INTRODUCTION
One of the most common health problems is usually associated with wound infection (Giacometti et al., 2000), commonly induced by the entry of the bacteria through contamination of breached skin. With the same importance as renal and cardiovascular disorders, wounds can also have a similar impact on the quality of life. The Global Wound Care Market report indicated that this sector has reached about 20 billion dollars and is estimated to exceed 25 billion dollars in 2023 worldwide (Weller et al., 2020). For this reason, and the pressure induced on both patients and the medical system, efforts have been made with the aim of managing wound infections and their associated pathological conditions, which in turn could improve the quality of life and increase life expectancy. In recent decades, different approaches have been developed to improve the rate and quality of chronic wound healing (Nosrati et al., 2021). Despite the multiple etiologies of wound infections, bacterial colonization usually characterizes their chronicity (Loesche et al., 2017). Multiple variable organisms concerning wound infection were documented in previous studies (Melling et al., 2001). Most wound culture isolation procedures implement swab cultures, the easiest trauma-free way, which avoid hazards of complications on the wound bed (Meyers, 2008). A variety of microbes such as bacteria, fungi, and parasites are associated with wound infection (Bowler et al., 2001). Wound microbiome composition could also be influenced by ecological processes, which in turn induces the emergence of polymicrobial infections exhibiting both synergistic effect and enhanced tolerance to antimicrobials (Baishya and Wakeman, 2019; Kalan and Brennan, 2019). In addition, stable microbial communities are usually correlated with delayed healing (Loesche et al., 2017; Sloan et al., 2019). Moreover, genotypic effects could also be the possible way to explain the recently observed patient-specific immunological responses to the same microbial exposure (Deusenbery et al., 2019). Both Gram-positive and Gram-negative bacteria are usually detected including Escherichia coli, Proteus spp., and Klebsiella spp., in addition to Enterobacter spp. and Staphylococcus aureus, which was also identified (El-Saed et al., 2020; Gupta et al., 2019). However, a challenging bacterium, Pseudomonas aeruginosa, exhibiting resistance to most antibacterial drugs, could also be detected (Bowler et al., 2001). In addition, the presence of anaerobic bacteria at baseline infection could also be associated with worse healing outcomes (Min et al., 2020). In contrast to the intestinal microbiota, normal skin is mainly colonized by only a few taxa including Corynebacterium, coagulase-negative Staphylococci, and Propionibacterium (Scharschmidt and Fischbach, 2013) in which sebaceous sites are dominated by lipophilic Propionibacterium species, while Corynebacterium and Staphylococci are preferentially abundant in moist areas (Costello et al., 2009; Grice et al., 2009; Oh et al., 2014). Wound infections are classified into two major classes, skin and soft tissue infections, which are usually undistinguished upon the progression of infection. In addition, in these infections when untreated, infecting bacteria affect healing and may also produce other signs and symptoms. Moreover, wound infections are considered among the most leading nosocomial infections and are associated with increased morbidity and other medical expenses (Cutting and White, 2004). For assessing and diagnosing infection, the most common signs usually include redness, swelling, increased drainage, and increased pain. Therefore, microbial etiology is one of the fundamental steps required before starting the treatment protocols. Most microbial identification is usually based on the bacterial separation by an enrichment culture, which favors the selection of some microbes, while excluding other species. For this reason, bacterial identification is now shifting away from metabolic biochemical testing toward genetic or molecular identification (Espy et al., 2006; Han et al., 2011; Seng et al., 2010). Molecular technologies have provided worldwide researchers with more rapid and sensitive tools to examine human microbiota, in comparison with the old culture-based testing (Bowler and Davies, 1999; Dowd et al., 2008a). In most studies, universal primers for 16S rRNA genes are used, followed by specific identification approaches such as polymerase chain reaction (PCR) (Hill et al., 2003) and denaturing gradient gel electrophoresis and sequencing (Dowd et al., 2008a, 2008b; Hill et al., 2003; Price et al., 2009). In addition, these methods exhibited successfulness in the detection of bacteria in burn wounds (Pirnay et al., 2000), blood (Rothman et al., 2002; Wellinghausen et al., 2004; Yang et al., 2002), cerebrospinal fluid (Poppert et al., 2005), joint fluid (Yang et al., 2008), and heart tissue (Breitkopf et al., 2005). Moreover, in most cases, each experiment can be executed in not more than a few hours (Ecker et al., 2010). In the same respect, for developing new therapeutic strategies, the use of these techniques speeds up the early detection of infecting microorganisms assisting clinicians and pathologists to find out the appropriate treatment of hospitalized patients.
For the detection of pathogens, DNA probes are immobilized in microarray experiments, in which more than one copy of each target gene could be included (Cannon et al., 2010; Shen et al., 2015). However, the detection of wound-associated bacterial pathogens was rarely described in previous studies. The aim of this study is the detection of wound pathogens in mixed samples and their application in samples from Egyptian hospitals.
