INTRODUCTION
The urinary system is among the human body’s most important systems, which keeps toxins and waste from accumulating in our blood and excreting them as pee [1]. Millions of persons worldwide are afflicted with urinary tract infections (UTIs), which are among the most prevailing bacterial illnesses. UTIs are among the most prevalent bacterial infections globally, affecting approximately 150 million people annually and imposing significant healthcare costs exceeding $6 billion [2].
The majority of the bacteria that cause UTIs are uropathogenic, which includes both Gram-negative and positive bacteria, for instance, Proteus mirabilis, Escherichia coli, Klebsiella pneumoniae, Enterococcus faecalis, and Staphylococcus saprophyticus [3,4].
UTIs are predominantly caused by bacterial pathogens, with E. coli being the most prevalent, reaching up to 75% of uncomplicated cases, followed by K. pneumoniae, P. mirabilis, then Pseudomonas aeruginosa, and Gram-positive organisms such as E. faecalis (5%–6%) and S. saprophyticus (3%–4%) [3,5,6].
These uropathogens exhibit significant resistance to commonly prescribed antibiotics, with E. coli showing high resistance to ampicillin, amoxicillin/clavulanic acid, and trimethoprim-sulfamethoxazole, while K. pneumoniae frequently harbors extended-spectrum beta-lactamases (ESBLs) and carbapenemases, leading to resistance against third-generation cephalosporins and carbapenems. Proteus mirabilis is notable for its intrinsic resistance to tetracyclines and increasing fluoroquinolone resistance due to mutations in DNA gyrase (gyrA). In addition, Enterococcus spp. exhibit resistance to fluoroquinolones (50%) and penicillin (39%), while S. saprophyticus remains susceptible to nitrofurantoin (86.6%) but resistant to beta-lactams [7–12].
The rising prevalence of multidrug-resistant (MDR) strains, particularly in hospital-acquired UTIs, underscores the urgent need for antimicrobial stewardship and region-specific resistance surveillance to guide empirical therapy [13].
Globally, antibiotic resistance linked to UTIs is rising quickly. UTIs are the most prevalent bacterial infection that necessitates medical attention, represent a severe public health problem, and account for 8.6 million ambulatory care visits annually [14,15].
Although the effectiveness of traditional treatments is seriously threatened by the emergence of antimicrobial resistance (AMR), antibiotics are still a major part of the clinical management of UTIs. It is becoming more difficult to treat UTIs successfully due to resistant microbes, especially those that produce carbapenemases and ESBLs [16].
Actinobacteria genus Streptomyces spp. is well known for producing a variety of bioactive substances, such as antibiotics, which have transformed contemporary medicine. Most (approximately 67%) of all microbial antibiotics are produced by actinomycetes, with the Streptomyces genus accounting for about 80% of these productions [17,18].
Because of the wide range of antibiotic activity exhibited by these secondary metabolites, Streptomyces is a significant source of possible medicinal medicines. Many antibiotics, including erythromycin, tetracycline, and streptomycin, that are derived from Streptomyces species have been used to treat a variety of bacterial infections. Nonetheless, research on Streptomyces for new antibacterial drugs that target resistant UTI infections is still ongoing [18,19].
Recent studies focused on the potential of Streptomyces-derived compounds in battling MDR bacteria. The bioactive substances that these microbes create can work in several ways, such as by interfering with nucleic acid metabolism, disrupting protein synthesis, or inhibiting the creation of bacterial cell walls. The effectiveness of Streptomyces metabolites against UTI-associated microorganisms, especially those resistant to several antibiotics, requires more research despite their encouraging antibacterial qualities [18,20].
Moreover, stress on microorganisms may enhance the secondary metabolites produced by many mechanisms. Therefore, an arid soil-derived Streptomyces species often produces novel secondary metabolites, as environmental stressors drive the evolution of diverse antimicrobial pathways [21,22]. Streptomyces geysiriensis is one of the Streptomyces spp., which is a promising candidate for this study due to its isolation from arid soils, a niche known to harbor actinobacteria with unique biosynthetic capabilities adapted to extreme conditions (e.g., drought, high salinity, and oligotrophy) [22–24]. Streptomyces geysiriensis was reported as having broad-spectrum antimicrobial activity, including against Gram-negative bacteria. Streptomyces geysiriensis IN7 extracts, or their bioactive compounds, showed promising activity against the tested multidrug-resistant standard pathogens, including Proteus vulgaris, E. coli, and Salmonella enterica [25], which aligns with the need for new agents against uropathogens.
Therefore, the study of antibacterial activities of Streptomyces active metabolites and their isolated compounds against UTI–causing bacteria is crucial due to the rising prevalence of antibiotic resistance among pathogens. UTIs, commonly caused by multidrug-resistant bacteria, pose significant challenges to global healthcare systems. Genus Streptomyces, especially novel strains, a well-known producer of bioactive secondary metabolites, offers a promising source of novel antibacterial compounds that can combat these resistant strains. By isolating and characterizing these isolated compounds, researchers aim to develop new, effective therapeutic agents to address the growing threat of antibiotic resistance, improve treatment outcomes for UTIs, and reduce the reliance on conventional antibiotics that are increasingly becoming ineffective.
The purpose of this study is to assess the antibacterial activity of active metabolites derived from native S. geysiriensis against bacteria that have been isolated from UTIs, with an emphasis on their potential as alternative treatment agents. In addition to helping combat the growing issue of antibiotic resistance in UTI treatment. This research aligns with the urgent need for innovative solutions to tackle AMR and safeguard public health. Extraction, purification, and identification of active compounds as antibacterials for UTI. The research findings may offer fresh perspectives on the creation of new antibiotics or finding new natural sources of isolated compounds as potential sources for pharmacological uses.
MATERIALS AND METHODS
Actinomycete strain
The actinomycetes isolate was obtained from the Desert Research Center (isolated from wild desert valleys belonging to Marsa Matrouh soil, Egypt, coordinates 31.472377°N, 26.662852°E) and established through molecular characterization and phylogeny.
Streptomyces geysiriensis was identified under accession number OQ389482, NCBI-GenBank [26].
Bacterial isolation from UTI
Collection of urine specimens
The UTI samples were selected according to the patient’s nonresponse to antibiotic treatment from June to December 2022. To collect the urine specimens, the patients were instructed to use liquid soap containing chlorhexidine to clean their external genitalia. Patients suffering from UTI are instructed to provide a mid-stream urine sample into a sterile urine container by voiding the first urine drops into the toilet [27]. This study was granted ethical approval and approved by the Research Ethics of the Botany Department, Faculty of Science, Port Said University.
Bacterial isolation
The urine samples were kept between 4°C and 8°C in a refrigerator until it was evaluated. Centrifuge tubes containing approximately 10 ml urine samples were centrifuged for 10 minutes at 2,000 rpm and sealed to prevent contamination. The urine sediment was separated by a sterile wire loop on nutrient agar media, and cystine–lactose–electrolyte-deficient agar media and incubated at 37°C for 24 hours. After incubation, the colonies’ purity is checked by colonial morphology and Gram stain. The pure bacterial colonies were kept for further studies.
