INTRODUCTION
Molecular modeling methods present useful tools in medicinal and biological research. Indeed, molecular modeling is very important and indispensable to understand the interaction between disease’s enzymes and inhibitors for the conception of new drugs; it permits to save time and financial spending. According to different research studies, natural molecules from thyme essential oil and flavonoids (Apigenine, Luteoline, Thymol, Carvacrol, Naringenine, and Chlorogenique) are extremely recommended to treat inflammation by inhibition responsible enzyme. Many inhibitors are used for Cyclooxygenase-2 (COX-2) inhibition but synthetic ones are the most used namely the non-steroidal anti-inflammatory drugs (NSAID) (Etoricoxib, Celecoxib, Ibuprofen, and Rofecoxib). Inflammation is a part of the complex biological response of body tissues to harmful stimuli, such as pathogens, damaged cells, or irritants. The function of inflammation is to eliminate the initial cause of cell injury, clear out necrotic cells and tissues damaged from the original insult and the inflammatory process, and to initiate tissue repair (Miliani, 2007). Thyme, known as a powerful antiseptic, is also an antibacterial, antiviral, anti-fungal, and major antiparasitic. As such, it is used against different infections, both of the otorhinolaryngology sphere and of the respiratory, genitourinary, and digestive systems. Thyme is also a major antiviral effective against herpes simplex. Several studies indicated that thyme can be useful for people suffering from inflammatory diseases (ID) (Kuete, 2017). Indeed, it has been proved that carvacrol, a component of thyme oil, activate PPARα and γ and suppresses COX-2 expression (Hotta et al., 2010). These results may be important in understanding the anti-inflammatory and anti-lifestyle-related disease properties of carvacrol. Also, it has been indicating that combined treatment with appropriate concentrations of thyme and oregano essential oils can reduce the production of pro-inflammatory cytokines, and thereby attenuate 2,4,6-trinitrobenzene sulfonic acid induced colitis in mice (Bukovska et al., 2007). In a study from Japan’s Nara Women’s University, researchers found that one of thyme oil’s constituents, carvacrol, actually inhibits the COX-2 enzyme part of the body’s inflammatory process that produces pain (Katsukawa et al., 2010). This strategy of inhibiting COX-2 has been utilized by pharmaceutical medications including NSAID. Until now, some of these NSAID and COX-inhibitor drugs come with side effects such as cardiovascular and digestive problems, which docile herbs like thyme don’t seem to come with (Salmalian et al., 2014). Actually, the comparisons between different ligands inhibition of the same enzyme can be done by means of molecular modeling; in fact, this technique is used widely in drug design. In this work, we aim to carry out a comparative study of the COX-2 inhibition between synthetic inhibitors namely (NSAID) and the natural inhibitors (thyme essential oil derivatives). In order to rationalize the properties of the inhibitors and to determine the reaction processes involving these compounds, we studied the interaction and binding of the complex formed with inhibitors of COX-2 (natural and synthetic), with better complementarities (better activity). Molecular modeling study is performed using molecular operating environment (MOE) software to advise among molecules contained in thyme (natural inhibitor) which is better for treatment of the Inflammation, and also what is the best synthetic inhibitor (NSAID) including solvatation parameter.
MATERIAL AND METHODS
Cyclooxygenase-2 enzyme
COX-2 is highly inducible in response to cellular activation by hormones, pro-inflammatory cytokines, growth factors, and tumor promoters. COX-2 activates procarcinogens, promotes angiogenesis, and indirectly increases free radical production (Picot and Loll, 1994).
Cyclooxygenase-2 synthetic and natural inhibitors
Some COX-2 inhibitors are used in a single dose to treat pain after surgery. COX-2 inhibitors have been found to be effective in suppressing inflammatory neurodegenerative pathways in mental illness, with beneficial results in trials for the major depressive disorder as well as schizophrenia (Hemler et al., 1976). Synthetic and natural inhibitors are reported in Tables 1 and 2.