MATERIALS AND METHODS
Specimens collection and processing
Hospitalized immunocompetent symptomatic patients, almost all adults suffering from wound infection, were included in this study. Control samples were taken from nonhospitalized individuals and those free from any symptoms of wound infection. Samples were collected from 20 different patients admitted to one of the Mansoura hospitals in Egypt. This study was approved and funded by the Competitive Funding Projects Postgraduate Research and Cultural Affairs Sector, Mansoura University. The experimental protocol conducted in this study was approved by the Research Ethics Committee, Faculty of Pharmacy, Mansoura University, and conducted in accordance with the Declaration of Helsinki involving the use and handling of human subjects.
Extraction of genomic DNA and DNA from Standard Cultures
Different bacterial cultures obtained from wound samples were prepared in LB broth, followed by incubation aerobically at 37°C and observation for the presence of visible microbial growth. Genomic DNAs were extracted from 37°C 24 hours cultures of wound samples using the genomic QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. By using a Nanodrop instrument (OPTIZEN NanoQ, Mecasys, Daejon, South Korea), the concentration of gDNA was determined. Purified DNAs were stored at −80°C.
PCR Amplification and Labeling to Get Hybridization Targets
By starting from isolated gDNA as a template, nearly full-length 16S rRNA was amplified in a PCR reaction. Two different biotin-labeled primers were used: forward primer 27F (5′-AGAGTTTGATCCTGGCTCAG) and reverse primer 1492R (5′-GGTTACCTTGTTACGACTT). Each PCR reaction is composed of 25 μl containing 2 μl of template DNA, 0.5 μM of each primer, 1.5 mM MgCl2, 0.2 mM dNTPs, 1 U Taq polymerase (Dream Taq Green DNA Polymerase, Fermentas), and to the final volume nuclease-free water. PCR reactions were carried out using Cycler 003 PCR Machine [A & E Lab (UK)] with 2 minutes initial denaturation at 94°C, followed by 35 cycles of heating at 94°C for 30 seconds, cooling to 52°C for 30 seconds in an annealing step, and heating at 72°C for 30 seconds in an extension step, and a final extension at 72°C for 10 minutes. The 1% agarose gel and electrophoresis-separated PCR products were visualized by staining with ethidium bromide and UV light exposure.
Strain-Specific Genes PCR Test
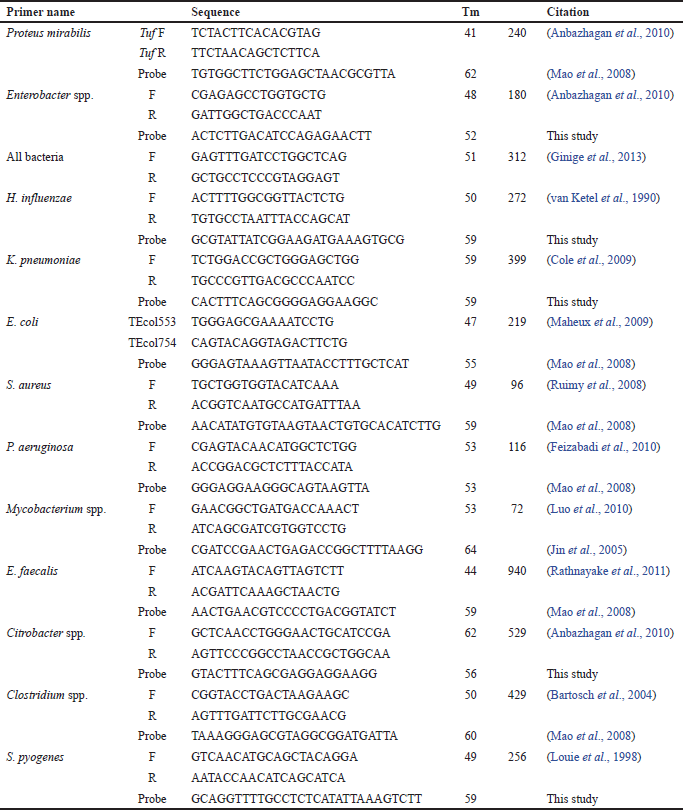
As described previously, amplification of both genomic DNA and standard DNA was carried out using primers (Biosearch Technologies) listed in Table 1. The diagnostic PCR screening consists of a panel of 12 taxa including S. aureus, Staphylococcus pyogenes, Mycobacterium spp., P. aeruginosa, Enterobacter spp., Klebsiella pneumoniae, Haemophilus influenzae, E. coli, Enterococcus faecalis, Citrobacter spp., Proteus spp., and Clostridium spp. PCR reactions began with heating at 94°C for 5 minutes, followed by 35 cycles starting with heating at 94°C for 30 seconds, then cooling to annealing temp (as listed in Table 1) for 30 seconds, and heating at 72°C for 30. Finally, each PCR reaction was ended with a final extension step at 72°C for 10 minutes.
| Table 1. Different oligonucleotides used in this work to detect different species of bacteria. |
Oligonucleotide Probes Design and synthesis
Segments of 20–30 mer oligonucleotides with similar lengths, melting, and GC content with the optimum BLAST hit were selected after comparison (http://www.ncbi.nlm.nih.gov/BLAST/) with all available sequences in the GenBank database. All oligonucleotides used in this study were listed in Table 1.