Identification of UTI-isolated bacteria
Biochemical and morphological characterization
Cell bacterial morphology and arrangements, and characteristics of their cell walls were decided after Gram staining involves a series of steps [28,29]. Different culture media and biochemical tests in the medical lab, including IMViC tests (indole, methyl-red, Voges–Proskauer, and citrate), triple sugar iron agar, oxidase test, urease test, catalase test, and other biochemical tests, were used to identify unbranched bacteria isolated from UTI.
Molecular biosystematics
The international standards for molecular biosystematics using 16S rRNA gene sequencing. DNA of the UTI-isolated bacteria was extracted by enzyme method according to the protocol prescribed by Marmur and Doty [30]. After that, purity was checked on agarose (0.8%) gel electrophoresis. To obtain the 16S rRNA gene, purified DNA was subjected to polymerase chain reaction (PCR) amplification for sequencing. A PCR kit containing Primer A-8.27f (5´-CCGTCGACGAGCTCAGAGTTTGATCCTGGCTCAG-3´) and Primer B-1573-1504-R(5´-CCCGGGTACCAAGATTAAGGAGGTGACCAGCCGCA-3´) was employed in the amplification. According to the standard protocol for PCR amplification [31]. After PCR amplification, isolated DNA fragments containing 16S rRNA genes of bacterial isolates were sent to Macrogen (Humanizing Genomics Macrogen, Seoul, Korea). The whole nucleotide profile of the 16S rRNA gene, which indicates complete sequencing, was obtained. Geneious software (Biomatters) was used to trim and assemble the resultant sequences. As a result, the exact sequences were found using GenBank’s basic local alignment search tool (BLAST).
Phylogenetic analysis
In addition to phylogenetic assessment, the sequenced 16S rRNA of UTI bacterial isolates corresponded with those in a known database using the EzBioCloud instrument. Nucleotide nucleic acid sequences were obtained from GenBank and aligned with the recognized sequences using MEGA-X, software used to understand molecular identification and polyphasic taxonomic approaches for genetic relatedness and phylogenetic relationships of microorganisms. Phylogenetic trees were constructed using the neighbor-joining method [32], employing the Tamura–Nei Model [33]. The trees were reviewed using 103 bootstrap replicates.
Extracellular bioactive metabolite(s)
Production
An agar disc was cut (with a Cork-borer) out of the actinomycete agar plate culture of the S. geysiriensis isolate, which was used to inoculate 250 ml Erlenmeyer flasks containing 100 ml of the sporulation medium (liquid starch nitrate at pH 7.0). The inoculated flask broth medium was incubated at 35°C for 3 days on a rotary shaker (150 rpm) to stimulate spore production. For active metabolite production, 20 flasks (250-ml Erlenmeyer flasks, 100 ml broth medium of starch nitrate, and pH 7.2 ± 2) were inoculated with 10 ml of the vegetative inoculum. The inoculated broth flasks were incubated at 35°C in a 250-rpm rotary shaker. After incubation, the collected broth culture media were filtered with cotton, centrifuged for 15 minutes at 4°C at 4,000 rpm to separate the mycelium from the flask, and stored in a refrigerator for further extraction procedures.
Active metabolite extraction
The cell-free filtrate (500 ml) was then concentrated using a rotary evaporator (Büchi, Flawil, Germany) at 35°C ± 5°C. The concentrated filtrate was then subjected to sequential solvent extraction in a separating funnel using the following gradient-polarity solvents (each 300 ml): hexane, diethyl ether, chloroform, and ethyl acetate (EA). After each extraction step, the solvent layer was separated, and the remaining aqueous phase was further extracted with the next solvent. The residue from the cell-free filtrate after organic solvent extraction was evaporated to dryness using a rotary evaporator and then re-extracted with 300 ml of absolute ethanol. All solvent extracts were filtered (by filter paper Whatman no. 1) and reduced using rotary evaporation and then dried to remove all solvent (complete dryness). The dried extracts were individually dissolved in 1 ml of dimethyl sulfoxide (DMSO) for solubility. The resulting solutions were screened for bioactivity against the test organisms.
Evaluation and purification of the active compound
Using a different system of toluene: EA: formic acid (5:4:1 V/V), the active solvent extract of S. geysiriensis was analyzed by Thin-layer chromatography (TLC) (silica gel 60, F254, aluminum sheet 20 × 20 cm, Merck KGOA, Germany) and detected at short and long wave UV. Column (Silica gel) chromatography was used for the fractionation and purification of the most potent EA extract. By visualization of TLC, similar chromatographic bands of successive extracts were added together. The pure bands on TLC were through the proper solvent solution, and the bands were separated using UV light. After scraping the bands off the plate, they were put in a Buckner funnel, eluted with the proper solvent, and filtered.
Determination of the active compounds of EA extract
To evaluate the active band of the most potent fraction, the activity at the partial purification stage was checked using the bioautographic method, in which the developed TLC strip was positioned on a UTI bacterial isolate that was seeded in an agar plate. Then, the prepared Petri dish was incubated for 24 hours at 37°C. The inhibition zone around the band was decided, and retention factor (RF) was calculated to determine the active band on the TLC strip.
Identification of active compound
Standard authentic compound 3,4-dihydroxybenzoic acid was obtained from Merck Chemical Co. It was used as an authentic compound for the proposed structure of an isolated compound, which was performed by co-chromatographic analysis.
Liquid chromatography-quadrupole time-of-flight tandem mass spectrometry analysis
The preparation of the sample, acquisition, and LC-MS data processing methods were performed according to Zhu et al. [34] and Badawy et al. [35]. A solvent for reconstitution (acetonitrile) was created using H2O: MeOH: MeCN 50:25:25 v/v. Fifty milligrams of the dry extract were dissolved in 1 ml of the reconstitution solvent. Solubility was reached after centrifuging at 104 rpm for 10 minutes, ultrasonicating for 10 minutes, and vortexing for 2 minutes. The stock solution was diluted using reconstitution solvent from 50 µl to 1,000 µl. The final injection concentration was 2.5 µg/µl. The injection sample was 10 µl. Solutions A, B, and C were prepared for positive mode, negative mode, and both negative and positive mode, respectively, then used in the acquisition Method. An X select HSS T3 (2.5 µm, 2.1 × 150 mm) column (Waters, USA) and in-line filter disks pre-column (0.5 µm × 3.0 mm, Phenomenex, USA) conditioned at 40ºC were used to separate the compounds. Chromatographic separation and information were carried out using Exion LCTM Series UHPLC equipment on a Triple TOF 5600+ (Sciex, USA).
Screening for antimicrobial activity
Mueller–Hinton agar (MHA) was used for antimicrobial assays, as per CLSI guidelines and according to the diffusion plate method [36–38], the inhibitory clear zones on agar plates around the holes or discs were measured in millimeters.
Agar well method
MHA was seeded with test bacteria before solidification under an aseptic condition and poured into sterilized plates. The inoculated seeded plates were punched by a sterile Cork borer to create 6 mm diameter holes. An exact 100 µl of the Streptomyces cell-free growth filtrate was filled in each hole aseptically. Then, the Petri dishes were kept in the fridge for 60 minutes before incubation (at 37°C for 24 hours) of each organism to acquire full diffusion. The antimicrobial activities of the actinomycetes isolates were detected by measuring a clear inhibition zone around holes. The experiment was carried out in triplicate.