Preparation and optimization of both enzyme and inhibitors
Download of COX-2 was done from PROTEIN DATA BANK (code 4PH9) with the three-dimensional structure obtained by X-ray diffraction (resolution 1.81 Å). We note that the COX-2 crystallizes as a monomer (Fig. 1) with residues and atoms. Compounds of inhibitors were downloaded from the PubChem database. Structures and CID code are reported in Tables 3 and 4. Using MOE software (MOE, 2013), we select the active site in the enzyme and we minimize the energy of both enzyme and molecules (a, b, and c). Energy minimizing was done under the following conditions: temperature = 300°K, pH = 7, the geometry was performed using the field strengths in the MMFF94x implanted in MOE and Hamiltonian AM1. Figure 2 shows the active site of the enzyme with a molecule of co-crystallization. Minimized energy of ligands and their toxicity are given in Table 5. Natural ligands present a very important biological activity in accordance with the Lipinski rule of 5 (Powers et al., 2006).
Docking and building complexes
After ligand building, we proceed to positioning it in the active site of COX-2. For this, we used the molecular docking module using MOE software. Once the ligand-receptor complex is formed, it will adopt the most stable conformation, i.e., the lowest energy level. The purpose of the dock application is looking at favorable conformational binding between medium-size ligands and a not so soft macromolecular target, which is usually a protein (Goto et al., 2008; Manikrao et al., 2011). For each compound, a number of conformations called poses were generated to identify favorable binding modes. The search for binding modes is generally constrained to a small specific region of the receptor called the active site. First docking is without the solvatation parameter (without H2O molecules), the second docking is done taking into account the presence of H2O molecules.
 | Table 1. Physico-chemical properties of synthetic inhibitors for cyclooxygenase. [Click here to view] |
 | Table 2. Physico-chemical properties of cyclooxygenase natural inhibitors. [Click here to view] |
 | Figure 1. Simplified model of COX-2 enzyme. [Click here to view] |
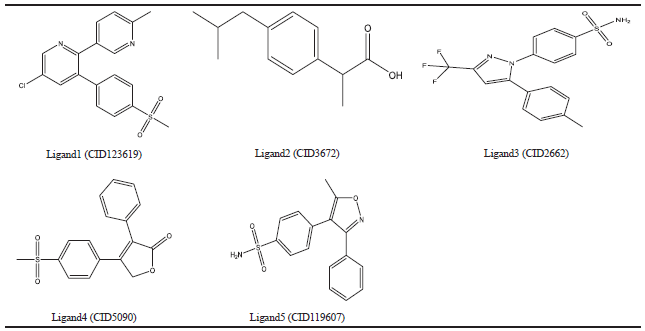 | Table 3. COX-2 synthetic inhibitors. [Click here to view] |
RESULTS
The obtained results are given in Tables 6–9 which showed that the orientation of the ligands plays a significant role in positioning the ligands in the active site of the enzyme; one can conclude that the introduction of bulky groups causes a rearrangement of conformation inside the cavity of the active site, which will be probably the complementarity and consequently the activity. Two-dimensional molecular method of the screen has been attributed to the MOE software, which is designed to visualize the active sites of the complex (protein-ligand). The ligand is prepared and made with an improved 2D depiction layout algorithm, and protein residues version is arranged around it to indicate links spatial proximity (Labute et al., 2001). Residues are marked with their amino acid code of three letters and job classification (Clark et al., 2006;2008). If there are multiple channels in the system, the positions are prefixed by the letters of the alphabet. Interactions between 2.5 Å and 3.1 Å are considered high and those between 3.1Å and 3.55Å are average. Greater than 3.55Å interactions are weak (Ritchie and Kemp, 2000).