Oligo Aarray Printing
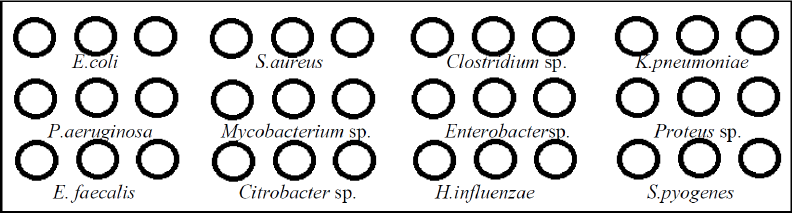
In a 384-well printing plate, using ArrayIt spotting buffer, each printing oligo solution was prepared to a final concentration of 50 pmol μl−1. Spotting was carried out onto microarray slides in triplicate for each probe (Scienion, Berlin, Germany) using the SpotBot microarrayer (ArrayIt, Sunnyvale, CA) spotting machine (Fig. 1).
| Figure 1. Different ssDNA probes spotted in the schematic representation of each microarray slide. |
Microarray Testing
Samples were labeled and then resuspended in a hybridization buffer (Scienion, Berlin, Germany). The method was carried out according to the recommendations of the manufacturer (Scienion, Berlin, Germany). In a final volume of 16, 8 μl of the PCR products was mixed with 8 μl of the hybridization buffer. The prehybridization step was carried out using 20 μl of the prehybridization solution (Scienion, Berlin, Germany). Labeled samples were then applied to a 42°C prewarmed microarray slide. Hybridizations were carried out in a hybridization station (ArrayIt, Sunnyvale, CA) using the hybridization mix, previously boiled for 2 minutes and cooled on ice. Hybridization was carried out by incubation at 42°C for 12 hours. Finally, at room temperature, using buffers I, II, and III (Scienion, Berlin, Germany), washing was carried out. Microarray staining was carried out by a streptavidin biotin color development system (Fermentas, Waltham, MA) using the manufacturer’s instructions. For documentation of the results, images were acquired using the ArrayIt Microarray Scanner (ArrayIt, Sunnyvale, CA). Signal intensities were recorded using the Spotware software after subtracting the local background values from the per-sample median.
RESULTS
Results of bacterial identification
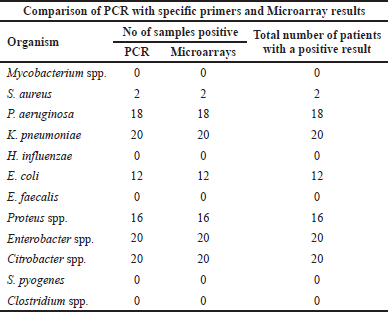
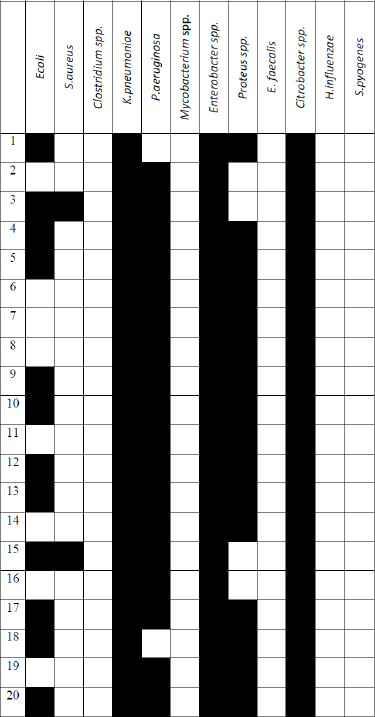
Twenty wound samples were analyzed by molecular testing. Overall, the most common organisms identified using molecular PCR testing were Citrobacter spp. (100%) in 20 samples, Enterobacter spp. (100%) in 20 samples, and K. pneumoniae (100%) in 20 samples, followed by P. aeruginosa (90%) in 18 samples, Proteus spp. in 16 samples (80%), and E. coli (60%) in 12 samples. However, S. aureus was rarely identified in two samples (10%). Other bacterial taxa such as E. faecalis, H. influenzae, and Mycobacterium spp. could not be detected in any isolate. Control samples were free from the tested pathogens. Concerning a mixed microbial infection, a total of eight wounds were found to have five different bacterial taxa; in addition, 10 of the total samples (50%) had six bacterial taxa. However, two of the total wounds (10%) had only four bacterial taxa (Table 2).