An agar-well diffusion experiment was used to screen different successive extracts. Different extracts were dissolved (50 mg/ml) in water or DMSO according to solubility. The antimicrobial activities of the S. geysiriensis isolate were detected by measuring a clear inhibition zone around holes. Distilled water and DMSO served as negative controls. The experiment was carried out in triplicate.
Disk diffusion method
The disc diffusion method was used for confirmation of the antibacterial assay of the purified isolated compound. MHA media were seeded with test bacteria before solidification in an aseptic condition and poured into sterilized plates. Antimicrobial disks were prepared by cutting a 6.0 mm diameter paper disk (Whatman No. 3). The discs were saturated with the actinomycete growth filtrate or filtrate fractions, allowed to dry, and then placed on the surface of an agar-seeded medium with the test organisms. The antimicrobial activities of the actinomycetes isolates were detected by measuring a clear inhibition zone around the discs. The experiment was carried out in triplicate.
Determination of minimal inhibitory concentrations
The minimum inhibitory concentration (MIC) of successive extracts was determined using the agar-well diffusion method. Different concentrations of 20, 10, 5, 2.5, and 1.25 ug/ml were achieved by two-fold serial dilution. Agar wells were made on each Petri plate seeded with test bacteria, and 100 µl of each extract was added to each well. The plates were refrigerated for 1 hour and then incubated at 37°C for 24 hours. MIC was originally thought to be the lowest concentration of an agent or extract at which bacterial growth is completely inhibited. Distilled water and DMSO served as negative controls. The experiment was carried out in triplicate.
Statistical analysis
The statistical software SPSS 19 was used to analyze the data. Every data point was shown as a mean ± SD.
RESULTS
UTI bacterial isolation and identification
The urine samples were collected and subjected to the isolation of bacterial pathogens. After incubation, the colonies’ purity is checked by colonial morphology and Gram stain, then subjected to biochemical and molecular tests for identification. Three bacterial isolates, including E. coli (Ur-33), P. aeruginosa (Ur-14), and Bacillus cereus (Ur-09) confirmed identification corresponding to 16S rRNA gene sequencing, and the results are as follows.
Identification of bacterial strain Ur-09
By the semi-quantitative culture technique, the urinary tract pathogenic bacteria were subjected to 16S rRNA gene sequencing, and the DNA amplification of the Ur-09 isolate represented around 1,386 bp. To complete the identification of isolate Ur-09, the sequence obtained was blasted to NCBI, and the result was aligned to the partial gene sequence of 16S rRNA of B. cereus with Gene Bank accession number PP732311. From the BLAST search, the homologous sequences were used to create a phylogenetic tree, revealing (100%) relatedness to the B. cereus strain (Fig. 1).
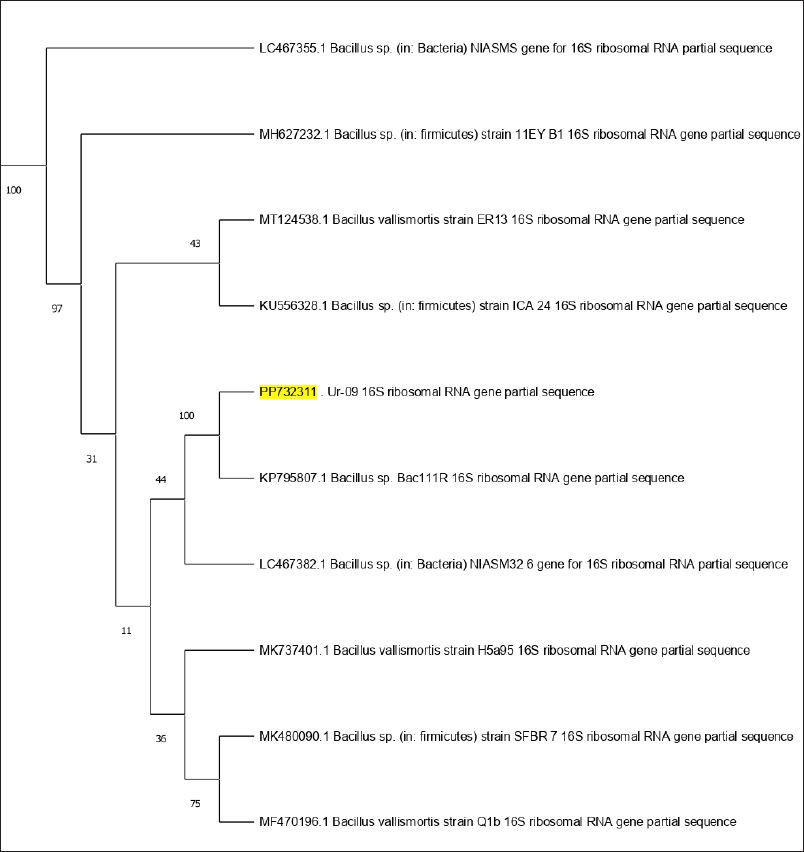 | Figure 1. Phylogenetic tree of bacterial evolutionary relationships inferred from aligned 16S rDNA sequences of coded bacterial isolate strain Ur-09. [Click here to view] |
Identification of bacterial strain Ur-14
In the second strain, the DNA PCR amplification and sequencing of the 16S rRNA gene yielded the results of the DNA sequence. The DNA sequence was then assessed using BLAST. According to BLAST pattern matching against database sequences, bacterial isolate Ur-14 was molecularly confirmed as P. aeruginosa with Gene Bank accession number PP732312. Phylogenetic tree of bacterial evolutionary relationships inferred from aligned 16S rDNA sequences of coded bacterial isolate strain Ur-14 (Fig. 2).
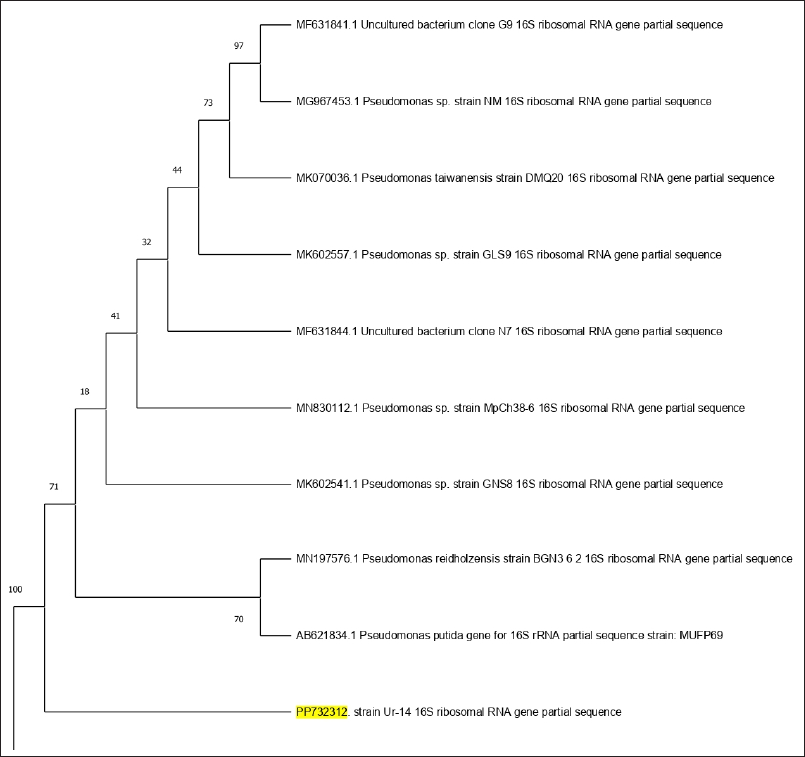 | Figure 2. Phylogenetic tree of bacterial evolutionary relationships inferred from aligned 16S rDNA sequences of coded bacterial isolate strain Ur-14. [Click here to view] |
Identification of bacterial strain Ur-33
The investigation of the 16S rRNA sequences of the bacterial isolate Ur-33, which contains 1,895 bp. It has been submitted to the NCBI BLAST engine for processing. A phylogenetic tree with similar sequences was subsequently generated using the neighbor-joining approach for an isolated bacterial strain (Fig. 3). From the BLAST search, the homologous sequences were used to create a phylogenetic tree, revealing that Ur-33 is 100% related to E. coli strain (with an accession number PP732310).
