Z, Liu H, Chen Liwen, Chen L, Zhou C, Hong P, Deng C. Characterization and comparison of collagen extracted from the skin of the Nile tilapia by fermentation and chemical pretreatment. Food Chem, 2021; 340:128139; doi:org/10.1016/j.foodchem.2020.128139 CrossRef
Tatulian SA. FTIR analysis of proteins and protein membrane interactions. Methods in molecular biology. Springer, New York, NY, pp 281–325, 2019; doi:org/10.1007/978-1-4939-9512-7_13 CrossRef
Tian J, Wang Y, Zhu Z, Zeng Q, Xin M. Recovery of tilapia (Oreochromis niloticus) protein isolate by high-intensity ultrasound-aided alkaline isoelectric solubilization/precipitation process. Food Bioproc Technol, 2014; 8:758–69; doi:org/10.1007/s11947-014-1431-6 CrossRef
van Doren SR. Matrix metalloproteinase interactions with collagen and elastin. Matrix Biol, 2015; 44–46:224–31; doi:org/10.1016/j.matbio.2015.01.005 CrossRef
Xiao Z, Liang P, Chen J, Chen M-F, Gong F, Li C, Zhou C, Hong P, Yang P, Qian Z-J. A peptide YGDEY from tilapia gelatin hydrolysates inhibits UVB-mediated skin photoaging by regulating MMP-1 and MMP-9 expression in HaCaT cells. Photochem Photobiol, 2019; 95:1424–32; doi:org/10.1111/php.13135 CrossRef
Yang G, Wu M, Yi H, Wang J. Biosynthesis and characterization of a non-repetitive polypeptide derived from silk fibroin heavy chain. Mater Sci Eng: C, 2016; 59:278–85; doi:org/10.1016/j.msec.2015.10.023 CrossRef
Yang H, Yang S, Kong J, Dong A, Yu S. Obtaining information about protein secondary structures in aqueous solution using fourier transform IR spectroscopy. Nat Protoc, 2015; 10:382–96; doi:org/10.1038/nprot.2015.024 CrossRef
Zhang Y, Tu D, Shen Q, Dai Z. Fish scale valorization by hydrothermal pretreatment followed by enzymatic hydrolysis for gelatin hydrolysate production. Molecules, 2019; 24:2998; doi:org/10.3390/molecules24162998 CrossRef
APPENDIX
A Optimization of enzymatic hydrolysis of Nile tilapia skin with natural protease from pineapple stem
Hydrolysis conditions were optimized in a stepwise manner. First, the temperature and time of hydrolysis were varied while the protease activity and STR ratio were kept constant. Afterward, the chosen temperature and time were used as constant variables to optimize protease activity, with the STR ratio kept at one value. Lastly, the preferred temperature and time of hydrolysis along with the chosen protease activity were applied for the optimization of the STR ratio. All variables were assessed in terms of protein and collagen yield. The step by step of the hydrolysis optimization experiment as explained above is summarized in Table 3 along with the preferred variation.
A.1 Hydrolysis temperature and time
Quantitative analysis of the protein hydrolysate reveals that the hydrolysis temperature of 60°C yields the highest protein content at every timepoint (Fig. A1a). The highest protein content is achieved just after 60 minutes of hydrolysis at this temperature, after which the protein level generally becomes stagnant. The stagnancy indirectly indicates that the hydrolysis process was completed. In addition, observation done by Jutamongkon and Charoenrein (2010) revealed that prolonged heat exposure resulted in instability and eventual loss of function in bromelain. Thus, a longer extraction period might have broken the bromelain and rendered it nonfunctional, triggering the halt of the reaction.
The collagen level throughout hydrolysis time shows a similar pattern of reaching its peak after 45–60 minutes of hydrolysis (Fig. A1b). However, the collagen content is higher with a hydrolysis temperature of 70°C. This discrepancy can be attributed back to the nature of the collagen assay that relies on HYP count. Higher heat may destroy the fish protein, resulting in lower protein content, but it will increase the number of HYP detected in the collagen test. Moreover, research on bromelain tolerance to high temperature revealed that the threshold is around 65°C, after which the activity would promptly decline (de Lencastre Novaes et al., 2016). If the temperature of 70°C was applied, that extraction and hydrolysis of fish protein might not be effective as bromelain from the pineapple stem might be broken. The optimized condition for hydrolysis with bromelain is therefore deemed satisfactory—hydrolysis temperature and time of 60°C for 60 minutes. This result is consistent with previous findings regarding the conditions for optimum activity of bromelain (Arshad et al., 2014; Bala et al., 2012).
A.2 Protease Activity of Bromelain from Pineapple Stem Solution
Protein profiling uncovers a stark difference between the hydrolysates obtained without and with a protease, in which there is still a lot of protein sized above 26.2 kDa in the hydrolysate without the protease (data not shown). The Lowry protein measurement backs this notion, displaying that the hydrolysate obtained without bromelain has significantly lower protein content (Fig. A2a). Thus, the bromelain obtained from the pineapple stem successfully hydrolyzes the protein in tilapia skin.