| Table 2. Comparison between PCR and microarray results. |
Microarray Testing of clinical samples
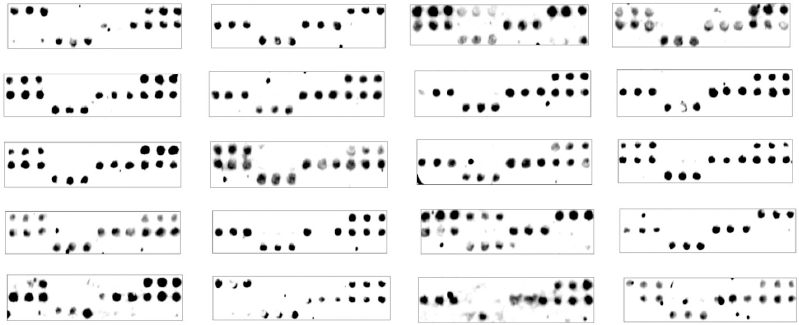
In this study, by DNA microarray testing, the proportions of bacteria identified from the 20 positive samples were Citrobacter spp. (100%), Enterobacter spp. (100%), and K. pneumoniae (100%), followed by P. aeruginosa (90%), Proteus (80%), and E. coli (60%). However, S. aureus was rarely identified (10%). A mixed bacterial infection could be identified in all different samples as four, five, or six different bacteria were identified in each sample as shown in Figure 2. No difference could be observed when compared to the results obtained by PCR (Table 3).
DISCUSSION
Microbial molecular diagnostic tests, in comparison with traditional culture methods, enable more patient care improvement. However, it is also necessary to continue to include all these introduced newer methods. In addition, it has been proven that culture-based methodologies in bacterial detection are markedly less sensitive to DNA detection methodologies (Han et al., 2011). Moreover, the results obtained using these traditional culture-based methods are usually associated with misidentification reflecting overestimation of the relative presence of easily cultured and identifiable microbes (Davies et al., 2004).
| Figure 2. Results of hybridization with the target wound sample DNAs from Mansoura Hospital. |
According to the recent reports, the results obtained using PCR and 16S sequencing commonly used have demonstrated the close correlation between most of these molecular methods. As previously documented, different molecular methods such as real-time, uniplex, and multiplex PCR, especially in mixed samples, have enhanced both the speed and the sensitivity of microbial detection (Gebert et al., 2008; Lehmann et al., 2008). However, multiplex PCR, in each reaction, can only detect a limited number of pathogens (Edin et al., 2015; Gadsby et al., 2015). For these reasons, when dealing with samples containing a mixed culture or mostly environmental samples, a comprehensive view of microbial communities is required. In the same respect, a better solution is provided by DNA microarrays, especially in mixed microbial infections.
Microarray testing is usually based on the panel of taxa selection, detection capacity, and probe coverage, which may differ between sequence targets. Microarray-detecting oligonucleotides are mainly designed based on the available sequence data and the variation between target organisms exhibiting reduced power for identifying other different organisms (Cannon et al., 2010; Shen et al., 2015).
As previously reported (Costello et al., 2009; Grice and Segre, 2011; Grice et al., 2009; Oh et al., 2014), in sequencing surveys of healthy adults, dry, moist, and sebaceous microenvironmental changes usually affect the relative abundance of bacterial taxa. However, a lower bacterial diversity is usually detected (Scharschmidt and Fischbach, 2013), as the normal skin is usually dominated by only a few taxa, mainly lipophilic Propionibacterium species of the phylum Actinobacteria thriving sebaceous sites, while Staphylococcus and Corynebacterium species of the phyla Firmicutes and Actinobacteria are preferentially abundant in moist areas (Costello et al., 2009; Grice et al., 2009). Interestingly, some members of the skin microbiota such as Staphylococcus epidermidis are nonpathogenic; however, when reaching the blood circulation, serious systemic diseases in some individuals can be caused (Blum and Rodvold, 1987). In this study, to distinguish some major bacterial species, frequently causing wound-associated infections including E. faecalis, Enterobacter spp., K. pneumoniae, H. influenzae, E. coli, S. pyogenes, Clostridium spp., S. aureus, Mycobacterium spp., Citrobacter spp., P. aeruginosa, and Proteus spp., 12 oligonucleotide probes, complementary to 16S PCR products, were selected for detection based on several species-specific regions of each pathogen.