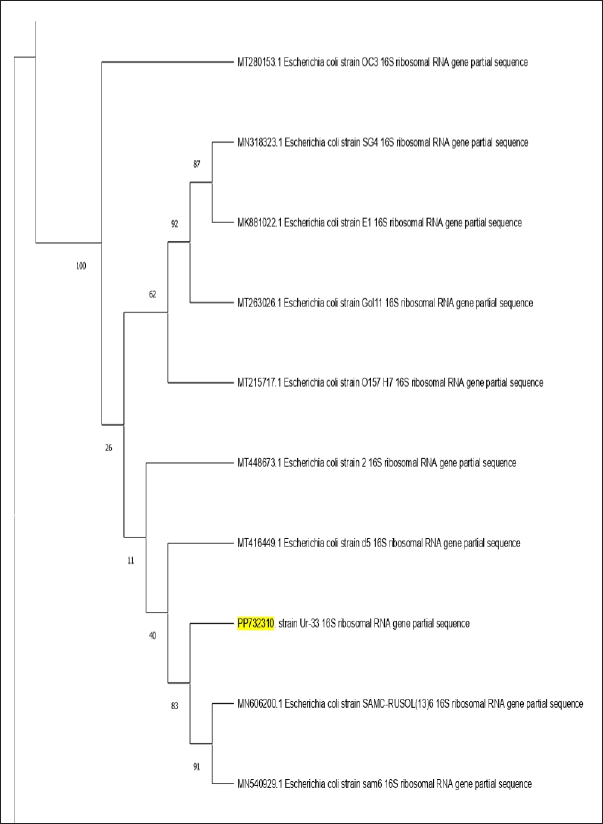 | Figure 3. Phylogenetic tree of bacterial evolutionary relationships inferred from aligned 16S rDNA sequences of coded bacterial isolate strain Ur-33. PP732310. [Click here to view] |
Morphology and antibacterial activity
Streptomyces geysiriensis growth and antibacterial activity screening
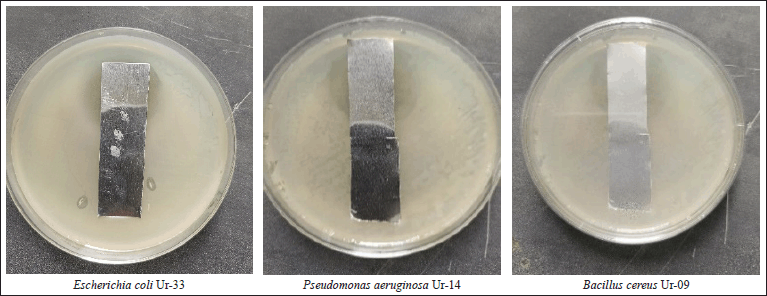
Morphological characteristics of S. geysiriensis on growth on yeast-malt extract agar medium and tryptone-yeast extract agar media showed creamy white aerial mycelium and grey–white substrate mycelium (Fig. 4). Streptomyces geysiriensis growth on starch nitrate culture media, the aerial mycelium had a creamy color, while the base mycelium had a greyish color. After incubation of inoculated starch nitrate broth medium, the antimicrobial activity of S. geysiriensis cell growth-free filtrate against E. coli Ur-33, B. cereus Ur-09, and P. aeruginosa Ur-14 showed promising activity, measuring 18, 19, and 20 mm, respectively.
Antibacterial activity of successive extracts
The antibacterial activity of various S. geysiriensis cell-free filtrate solvent extracts (hexane, chloroform, EA, and ethanol) against three UTI bacterial isolates (B. cereus Ur-09, E. coli Ur-33, and P. aeruginosa Ur-14). All successive solvents showed variant antibacterial activity against UTI bacterial isolates. The results indicate that each of the three bacterial strains was targeted by EA with inhibition zones of 19.67, 21.67, and 20.33 mm against E. coli Ur-33, B. cereus Ur-09, and P. aeruginosa Ur-14, respectively. According to these results, EA extract was the most active among the successive solvents (Table 1).
Table 1. Effect of different successive solvent extracts on UTI-isolated bacteria.
| Solvent extract | Escherichia coli Ur-33 | Bacillus cereus Ur-09 | Pseudomonas aeruginosa Ur-14 |
|---|---|---|---|
| Hexane | NA | NA | NA |
| Chloroform | 10.67 ± 0.27 | NA | 12.67 ± 0.47 |
| E. acetate | 19.67 ± 0.94 | 21.67 ± 0.54 | 20.33 ± 0.72 |
| Ethanol | 14.67 ± 0.54 | 12.33 ± 0.27 | 12.67 ± 0.72 |
| Erythromycin | 15.89 ± 0.64 | 16.34 ± 0.66 | 15.56 ± 0.87 |
NA = no activity.
Minimal inhibitory concentration determination
MICs of the most potent EA successive solvent of S. geysiriensis cell-free filtrate against urinary tract bacterial strains were determined (Table 2). MICs of EA were recorded at 5 µg/ml for E. coli Ur-33 and B. cereus Ur-09, with inhibition zones 15.60 ± 0.12 and 13.67 ± 0.29 mm, respectively, while 10 µg/ml for P. aeruginosa Ur-14 with an inhibition zone 12.59 ± 0.46 mm. Erythromycin at a concentration of 10 µg/ml (control) showed inhibition zone 9.67 ± 0.07, 10.19 ± 0.06, and 11.3 ± 0.33 mm against E. coli Ur-33, P. aeruginosa Ur-14, and B. cereus Ur-09, respectively.
Table 2. Effect of different concentrations of EA S. geysiriensis extract on the UTI bacterial isolates.
| µg/ml | Escherichia coli Ur-33 | Bacillus cereus Ur-09 | Pseudomonas aeruginosa Ur-14 |
|---|---|---|---|
| 1.25 | NA | NA | NA |
| 2.5 | NA | NA | NA |
| 5 | 15.60 ± 0.12 | 13.67 ± 0.29 | NA |
| 10 | 20.33 ± 0.11 | 20.63 ± 0.63 | 12.59 ± 0.46 |
| 20 | 26.27 ± 0.07 | 24.57 ± 0.45 | 16.91 ± 0.28 |
| Erythromycin 10 µg/ml | 9.67 ± 0.07 | 11.3 ± 0.33 | 10.19 ± 0.06 |
NA = no activity.