 | Figure 2. Isolated active site of COX-2 enzyme. [Click here to view] |
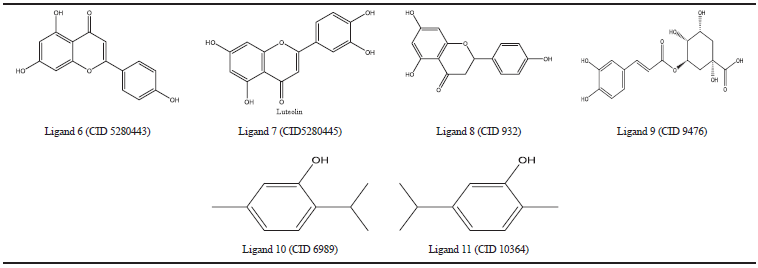 | Table 4. COX-2 natural inhibiros. [Click here to view] |
 | Table 5. Energy minimization of synthetic and natural molecules (Kcal/mol). [Click here to view] |
 | Table 6. Energy balance of five synthetic complexes without water (Kcal/mol). [Click here to view] |
 | Table 7. Energy balance of five synthetic complexes in water (Kcal/mol). [Click here to view] |
 | Table 8. Energy balance of six natural complexes without water (Kcal/mol). [Click here to view] |
 | Table 9. Energy balance of six natural complexes in water (Kcal/mol). [Click here to view] |
Docking interpretation of Synthetic inhibitors without water
Results given in Table 6 (Fig. 3a,b) show that the complex-5 has the lowest energy (−5.99548626 Kcal/mol) and is more active than complex-3 (−5.81052923 Kcal/mol).
For complex 5: Valdecoxib interacts with the amino acids [ARG 121 (A) H-acceptor N6 (NE; NH2); ARG 121 (A) ionic [N6 (NE; NH2), (O3, NH2), and LYS 83 (A) pi-cation] at a distance of 3.13 Å, 3.01 Å, 3.58 Å, and 4.87 Å, respectively (for the 1st, 2nd strong interaction, 3th and 4th weak interaction), with the existence of electric force PRO 84 this suggests that Valdecoxib can inhibit COX-2 and interfere with [ARG 121 (A) H-acceptor N6 (NE; NH2); ARG 121 (A) ionic [N6 (NE; NH2), (O3, NH2) and LYS 83 (A) pi-cation] (Yamaguchi et al., 2014).
For complex 3: Celecoxib interacts with the amino acids [ARG 121 (A) H-acceptor O5 (NE, NH2) ARG 121 (A) ionic O5 (NE, NH2), (O6, NH2); LYS 83 (A) pi-cation; TYR 116 (A) pi-H] at a distance of 2.87 Å, 2.79 Å , 3.89 Å, and 3.40 Å, respectively (for the 1st, 2nd strong interaction, 4th average interaction, and 3rd weak interaction), with the existence of three electric force PRO 84 wich suggesting that Celecoxib can inhibit COX-2 and interfere with [ARG 121 (A) H-acceptor O5 (NE, NH2) ARG 121 (A) ionic O5 (NE, NH2), (O6, NH2); LYS 83 (A) pi-cation; TYR 116 (A) pi-H] (Yamaguchi et al., 2014).
Docking interpretation of Synthetic inhibitors with water
Our results given in Table 7 and Figures 4a and 4b show that the complex-2 presents the best score (−6.95738792 Kcal/mol) succeeded by complex-4 (−6.59314966 Kcal/mol). For this complex, Ibuprofen interacts with the amino acids [LYS 83 (A) (H-acceptor 83, (A) ionic)] at a distance of 2.84 Å (for the 1st, 2nd strong interaction) with the existence of electric force PRO 84 wich suggesting that Ibuprofen can inhibit COX-2 and interfere with LYS 83 (A) (H-acceptor 83, (A) ionic)] (Yamaguchi et al., 2014).
For complex 4: Rofecoxib interacts with the amino acid [LYS 83 (A) H-acceptor] at a distance of 2.68 Å (for the 1st) with the existence of electric force PRO 84 witch suggesting that Rofecoxib can inhibit COX-2 and interfere with [LYS 83 (A) H-acceptor] (Yamaguchi et al., 2014).
 | Figure 3(a). Diagram interaction of complex-5 (COX-2 + Valdecoxib). [Click here to view] |
 | Figure 3(b). Diagram interaction of complex-3 (COX-2 + Celecoxib). [Click here to view] |
Docking interpretation of natural inhibitors without water
Obtained results (Table 8 and Fig. 5a,b) show that the complex-8 has the lowest energy (−6.11540222 Kcal/mol) and is more active than complex-7 (−6.05466223 Kcal/mol).