In samples obtained in the current study, by DNA microarray testing, all samples (100%) were found to be positive and identified mainly as Gram-negative bacteria, which is consistent with some previous studies in Saudi Arabia (Alkaaki et al., 2019; Al-Mulhim et al., 2014) and other countries (Allegranzi et al., 2011). The percentages of identified bacteria were Citrobacter spp. (100%), Enterobacter spp., (100%), and K. pneumoniae (100%), followed by P. aeruginosa (90%), Proteus (80%), and E. coli (60%). Similarly, in a previous study, the same bacterial taxa were detected, but at lower percentages, 56.7%, 6.7%, and 3.3% for E. coli, Klebsiella spp., and Enterobacter spp., respectively (Adwan et al., 2016). In addition, an increased level of Pseudomonas species (43%) was detected in a previous study (Gupta et al., 2019), compared to K. pneumoniae and Acinetobacter baumannii, which were detected at the lower prevalence of 28% and 14.83%, respectively. Moreover, other studies in a tertiary care hospital in India reported the predominance of P. aeruginosa, followed by K. pneumoniae and A. baumannii (Dash et al., 2013; Singh et al., 2003). Interestingly, the wound microbiome, in which Pseudomonas dominates, exhibited increased temporal stability against multiantibiotic therapy in comparison to staphylococcal infections (Tipton et al., 2017). However, other different results were obtained in another study on surgical site infections (Negi et al., 2015), as the commonest organism identified was S. aureus (50.4%), followed by E. coli (23.02%), P. aeruginosa (7.9%), and Citrobacter spp. (7.9%). In addition, in another study (Be et al., 2014), in wound samples, A. baumannii was mainly detected (23%) in 28 samples. In this current study, S. aureus was rarely identified in 10% of total isolates. A similar observation was previously reported, as S. aureus was rarely detected (two wound samples) in one study using a microarray (Be et al., 2014) and in another study on combat wound colonization (Brown et al., 2011). On the contrary, a higher prevalence of S. aureus (30%) could be detected in one previous study (Adwan et al., 2016). In addition, in another study, the most commonly isolated organism was S. aureus, followed by coagulase-negative Staphylococci (CNS), Enterococcus spp., and E. coli, while the pathogen detection was mainly dependent on the surgical procedure (Owens and Stoessel, 2008). Moreover, in another study, S. aureus was the most frequent pathogen detected (22.8%), while other pathogens, namely, P. aeruginosa (20.1%), E. coli (12.2%), Klebsiella spp., (12.2%), Enterobacter spp., (7.7%), and Enterococcus spp. (5.9%), were identified at lower percentages (El-Saed et al., 2020).
| Table 3. Schematic representation of hybridization results obtained with the target DNAs from wound samples obtained from Mansoura Hospital. |
Interestingly, in previous studies, not so much data were found concerning the detection of mixed wound infection. In this current study, four, five, or six different bacteria were identified in each sample, representing mixed microbial infections. K. pneumoniae, P. aeruginosa, Enterobacter spp., Proteus spp., and Citrobacter spp. were the most identified mixed infections.
CONCLUSION
In the present study, by PCR detection, Citrobacter spp., Enterobacter spp., K. pneumoniae, P. aeruginosa, Proteus spp., and E. coli were mainly detected. In addition, pathogen detection with both PCR and the DNA chip indicated 100% matching. Moreover, the use of a DNA chip in a mixed culture facilitates the fast identification of wound-infecting pathogens and the proper treatments strategies.
CONFLICTS OF INTEREST
The author declares that there are no conflicts of interest regarding the publication of this paper.
FUNDING
This study was funded by the Competitive Funding Projects Postgraduate Research and Cultural Affairs Sector, Mansoura University.
There were no financial interests related to the material in the manuscript.
REFERENCES
Adwan G, Hasan NA, Sabra I, Sabra D, Al-butmah S, Odeh S, Abd Albake Z, Badran H. Detection of bacterial pathogens in surgical site infections and their antibiotic sensitivity profile. Int J Med Res Health Sci, 2016; 5(5):75–82.
Alkaaki A, Al-Radi OO, Khoja A, Alnawawi A, Maghrabi A, Altaf A, Aljiffry M. Surgical site infection following abdominal surgery: a prospective cohort study. Can J Surg, 2019;62(2):111–7. CrossRef
Allegranzi B, Bagheri Nejad S, Combescure C, Graafmans W, Attar H, Donaldson L, Pittet D. Burden of endemic health-care-associated infection in developing countries: systematic review and meta-analysis. Lancet, 2011; 377(9761):228–41. CrossRef
Al-Mulhim F, Baragbah M, Sadat-Ali M, Alomran A, Azam M. Prevalence of surgical site infection in orthopedic surgery: a 5-year analysis. Int Surg, 2014; 99(3):264–8. CrossRef
Anbazhagan D, Kathirvalu GG, Mansor M, Yan GOS, Yasim M, Devi Sekaran S. Multiplex polymerase chain reaction (PCR) assays for the detection of Enterobacteriaceae in clinical samples. Afr J Microbiol Res, 2010; 4(11):1186–91.
Baishya J, Wakeman C. Selective pressures during chronic infection drive microbial competition and cooperation. NPJ Biofilms Microbiomes, 2019; 5:16. CrossRef
Bartosch S, Fite A, Macfarlane G, McMurdo M. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Microbiol, 2004; 70:3575–81. CrossRef
Be NA, Allen JE, Brown TS, Gardner SN, McLoughlin KS, Forsberg JA, Kirkup BC, Chromy BA, Luciw PA, Elster EA, Jaing CJ. Microbial profiling of combat wound infection through detection microarray and next-generation sequencing. J Clin Microbiol, 2014; 52(7):2583–94. CrossRef
Blum R, Rodvold K. Recognition and importance of Staphylococcus epidermidis infections. Clin Pharm, 1987; 6:464–75.