Biological activity of active compounds
The most potent EA extract was subjected to column fractionation. After fractionation, five major TLC plate bands were screened against E. coli Ur-33, B. cereus Ur-09, and P. aeruginosa Ur-14. The active compounds of the EA extract using TLC diffusion on the surface of seeded bacteria were determined (Fig. 5), which demonstrates that one spot is active in the TLC chromatogram of the EA successive extract. This pure compound loaded with filter paper disc showed the inhibiting activity on three UTI test bacteria: E. coli Ur-33, B. cereus Ur-09, and P. aeruginosa Ur-14, with inhibition zones of 15.67, 17.66, and 15.33 mm, respectively.
 | Figure 5. Determination of the active compounds of EA extract using TLC band diffusion. [Click here to view] |
Identification of purified active compound
The major crude compound was identified by the Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) tools. Positive mode LC-MS analysis of crude extract showed that 3,4-dihydroxybenzoic acid (Figs. 6 and 7) was the major compound. Liquid chromatography-MS/MS assessment (positive mode) of the S. geysiriensis EA extract was used to anticipate the chemical formula of a molecular compound in accordance with m/z 155.1177 by using the Peak View software, which recommended the molecular formula of C7H6O4, [M+H+]. Liquid chromatography-MS/MS decided the major metabolite (labeled by arrow), with antibacterial assay defined as 3,4-dihydroxybenzoic acid. Further confirmation for the proposed structure was performed by co-chromatographic analysis of authentic 3,4-dihydroxybenzoic acid with an isolated compound, which reveals that both have the same RF value, which is further evidence for the identity of the proposed structure.
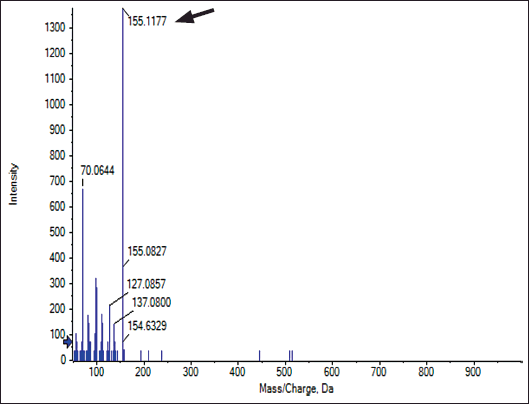 | Figure 6. Liquid chromatography-MS/MS assessment of crude Streptomyces geysiriensis EA extract. [Click here to view] |
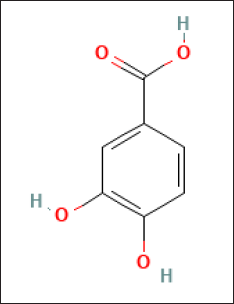 | Figure 7. The suggested chemical structure of isolated compound 3,4-dihydroxybenzoic acid. [Click here to view] |
DISCUSSION
Millions of persons worldwide are afflicted with UTIs, which are among the most prevailing bacterial illnesses. Uropathogenic bacteria are the main cause of UTIs, with E. coli being the most common pathogen [3]. One of the most prevalent bacterial illnesses in the world. UTIs can occur in both public and medical settings [4]. Despite advances in antibacterial therapy, UTIs continue a significant cause of morbidity [39,40]. Long-term usage of antibiotics results in adverse effects in humans as well as plasmid and/or mutational alterations that provide bacteria resistance. There is a growing need for functional actinobacteria and other natural products due to environmental concerns and MDR infections. Nowadays, finding novel sources and bioactive substances for antibiotics is one of the most important challenges [41–43]. Streptomyces spp. is one of the most important sources of antibiotics, and its soils are rich with it. This bacterial genus produces around 6,000 distinct types of bioactive secondary metabolites that are generated from different Streptomyces species. Many of these drugs are antibiotics, herbicides, and insecticides [44].
In previous studies, the most common UTI bacteria, including E. coli, K. pneumoniae, P. mirabilis, and E. faecalis are common intestinal bacteria that cause UTIs. UTIs are also caused by S. saprophyticus, P. aeruginosa, and Group B Streptococcus. Rarely occurring bacterial species are also linked to UTIs, including Salmonella species, Acinetobacter, B. cereus, Enterococcus hirae, and Staphylococcus spp. (sciuri, vitulinus, lentus, xylosus, and pulvereri) [3,45–52]. Gram-negative bacteria are the primary cause of UTIs because they can acquire plasmids with genes ESBLs or other resistant genes that plasmids can transport [53]. In addition, the most prevalent bacterial Gram-negative resistance mechanism involves the active efflux of antibiotics from cells through membrane transporters [54]. Furthermore, Gram-negative bacteria have an outer membrane that acts as a natural barrier and controls antimicrobial entrance or increases resistance [6].
In the current study, three bacteria, including two Gram-negative E. coli (Ur-33), P. aeruginosa (Ur-14), and one Gram-positive B. cereus (Ur-09), were isolated from UTI patients and identified according to biochemical and confirmed identification according to 16S rRNA gene sequencing. Streptomyces geysiriensis isolated strain (under accession number OQ389482, NCBI-GenBank) was cultivated on starch–nitrate broth at 37°C for 12 days and under other suitable cultivation conditions for Streptomyces spp. to produce bioactive metabolites [26].
Previous studies have shown that S. geysiriensis was reported as having broad-spectrum antimicrobial activity, including against Gram-negative bacteria. Streptomyces geysiriensis IN7 (isolated from marine sediments on Starch Casein Agar) extracts, or their bioactive compounds, showed promising activity against the tested multidrug-resistant standard pathogens, including P. vulgaris, E. coli, and S. enterica [25].
Several methods are used to extract the bioactive chemicals from natural sources. Secondary metabolites are typically extracted from the culture filtrates using solvent extraction [55,56]. Bioactive substances from actinobacteria have been extracted using organic solvents with varying polarity [57]. The same thing was detected in the current study; it was also found that the extraction of active substances from S. geysiriensis cell-free filtrate, which was subjected to the process of extraction by different solvents (ethanol, EA, chloroform, and n-hexane), showed that EA exhibited respectable antimicrobial activity against the tested microorganisms. These results are very similar to a study conducted by Rammali et al. [56], who proved that potent secondary metabolites, especially those with antibacterial and antifungal activities, were produced by the isolated Streptomyces strain, and the EA extract demonstrated notable antimicrobial activity against both drug-resistant bacteria and phytopathogenic fungi. In addition, the Karthick et al. [25] study uses different solvents, methanol, hexane, and EA, in extracting bioactive compounds against the tested multidrug-resistant standard pathogens, including P. vulgaris, E. coli, and S. enterica.
In addition, according to a previous study conducted by Rante et al. [58], the same results were largely similar to our study; it was found that the EA extract from S. geysiriensis showed antimicrobial efficacy against Candida albicans, E. coli, and multidrug-resistant Staphylococcus aureus at a concentration of 2 mg/20 μl. The EA extract was found to be superior when the identical extracts of organic solvents were investigated on bacteria isolated from the urinary system, as it provided the highest rate of inhibition.
In addition to Apsari et al. [59] study, it was also found that the supernatant of Streptomyces spp. was extracted by using EA, and crude extract was tested by the paper disc method on six test bacteria isolated from UTI patients, and gave a higher inhibition zone among others. In the current study, the most active EA extracts were subjected to column fractionation and preparative TLC, and one spot out of five spots was more active against UTI bacteria. The same steps in another study demonstrated that eight spots of the EA supernatant of Streptomyces sp. CRB46 was separated on the TLC chromatogram and inhibited Gram-positive (S. aureus, Bacillus subtilis, and Bacillus licheniformis) and Gram-negative bacteria (Salmonella typhimurium and P. aeruginosa) standard test bacteria with a large inhibition zone [60,61].