Generally, when the substrate quantity is kept constant, the rate of reaction, and eventually the reaction yield, substantially increases as the amount of enzyme increases. The velocity shows a linear relationship with the concentration of enzymes used for the reaction. However, maximum velocity will be reached after a certain point where the addition of more enzymes has no effect as there is no more substrate available to break down (Robinson, 2015). Based on the Lowry protein measurement (Fig. A2a), the maximum yield is achieved with the bromelain solution with a protease activity of 60 U/ml. This data is supported by the collagen measurement, in which the amount of collagen obtained with 60 U/ml of bromelain was second only to the hydrolysate obtained with 15 U/ml of bromelain (Fig. A2b). Thus, it is confirmed that the optimum bromelain activity for hydrolysis is 60 U/ml.
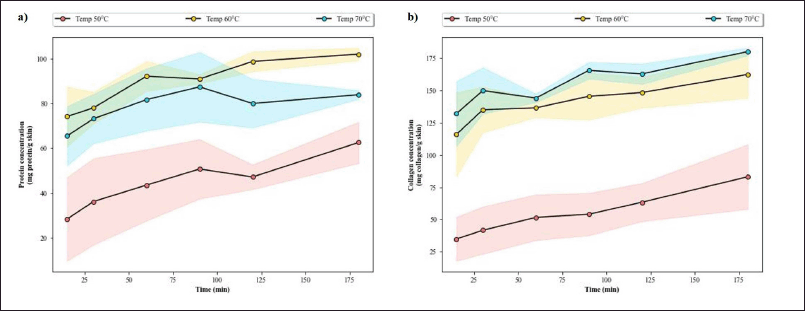 | Figure A1. Assessment of hydrolysates obtained from various hydrolysis temperatures and times. (a) The protein yield (mg protein/g skin) along with (b) the collagen yield (mg collagen/g skin) according to Lowry and HYP method, respectively. The SD of the data was shown as shaded areas across the line. [Click here to view] |
A.3 Solid-to-Liquid Ratio
As the reaction mixture became more saturated with the substrate, the concentration of protein and collagen in the solution increased yet their absolute yields decreased (Fig. A3a and b). This is to be expected as there would be less room for the interaction of the enzyme solution and the substrate with a higher STRratio (Jafari et al., 2020). Yet, it is also worth noting that the hydrolysate more saturated with the substrate (i.e., 1:3 ratio) still produced a similar protein profile to the less saturated hydrolysate. This means that, in this case, high saturation of the substrate only alters the quantity of the protein and collagen extracted and not the end product of the hydrolysis (i.e., hydrolyzed peptides).
On the other hand, if the lower STR ratios were implemented, the low protein and collagen concentration would be of concern. A stronger evaporation step would have to be implemented after hydrolysis to compensate for this dilution effect. This might destroy the protein and collagen; hence, the 1:5 ratio which offered balanced concentration and yield was preferred. This ratio yields significantly more protein and collagen than the larger ratios without the need for more extreme evaporation.
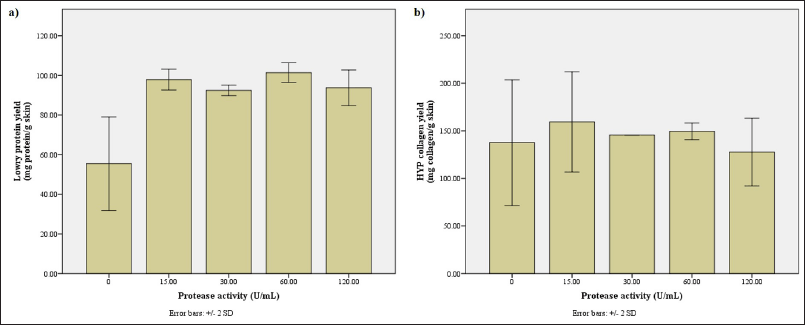 | Figure A2. Assessment of hydrolysates obtained with bromelain solution of various protease activities. (a) The protein yield (mg protein/g skin) along with (b) the collagen yield (mg collagen/g skin) according to Lowry and HYP method, respectively. [Click here to view] |
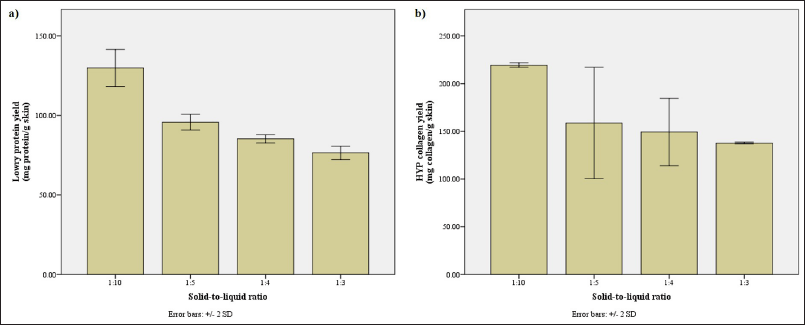 | Figure A3. Assessment of hydrolysates obtained with various STR ratios. (a) The protein yield (mg protein/g skin) along with (b) the collagen yield (mg collagen/g skin) according to Lowry and HYP method, respectively. [Click here to view] |