Bowler PG, Davies BJ. The microbiology of infected and noninfected leg ulcers. Int J Dermatol, 1999; 38(8):573–8. CrossRef
Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev, 2001; 14(2):244–69. CrossRef
Breitkopf C, Hammel D, Scheld HH, Peters G, Becker K. Impact of a molecular approach to improve the microbiological diagnosis of infective heart valve endocarditis. Circulation, 2005; 111(11):1415–21. CrossRef
Brown T, Hawksworth J, Sheppard F, Tadaki D, Elster E. Inflammatory response is associated with critical colonization in combat wounds. Surg Infect (Larchmt), 2011; 12:351–7. CrossRef
Cannon GA, Carr MJ, Yandle Z, Schaffer K, Kidney R, Hosny G, Doyle A, Ryan J, Gunson R, Collins T, Carman WF, Connell J, Hall WW. A low density oligonucleotide microarray for the detection of viral and atypical bacterial respiratory pathogens. J Virol Methods, 2010; 163(1):17–24. CrossRef
Cole JM, Schuetz AN, Hill CE, Nolte, FS. Development and evaluation of a real-time PCR assay for detection of K. pneumoniae carbapenemase genes. J Clin Microbiol, 2009; 47(2):322–6. CrossRef
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science, 2009; 326(5960):1694–7. CrossRef
Cutting KF, White R. Defined and refined: criteria for identifying wound infection revisited. Br J Community Nurs, 2004; 9(3):S6–15. CrossRef
Dash M, Mishra P, Routray S. Bacteriological profile and antibiogram of aerobic burn wound isolates in a tertiary care hospital, Odisha, India. Int J Med Med Sci, 2013; 3.
Davies C, Hill K, Wilson M, Stephens P, Hill C, Harding K, Thomas D. Use of 16S ribosomal DNA PCR and denaturing gradient gel electrophoresis for analysis of the microfloras of healing and nonhealing chronic venous leg ulcers. J Clin Microbiol, 2004; 42:3549–57. CrossRef
Deusenbery C, Kalan L, Meisel J, Gardner S, Grice E, Spiller K. Human macrophage response to microbial supernatants from diabetic foot ulcers. Wound Repair Regen, 2019; 27(6):598–608. CrossRef
Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol, 2008b; 8:43. CrossRef
Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One, 2008a; 3(10):e3326. CrossRef
Ecker DJ, Sampath R, Li H, Massire C, Matthews HE, Toleno D. New technology for rapid molecular diagnosis of bloodstream infections. Expert Rev Mol Diagn, 2010; 10(4):399–415. CrossRef
Edin A, Granholm S, Koskiniemi S, Allard A, Sjostedt A, Johansson A. Development and laboratory evaluation of a real-time PCR assay for detecting viruses and bacteria of relevance for community-acquired pneumonia. J Mol Diagn, 2015; 17(3):315–24. CrossRef
El-Saed A, Balkhy HH, Alshamrani MM, Aljohani S, Alsaedi A, Al Nasser W, El Gammal A, Almohrij SA, Alyousef Z, Almunif S, Alzahrani M. High contribution and impact of resistant gram negative pathogens causing surgical site infections at a multi-hospital healthcare system in Saudi Arabia, 2007–2016. BMC Infect Dis, 2020; 20(1):275. CrossRef
Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JD, Wengenack NL, Rosenblatt JE, Cockerill FR, Smith, TF. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev, 2006; 19(1):165–256. CrossRef
Feizabadi MM, Majnooni A, Nomanpour B, Fatolahzadeh B, Raji N, Delfani S, Habibi M, Asadi S, Parvin M. Direct detection of Pseudomonas aeruginosa from patients with healthcare associated pneumonia by real time PCR. Infect Genet Evol, 2010; 10(8):1247–51. CrossRef
Gadsby NJ, McHugh MP, Russell CD, Mark H, Conway Morris A, Laurenson IF, Hill AT, Templeton KE. Development of two real-time multiplex PCR assays for the detection and quantification of eight key bacterial pathogens in lower respiratory tract infections. Clin Microbiol Infect, 2015; 21(8):788.e1–13. CrossRef