The MIC provides an idea of the in vitro effectiveness of an active extract or compound against a microorganism, a quantitative assay against tested bacteria [62]. In the current study, the lowest MIC of successive EA extract of S. geysiriensis medium was recorded against E. coli and B. cereus. In other studies, the lowest activity of Streptomyces extract was against Gram-positive bacteria than Gram-negative bacteria [63,64].
In the current study, the major and active compound has been strongly suggested its identity as 3,4-dihydroxybenzoic acid by using LC-MS/MS and co-chromatography. The active major compound isolated from S. geysiriensis has activity against UTI bacterial isolates.
While LC-MS/MS and co-chromatography strongly suggest the identity of the major compound as 3,4-dihydroxybenzoic acid, nuclear magnetic resonance (NMR) analysis would be required for unequivocal confirmation. Future studies will prioritize NMR validation.
A few studies indicated that active isolated compound (3,4-dihydroxybenzoic acid) formation is a relatively common natural source, including algae Cladophora wrightiana [65], bacterial nitrogen-fixing Azomonas macrocytogenes [66], and leaves of Ageratum conyzoides L. [67]. 3,4-dihydroxybenzoic acid, an isolated compound from Streptomyces species, is effective against Rhizoctonia solani plant pathogens [68]. In addition, 3,4-dihydroxybenzoic acid is isolated from the rhizome of Drynaria quercifolia, which is active against several Gram-positive and negative bacteria [69]. Furthermore, from the EA fraction of the fern Trichomanes chinense crude extract, 3,4-dihydroxybenzoic acid has been identified. In addition, this molecule demonstrated strong antibacterial and antioxidant properties [70].
One common natural phenolic compound is 3,4-dihydroxybenzoic acid, often known as protocatechuic acid (PCA). PCA exhibits a wide range of pharmacological characteristics and may function as anticancer, anti-inflammatory, neuroprotective, antibacterial, antidiabetic, antiapoptotic, antitumor, and antiasthmatic drugs. Along with being an antioxidant, they may also promote cell proliferation and apoptosis, inhibit cell apoptosis, promote the autophagy-lysosome pathway, suppress oxidative stress damage, inhibit inflammatory responses, improve synaptic plasticity dysfunction, and provide neuroprotective factors. Atherosclerosis, diabetes, Alzheimer’s disease, cancer, and other illnesses may all benefit from PCA’s prevention and therapy. One kind of naturally occurring phenolic acid that is extensively distributed is PCA. PCA shares structural similarities with well-known antioxidant chemicals such as gallic acid, caffeic acid, vanillic acid, and syringic acid. Together with other potential mechanisms, such as anti-inflammatory qualities and interactions with many enzymes, PCA is an active ingredient found in over 500 plants that impart a variety of pharmacological activities [71–73]. An iron-chelating dihydrobenzoic acid called 2,3-dihydroxybenzoic acid was shown to have significant anti-biofilm activity against one of the tested Methicillin-Resistant Staphylococcus aureus strains, significantly reducing the biofilm formation [74]. The mechanism of action of 3,4-dihydroxybenzoic acid as an antimicrobial or antibacterial is understood. The modes of action of natural polyphenol antimicrobials include adenosine triphosphate depletion, proton motive force decay, rupture of cell membranes, and malfunctioning nucleic acid processes [75].
CONCLUSION
This study concludes by highlighting the biological activity and main active compound and S. geysiriensis strains that have shown activity against a variety of bacteria that cause UTI diseases. The findings suggest that the S. geysiriensis active compound (3,4-dihydroxybenzoic acid named PCA) acts on UTI-isolated bacteria after isolation and identification of these bacteria from clinical specimens. Since most germs can develop resistance to commercial antibiotics, this represents a novel pharmacological target that may not result in resistance. Determining the compounds’ structures in the active fractions is essential to increasing the output of extracts that include these chemicals. These results lay a strong basis for future studies investigating the therapeutic potential of S. geysiriensis metabolites and its isolated compounds in the treatment of UTIs, especially in light of the pressing need for novel antibiotics to fight antibiotic-resistant bacteria. Determining their significance in tackling the increasing problem of resistant bacterial infections will require further research into their antimicrobial range, method of action, and clinical usefulness. In addition, this article highlighted the new natural source of PCA, which has several pharmacological uses.
RECOMMENDATION
Streptomyces geysiriensis could be a promising microorganism for the improvement of antibacterial drugs against a wide range of UTI-pathogenic bacteria. Approving of S. geysiriensis as a natural source of PCA (3,4-dihydroxybenzoic acid), which has several pharmacological uses. Further clinical studies to find out the side effects of the separated compound are needed to confirm clinical uses.
ACKNOWLEDGMENTS
This study is supported via funding from Prince Sattam bin Abdulaziz University project number (PSAU/2025/R/1446).
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
Ethical approval details are given in the 'Materials and Methods' section.
DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Mitrea L, Medeleanu M, Pop CR, Rotar AM, Vodnar DC. Biotics (Pre-, pro-, post-) and uremic toxicity: implications, mechanisms, and possible therapies. Toxins 2023;15(9):548. CrossRef
2. Amiri F, Safiri S, Aletaha R, Sullman MJM, Hassanzadeh K, Kolahi AA, et al. Epidemiology of urinary tract infections in the Middle East and North Africa, 1990–2021. Trop Med Health 2025;53:16. CrossRef
3. Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269–84. CrossRef
4. Mancuso G, Midiri A, Gerace E, Marra M, Zummo S, Biondo C. Urinary tract infections: the current scenario and future prospects. Pathogens 2023;12(4):623. CrossRef
5. Ahmed SS, Shariq A, Alsalloom AA, Babikir IH, Alhomoud BN. Uropathogens and their antimicrobial resistance patterns: relationship with urinary tract infections. Int J Health Sci (Qassim). 2019;13(2):48–55.