The energy of the reference ligand is important in comparison with that obtained by the Naringenine natural ligand. Therefore, we can validate Naringenine as a reference inhibitor. Indeed the corresponding complex energies are successively (ref: −5.98953867 Kcal/mol and Naringenine: −6.11540222 Kcal/mol).
In interaction between enzyme COX-2 and Naringenine, we did not find any bonding, only possible forces are electric (PRO 84 and LYS 83) with the existence of the Van der Walls forces, but the total energy of complex is very low comparing to other ligands in interactions.
For the interaction of Luteoline with COX-2, we get one bonde between PRO 86 (A) H-donor (O4, O) with the length of 3.08 Å; it is a strong interaction but two electric forces with (PRO 84 and LYS 83) with the existence of the Van der Walls forces.
 | Figure 4(a). Diagram interaction of complex-2 (COX-2 + Ibuprofen). [Click here to view] |
 | Figure 4(b). Diagram interaction of complex-4 (COX-2 + Rofecoxib). [Click here to view] |
 | Figure 5(a). Diagram interaction of complex-8 (COX-2 + Naringenine). [Click here to view] |
 | Figure 5(b). Diagram interaction of complex-7 (COX-2 + Luteoline). [Click here to view] |
Docking interpretation of natural inhibitors with water
Table 9 and Figures 6a,b show that the complex-9 has the lowest energy (−9.11451435 Kcal/mol) and is more active than complex-8 (−6.10765457 Kcal/mol).
On the other hand, the reference ligand complex energy is greater comparing with that obtained for the natural ligand Naringenine. Therefore, we can validate Chlorogenique as a reference inhibitor. Complex energies (ref: −5.98953867 Kcal/mol and Chlorogenique: −9.11451435 Kcal/mol).
For complex 9: Chlorogenique interacts with the amino acids [LYS 83 (A) (H-acceptor 83, (A) ionic)] at a distance of 3.04 Å (for the 1st strong interaction) and interaction with water HOH 0 (N4; N5) H-donor at a distance of 3.01, 2.71 Å (for the 1st and 2nd strong interaction) with the existence of electric force PRO 84 wich suggesting that Chlorogenique can inhibit COX-2 and interfere with LYS 83 (A) (H-acceptor 83, (A) ionic)] (Yamaguchi et al., 2014).
For complex 8: Naringenine interacts with the amino acid [PRO 86 (A) H-acceptor] at a distance of 3.30 Å (for the 1st average interaction with the existence of electric forces PRO 84 and PRO 86 witch suggesting that Naringenine can inhibit cyclooxygenase-2 and interfere with [PRO 86 (A) H-acceptor] (Yamaguchi et al., 2014). We can conclude from our obtained results that Ibuprofen and Chlorogenique would be the best to slow down the evolution of the treatment ID. This is confirmed by comparing their energies: Energy [Ibuprofen (−6.95738792 Kcal/mol) Ë‚ Rofecoxib (−6.59314966 Kcal/mol). Energy (Chlorogenique (−9.11451435 Kcal/mol) Ë‚ Naringenine (−6.10765457Kcal/mol)].
 | Figure 6(a). Diagram interaction of complex-9 (COX-2 + Chlorogenique). [Click here to view] |
 | Figure 6(b). Diagram interaction of complex-8 (COX-2 + Naringenine). [Click here to view] |
CONCLUSION
In this work, we studied the interaction of cyclooxygenase-2 (inflammation enzyme) by molecular docking taking into account solvatation parameter (presence of water molecules). Obtained results allow us to conclude that the synthetic NSAID (Ibuprofen) and also the natural flavonoid inhibitor (chlorogenique) present a more optimized interaction for better inhibition study of COX-2 in purpose to treat ID. Obtained results allow us to propose a natural and reliable treatment with natural products containing Chlorogenique during the first stage of the inflammatory disease. We also propose further studies to develop chlorogenique into a new drug.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
Bukovska A, Cikos S, Juhas S, IL’Kova G, Rehak P, Koppel J. Effects of a combination of thyme and oregano essential oils on TNBS-induced colitis in mice. Mediat Inflamm, 2007; ID 23296:9.