Gebert S, Siegel D, Wellinghausen N. Rapid detection of pathogens in blood culture bottles by real-time PCR in conjunction with the pre-analytic tool MolYsis. J Infect, 2008; 57(4):307–16. CrossRef
Giacometti A, Cirioni O, Schimizzi AM, Del Prete MS, Barchiesi F, D’Errico MM, Petrelli E, Scalise G. Epidemiology and microbiology of surgical wound infections. J Clin Microbiol, 2000; 38(2):918–22. CrossRef
Ginige MP, Kaksonen, AH, Morris C, Shackelton M, Patterson BM. Bacterial community and groundwater quality changes in an anaerobic aquifer during groundwater recharge with aerobic recycled water. FEMS Microbiol Ecol, 2013; 85(3):553–67. CrossRef
Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. Topographical and temporal diversity of the human skin microbiome. Science, 2009; 324(5931):1190–2. CrossRef
Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol, 2011; 9:244–53. CrossRef
Gupta M, Naik AK, Singh SK. Bacteriological profile and antimicrobial resistance patterns of burn wound infections in a tertiary care hospital. Heliyon, 2019; 5(12):e02956. CrossRef
Han A, Zenilman JM, Melendez JH, Shirtliff ME, Agostinho A, James G, Stewart PS, Mongodin EF, Rao D, Rickard AH, Lazarus GS. The importance of a multifaceted approach to characterizing the microbial flora of chronic wounds. Wound Repair Regen 2011; 19(5):532–41. CrossRef
Hill KE, Davies CE, Wilson MJ, Stephens P, Harding KG, Thomas DW. Molecular analysis of the microflora in chronic venous leg ulceration. J Med Microbiol, 2003; 52(Pt 4):365–9. CrossRef
Jin LQ, Li JW, Wang SQ, Chao FH, Wang XW, Yuan ZQ. Detection and identification of intestinal pathogenic bacteria by hybridization to oligonucleotide microarrays. World J Gastroenterol, 2005; 11(48):7615–9. CrossRef
Kalan L, Brennan M. The role of the microbiome in nonhealing diabetic wounds. Ann N Y Acad Sci, 2019; 1435(1):79–92. CrossRef
Lehmann LE, Hunfeld KP, Emrich T, Haberhausen G, Wissing H, Hoeft A, Stuber F. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med Microbiol Immunol, 2008; 197(3):313–24. CrossRef
Loesche M, Gardner S, Kalan L, Horwinski J, Zheng Q, Hodkinson BP, Tyldsley AS, Franciscus CL, Hillis SL, Mehta S, Margolis DJ, Grice EA. Temporal stability in chronic wound microbiota is associated with poor healing. J Invest Dermatol, 2017; 137(1):237–44. CrossRef
Louie L, Simor AE, Louie M, McGeer A, Low DE. Diagnosis of group A streptococcal necrotizing fasciitis by using PCR to amplify the streptococcal pyrogenic exotoxin B gene. J Clin Microbiol, 1998; 36:1769–71. CrossRef
Luo RF, Scahill MD, Banaei N. Comparison of single-copy and multicopy real-time PCR targets for detection of Mycobacterium tuberculosis in paraffin-embedded tissue. J Clin Microbiol, 2010; 48(7):2569–70. CrossRef
Maheux AF, Picard FJ, Boissinot M, Bissonnette L, Paradis S, Bergeron MG. Analytical comparison of nine PCR primer sets designed to detect the presence of Escherichia coli/Shigella in water samples. Water Res, 2009; 43(12):3019–28. CrossRef
Mao Z, Zheng H, Wang X, Lin S, Sun Y, Jiang B. DNA microarray for direct identification of bacterial pathogens in human stool samples. Digestion, 2008; 78(2–3):131–8. CrossRef
Melling AC, Ali B, Scott EM, Leaper DJ. Effects of preoperative warming on the incidence of wound infection after clean surgery: a randomised controlled trial. Lancet, 2001; 358(9285):876–80. CrossRef
Meyers B. Wound management: principles and practice: principles and practice. 2nd edition, Pearson Prentice Hall. Upper Saddle River, NJ, pp 100–102, 2008.