6. Zhou Y, Zhou Z, Zheng L, Gong Z, Li Y, Jin Y, et al. Urinary tract infections caused by uropathogenic Escherichia coli: mechanisms of infection and treatment options. Int J Mol Sci. 2023;24(13):10537. CrossRef
7. Weigel LM, Anderson GJ, Tenover FC. DNA gyrase and topoisomerase IV mutations associated with fluoroquinolone resistance in Proteus mirabilis. Antimicrob Agents Chemother. 2002;46(8):2582–7. CrossRef
8. Erdem I, Kara Ali R, Ardic E, Elbasan Omar S, Mutlu R, Topkaya AE. Community-acquired lower urinary tract infections: etiology, antimicrobial resistance, and treatment results in female patients. J Glob Infect Dis. 2018;10(3):129–32. CrossRef
9. Al-Qurashi E, Elbnna K, Ahmad I, Abulreesh HH. Antibiotic resistance in Proteus mirabilis: mechanism, status, and public health significance. J Pure Appl Microbiol. 2022;16(3):1550–61. CrossRef
10. Kraszewska Z, Skowron K, Kwiecinska-Piróg J, Grudlewska-Buda K, Przekwas J, Wiktorczyk-Kapischke N, et al. Antibiotic resistance of Enterococcus spp. isolated from the urine of patients hospitalized in the University Hospital in North-Central Poland, 2016-2021. Antibiotics (Basel). 2022;11(12):1749. CrossRef
11. Li Y, Kumar S, Zhang L, Wu H, Wu H. Characteristics of antibiotic resistance mechanisms and genes of Klebsiella pneumoniae. Open Med (Wars). 2023;18(1):20230707. CrossRef
12. Mares C, Petca RC, Popescu RI, Petca A, Multescu R, Bulai CA, et al. Update on urinary tract infection antibiotic resistance-a retrospective study in females in conjunction with clinical data. Life (Basel). 2024;14(1):106. CrossRef
13. Salam MA, Al-Amin MY, Salam MT, Pawar JS, Akhter N, Rabaan AA, et al. Antimicrobial resistance: a growing serious threat for global public health. Healthcare (Basel). 2023;11(13):1946. CrossRef
14. Cag Y, Haciseyitoglu D, Ozdemir AA, Cag Y. Antibiotic resistance and bacteria in urinary tract infections in pediatric patients. Medeni Med J. 2021;36(3):217–24. CrossRef
15. Mechal T, Hussen S, Desta M. Bacterial profile, antibiotic susceptibility pattern and associated factors among patients attending adult OPD at Hawassa University comprehensive specialized hospital, Hawassa, Ethiopia. Infect Drug Resist. 2021;14:99–110. CrossRef
16. Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–18. CrossRef
17. Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Meier-Kolthoff JP, et al. Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol Rev. 2015;80(1):1–43. CrossRef
18. Alam K, Mazumder A, Sikdar S, Zhao YM, Hao J, Song C, et al. Streptomyces: the biofactory of secondary metabolites. Front Microbiol. 2022;13:968053. CrossRef
19. Uddin TM, Chakraborty AJ, Khusro A, Zidan BR, Mitra S, Emran TB, et al. Antibiotic resistance in microbes: history, mechanisms, therapeutic strategies and future prospects. J Infect Public Health 2021;14(12):1750–66. CrossRef
20. Khameneh B, Eskin NM, Iranshahy M, Fazly Bazzaz BS. Phytochemicals: a promising weapon in the arsenal against antibiotic-resistant bacteria. Antibiotics 2021;10(9):1044. CrossRef
21. Pradeepa P. Secondary metabolites synthesis in microorganisms. Indian J Appl Microbiol. 2019;22(2):26–32. CrossRef
22. Donald L, Pipite A, Subramani R, Owen J, Keyzers RA, Taufa T. Streptomyces: still the biggest producer of new natural secondary metabolites, a current perspective. Microbiol Res. 2022;13(3):418–65. CrossRef
23. Mohammadipanah F, Wink J. Actinobacteria from arid and desert habitats: diversity and biological activity. Front Microbiol. 2016;6:1541. CrossRef
24. Yaradoddi JS, Kontro MH, Banapurmath NR, Ganachari SV, Sulochana MB, Hungund BS, et al. Extremophilic actinobacteria. In: Yaradoddi JS, Kontro MH, Ganachari SV, editors. Actinobacteria. Rhizosphere biology. Singapore: Springer; 2021. CrossRef
25. Karthick Kumar SB, Akilandeswari P, Pradeep BV, Begam MS, Julbiharahamed K, Vijayaselvendran RK. Isolation and identification of Streptomyces geysiriensis IN7 from marine water and evaluating the efficiency of their bioactive compounds against commonly exposed multidrug-resistant pathogens. Biol Bull Rev. 2024;14(Suppl 3):S254–64. CrossRef
26. Abdel Bast RA, Hanora A, Zaky M, Kobisi A. Antibacterial activity of Streptomyces sp. AMM1 metabolites isolated from Marsa Matrouh soil. Alfarama J Basic Appl Sci. 2024;5(3):320–32. CrossRef
27. Graham JC, Galloway AA. ACP best practice No 167: the laboratory diagnosis of urinary tract infection. J Clin Pathol. 2001;54(12):911–9. CrossRef
28. Rohde M. The Gram-positive bacterial cell wall. Microbiol Spectr. 2019;7(3):10–128. CrossRef
29. Paray AA, Singh M, Mir MA, Kaur A. Gram staining: a brief review. Int J Res Rev. 2023;10(9):336–41. CrossRef
30. Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962;5(1):109–18. CrossRef
31. Cui XL, Mao PH, Zeng M, Li WJ, Zhang LP, Xu LH, et al. Streptimonospora salina gen. nov., sp. nov., a new member of the family Nocardiopsaceae. Int J Syst Evol Microbiol. 2001;51(2):357–63. CrossRef
32. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25.
33. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–26.
34. Zhu ZJ, Schultz AW, Wang J, Johnson CH, Yannone SM, Patti GJ, et al. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat Protoc. 2013;8(3):451–60. CrossRef
35. Badawy SA, Hassan AR, Abu Bakr MS, Mohammed AEI. UPLC-qTOF-MS/MS profiling of phenolic compounds in Fagonia arabica L. and evaluation of their cholinesterase inhibition potential through in-vitro and in-silico approaches. Sci Rep. 2025;15:5244. CrossRef
36. Hudzicki J. Kirby-Bauer disk diffusion susceptibility test protocol. Am Soc Microbiol. 2009;15(1):1–23.
37. Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal. 2016;6(2):71–9. CrossRef
38. Scorzoni L, Sangalli-Leite F, de Lacorte Singulani J, Costa-Orlandi CB, Fusco-Almeida AM, Mendes-Giannini MJ. Searching new antifungals: the use of in vitro and in vivo methods for evaluation of natural compounds. J Microbiol Methods. 2016;123:68–78. CrossRef
39. Bader MS, Loeb M, Brooks AA. An update on the management of urinary tract infections in the era of antimicrobial resistance. Postgrad Med. 2017;129(2):242–58. CrossRef
40. Bader MS, Loeb M, Leto D, Brooks AA. Treatment of urinary tract infections in the era of antimicrobial resistance and new antimicrobial agents. Postgrad Med. 2020;132(3):234–50. CrossRef
41. Monciardini P, Iorio M, Maffioli S, Sosio M, Donadio S. Discovering new bioactive molecules from microbial sources. Microb Biotechnol. 2014;7(3):209–20. CrossRef
42. Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645–58. CrossRef
43. El-Sakhawy MA, Soliman GA, El-Sheikh HH, Ganaie MA. Anticandidal effect of Eucalyptus oil and three isolated compounds on cutaneous wound healing in rats. Eur Rev Med Pharmacol Sci. 2023;27(1):26–37.
44. Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981−2002. J Nat Prod. 2003;66(7):1022–37. CrossRef
45. Sato K, Ichiyama S, Ohmura M, Takashi M, Agata N, Ohta M, et al. A case of urinary tract infection caused by Bacillus cereus. J Inf Secur. 1998;36:247–8. CrossRef
46. Çoban B, Ülkü N, Kaplan H, Topal B, Erdogan H, Baskin E. Five-year assessment of causative agents and antibiotic resistances in urinary tract infections. Turk Pediatri Arsivi. 2014;49:124–9. CrossRef
47. Vasudevan R. Urinary tract infection: an overview of the infection and the associated risk factors. J Microbiol Exp. 2014;1:42–54. CrossRef
48. Nakamura T, Ishikawa K, Matsuo T, Kawai F, Uehara Y, Mori N. Enterococcus hirae bacteremia associated with acute pyelonephritis in a patient with alcoholic cirrhosis: a case report and literature review. BMC Infect Dis. 2021;21:999. CrossRef
49. Amaya-Tapia G, Ibarra-Nieto G, Rivas OC, Sánchez JLG. Urinary tract infection in HIV/AIDS patients. In: Hegazy DW, El FDAA, Lwegasila DLJ, editors. Urinary tract infections. Rijeka, Croatia: IntechOpen; 2023. CrossRef
50. Zhou G, Wang Q, Wang Y, Wen X, Peng H, Peng R, et al. Outer membrane porins contribute to antimicrobial resistance in gram-negative bacteria. Microorganisms. 2023;11(7):1690. CrossRef
51. Gebremedhin KB, Yisma E, Alemayehu H, Medhin G, Belay G, Bopegamage S, et al. Urinary tract infection among people living with human immunodeficiency virus attending selected hospitals in Addis Ababa and Adama, central Ethiopia. Front Public Health 2024;12:1394842. CrossRef
52. Al-Mathkhury H, Flayyih M, Alghrair Z. Pathological study on Staphylococcus xylosus isolated from patients with urinary tract infections. J Al-Nahrain Univ. 2008;11:123–30. CrossRef
53. Husna A, Rahman MM, Badruzzaman ATM, Sikder MH, Islam MR, Rahman MT, et al. Extended-spectrum β-lactamases (ESBL): challenges and opportunities. Biomedicines. 2023;11(11):2937. CrossRef
54. Gaurav A, Bakht P, Saini M, Pandey S, Pathania R. Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitors. Microbiology (Reading) 2023;169(5):001333. CrossRef
55. Khattab AI, Babiker EH, Saeed HA. Streptomyces: isolation, optimization of culture conditions and extraction of secondary metabolites. Int Curr Pharm J. 2016;5(3):27–32. CrossRef
56. Rammali S, Rahim A, El Aalaoui M, Bencharki B, Dari K, Habach A, et al. Antimicrobial potential of Streptomyces coeruleofuscus SCJ isolated from microbiologically unexplored garden soil in Northwest Morocco. Sci Rep. 2024;14(1):3359. CrossRef
57. Selvameenal L, Radhakrishnan M, Balagurunathan R. Antibiotic pigment from desert soil actinomycetes; biological activity, purification and chemical screening. Indian J Pharm Sci. 2009;71(5):499. CrossRef
58. Rante H, Alam G, Usmar U, Zahra S, Kurniawati A, Ali A. Antimicrobial activity of Streptomyces spp. sponge-associated isolated from Samalona Island of South Sulawesi, Indonesia. Biodiv J Biol Diversity. 2022 Mar 2;23(3):1392–8. CrossRef
59. Apsari PP, Budiarti SR, Wahyudi AT. Actinomycetes of rhizosphere soil producing antibacterial compounds against urinary tract infection bacteria. Biodiv J Biol Diversity. 2019;20(5):1259–65. CrossRef
60. Ambarwati A, Wahyuono S, Moeljopawiro S, Yuwono T. Antimicrobial activity of ethyl acetate extracts of Streptomyces sp. CRB46 and the prediction of their bioactive compounds chemical structure. Biodiv J Biol Diversity. 2020;21(7):3380–90. CrossRef
61. Shah I, Uddin Z, Hussain M, Khalil AAK, Amin A, Hanif F, et al. Streptomyces sp. from desert soil as a biofactory for antioxidants with radical scavenging and iron chelating potential. BMC Microbiol. 2024;24(1):419. CrossRef
62. Kowalska-Krochmal B, Dudek-Wicher R. The minimum inhibitory concentration of antibiotics: methods, interpretation, clinical relevance. Pathogens 2021;10(2):165. CrossRef
63. Maiti PK, Das S, Sahoo P, Mandal S. Streptomyces sp SM01 isolated from Indian soil produces a novel antibiotic picolinamycin effective against multi drug resistant bacterial strains. Sci Rep. 2020;10(1):10092. CrossRef
64. Chanthasena P, Hua Y, Rosyidah A, Pathom-Aree W, Limphirat W, Nantapong N. Isolation and identification of bioactive compounds from Streptomyces actinomycinicus PJ85 and their in vitro antimicrobial activities against methicillin-resistant Staphylococcus aureus. Antibiotics (Basel) 2022;11(12):1797. CrossRef
65. Cha JW, Piao MJ, Kim KC, Zheng J, Yao CW, Hyun CL, et al. Protective effect of 3,4-dihydroxybenzoic acid isolated from Cladophora wrightiana harvey against ultraviolet B radiation-induced cell damage in human HaCaT keratinocytes. Appl Biochem Biotechnol. 2014;172:2582–92. CrossRef
66. Westervelt P, Bloom ML, Mabbott GA, Fekete FA. The isolation and identification of 3, 4-dihydroxybenzoic acid formed by nitrogen-fixing Azomonas macrocytogenes. FEMS Microbiol Lett. 1985;30(3):331–5. CrossRef
67. Guria M, Mitra P, Ghosh T, Gupta S, Basu B, Mitra PK. 3, 4-dihydroxybenzoic acid Isolated from the leaves of Ageratum conyzoides L. Eur J Biotechnol Biosci. 2013;1:25–8.
68. Ebrahimi-Zarandi M, Bonjar GH, Riseh RS, El-Shetehy M, Saadoun I, Barka EA. Exploring two Streptomyces species to control Rhizoctonia solani in tomato. Agronomy. 2021;11(7):1384. CrossRef
69. Khan A, Haque E, Mukhlesur RM, Mosaddik A, Rahman M, Sultana N. Isolation of antibacterial constituent from rhizome of Drynaria quercifolia and its sub-acute toxicological studies. Daru J Pharm Sci. 2007;15:205–11.
70. Syafni N, Putra DP, Arbain D. 3, 4-dihydroxybenzoic acid and 3, 4-dihydroxybenzaldehyde from the fern Trichomanes chinense L.; isolation, antimicrobial and antioxidant properties. Indones J Chem. 2012;12(3):273–8. CrossRef
71. Kakkar S, Bais S. A review on protocatechuic acid and its pharmacological potential. ISRN Pharmacol. 2014:952943. CrossRef
72. Song J, He Y, Luo C, Feng B, Ran F, Xu H, et al. New progress in the pharmacology of protocatechuic acid: a compound ingested in daily foods and herbs frequently and heavily. Pharmacol Res. 2020;161:105109. CrossRef
73. Liang S, Zhao Z, Liu L, Zhang Y, Liu X. Research progress on the mechanisms of protocatechuic acid in the treatment of cognitive impairment. Molecules 2024;29(19):4724. CrossRef
74. Mangzira Kemung H, Tan LT, Chan KG, Ser HL, Law JW, Lee LH, et al. Streptomyces sp. strain MUSC 125 from mangrove soil in Malaysia with anti-MRSA, anti-biofilm and antioxidant activities. Molecules. 2020;25(15):3545. CrossRef
75. Panda L, Duarte-Sierra A. Recent advancements in enhancing antimicrobial activity of plant-derived polyphenols by biochemical means. Horticulturae 2022;8(5):401. CrossRef