Clark AM, Labute P. Detection and assignment of common scaffolds in project databases of lead molecules. J Med Chem, 2008; 52:469–83.
Clark AM, Labute P, Santavy M. 2D structure depiction. J Chem Inf Model, 2006; 46:1107–23.
Goto J, Kataoka R, Muta H, Hirayama N. ASEDock-docking based on alpha spheres and excluded volumes. J Chem Inf Model, 2008; 48:583–90.
Hemler M, Lands WE, Smith WL. Purification of the cyclooxygenase that forms prostaglandins: demonstration of two forms of iron in the holoenzyme. J Biol Chem, 1976; 251:5575–79.
Hotta M, Nakata R, Katsukawa M, Hori K, Takahashi S, Inoue H. Carvacrol, a component of thyme oil, activates PPAR α and γ, and suppresses COX-2 expression. J Lipid Res, 2010; 51:132–9.
Katsukawa M, Nakata R, Takizawa Y, Hori K, Takahashi S, Inoue. H. Citral a component of lemongrass oil activates PPARα and γ and suppresses COX-2 expression. Biochim Biophys Acta, 2010; 1801:1214–20.
Kuete V (ed.). Thymus Vulgaris. In: Medicinal spices and vegetables in Africa, Academic Press, pp 599–609, 2017.
Labute P, Williams C, Feher M, Sourial E, Schmidt JM. Flexible alignment of small molecules. J Med Chem, 2001; 44:1483–90.
Manikrao AM, Mahajan1 NS, Jawarkar RD, Mahajan DT, Masand VH, Hadda TB. Docking studies of few C-3 substituted azapteridines as hepatitis C virus RNA-dependent RNA polymerase inhibitors. J Comput Method Mol Design, 2011; 1:35–45.
Miliani LF, Nielsen OH, Andersen PS, Girardin SE. Chronic inflammation: importance of NOD2 and NALP3 in interleukin-1β generation. Clin Exp Immunol, 2007; 147:227–35.
Molecular Operating Environment (MOE). 2013.08; Chemical Computing Group ULC, 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2018.
Picot D, Loll PJ, Garavito RM. The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1. Nature, 1994; 367:243–9.
Powers JP, Piper DE, Li Y, Mayorga V, Anzola J, Chen JM, Jaen JC, Lee G, Liu J, Peterson MG, Tonn GR, Ye Q, Walker NPC, Wang Z. SAR and mode of action of novel non-nucleoside inhibitors of hepatitis C NS5b RNA polymerase. J Med Chem, 2006; 49:1034–46.
Ritchie DW, Kemp GJL. Protein docking using spherical polar Fourier correlations. Proteins Struct Funct Bioinform, 2000; 39:178–94.
Salmalian H, Saghebi R, Moghadamnia AA, Bijani A, Faramarzi M, Nasiri FA, Bakouei F, Behmanesh F, Bekhradi R. Comparative effect of thymus vulgaris and ibuprofen on primary dysmenorrhea: a triple-blind clinical study. Caspian J Intern Med, 2014; 5:82–8.
Yamaguchi H, Kamiie K, Kidachi Y, Noshita T, Umetsu H, Fuke Y, Ryoyama K. Structural insight into the ligand-receptor interaction between 6-(methylsulfinyl)hexyl isothiocyanate and multidrug resistance-associated protein 1 nucleotide-binding domain 1. Int J Comput Bioinform In Silico Modeling, 2014; 3:310–4.