Min KR, Galvis A, Nole KLB, Sinha R, Clarke J, Kirsner RS, Ajdic D. Association between baseline abundance of Peptoniphilus, a Gram-positive anaerobic coccus, and wound healing outcomes of DFUs. Plos One, 2020; 15(1):e0227006. CrossRef
Negi V, Pal S, Juyal D, Sharma MK, Sharma N. Bacteriological profile of surgical site infections and their antibiogram: a study from resource constrained rural setting of Uttarakhand State, India. J Clin Diagn Res, 2015; 9(10):DC17–20. CrossRef
Nosrati H, Khouy RA, Nosrati A, Khodaei M, Banitalebi-Dehkordi M, Ashrafi-Dehkordi K, Sanami S, Alizadeh Z. Nanocomposite scaffolds for acceleratingchronic wound healing by enhancing Angiogenesis. J Nanobiotechnol, 2021; 19:1. CrossRef
Oh J, Byrd AL, Deming C, Conlan S, Kong, HH, Segre JA. Biogeography and individuality shape function in the human skin metagenome. Nature, 2014; 514(7520):59–64. CrossRef
Owens CD, Stoessel K. Surgical site infections: epidemiology, microbiology and prevention. J Hosp Infect, 2008; 70(Suppl 2):3–10. CrossRef
Pirnay JP, De Vos D, Duinslaeger L, Reper P, Vandenvelde C, Cornelis P, Vanderkelen A. Quantitation of Pseudomonas aeruginosa in wound biopsy samples: from bacterial culture to rapid “real-time” polymerase chain reaction. Crit Care, 2000; 4(4):255–61. CrossRef
Poppert S, Essig A, Stoehr B, Steingruber A, Wirths B, Juretschko S, Reischl U, Wellinghausen, N. Rapid diagnosis of bacterial meningitis by real-time PCR and fluorescence in situ hybridization. J Clin Microbiol, 2005; 43(7):3390–7. CrossRef
Price LB, Liu CM, Melendez JH, Frankel YM, Engelthaler D, Aziz M, Bowers J, Rattray R, Ravel J, Kingsley C, Keim PS, Lazarus GS, Zenilman JM. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One, 2009; 4(7):e6462. CrossRef
Rathnayake IU, Hargreaves M, Huygens F. Genotyping of Enterococcus faecalis and Enterococcus faecium isolates by use of a set of eight single nucleotide polymorphisms. J Clin Microbiol, 2011; 49:367–72. CrossRef
Rothman RE, Majmudar MD, Kelen GD, Madico G, Gaydos CA, Walker T, Quinn TC. Detection of bacteremia in emergency department patients at risk for infective endocarditis using universal 16S rRNA primers in a decontaminated polymerase chain reaction assay. J Infect Dis, 2002; 186(11):1677–81. CrossRef
Ruimy R, Dos-Santos M, Raskine L, Bert F, Masson R, Elbaz S, Bonnal C, Lucet JC, Lefort A, Fantin B, Wolff M, Hornstein M, Andremont A. Accuracy and potential usefulness of triplex real-time PCR for improving antibiotic treatment of patients with blood cultures showing clustered gram-positive cocci on direct smears. J Clin Microbiol, 2008; 46(6):2045–51. CrossRef
Scharschmidt T, Fischbach M. What lives on our skin: ecology, genomics and therapeutic opportunities of the skin microbiome. Drug Discov Today Dis Mech, 2013; 10:3–4. CrossRef
Seng P, Rolain JM, Fournier PE, La Scola B, Drancourt M, Raoult D. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol, 2010; 5(11):1733–54. CrossRef
Shen H, Zhu B, Wang S, Mo H, Wang J, Li J, Zhang C, Zeng H, Guan L, Shi W, Zhang Y, Ma X. Association of targeted multiplex PCR with resequencing microarray for the detection of multiple respiratory pathogens. Front Microbiol, 2015; 6:532. CrossRef
Singh NP, Goyal R, Manchanda V, Das S, Kaur I, Talwar V. Changing trends in bacteriology of burns in the burns unit, Delhi, India. Burns, 2003; 29(2):129–32. CrossRef
Sloan TJ, Turton JC, Tyson J, Musgrove A, Fleming VM, Lister MM, Loose MW, Sockett RE, Diggle M, Game FL, Jeffcoate W. Examining diabetic heelulcers through an ecological lens: microbial community dynamics associated with healing and infection. J Med Microbiol, 2019; 68(2):230–40. CrossRef
Tipton C, Mathew M, Wolcott R, Wolcott R, Kingston T, Phillips C. Temporal dynamics of relativeabundances and bacterial succession in chronic wound communities. Wound Repair Regen, 2017; 25(4):673–9. CrossRef
van Ketel RJ, de Wever B, van Alphen L. Detection of Haemophilus influenzae in cerebrospinal fluids by polymerase chain reaction DNA amplification. J Med Microbiol, 1990; 33(4):271–6. CrossRef
Weller C, Team V, Sussman G. First-line interactive wound dressingupdate: a comprehensive review of the evidence. Front Pharmacol, 2020; 11:155. CrossRef
Wellinghausen N, Wirths B, Franz AR, Karolyi L, Marre R, Reischl U. Algorithm for the identification of bacterial pathogens in positive blood cultures by real-time LightCycler polymerase chain reaction (PCR) with sequence-specific probes. Diagn Microbiol Infect Dis, 2004; 48(4):229–41. CrossRef
Yang S, Lin S, Kelen GD, Quinn TC, Dick JD, Gaydos CA, Rothman RE. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. J Clin Microbiol, 2002; 40(9):3449–54. CrossRef
Yang S, Ramachandran P, Hardick A, Hsieh YH, Quianzon C, Kuroki M, Hardick J, Kecojevic A, Abeygunawardena A, Zenilman J, Melendez J, Doshi V, Gaydos C, Rothman RE. Rapid PCR-based diagnosis of septic arthritis by early Gram-type classification and pathogen identification. J Clin Microbiol. 2008; 46(4):1386–90. CrossRef