INTRODUCTION
Indonesia is famous for its abundant natural and marine biodiversity due to its distinct coastal and marine ecosystems. Around 25,000 square kilometers of coral reefs are mapped in its waters. There is a variety of life, including around 8,500 species of fish, 555 species of seaweed, and 950 coral reef biotas. In the waters of Podang Island, South Sulawesi, various types of sea sponges are found. The bioactive compounds contained are reported to have various potential activities, especially the alkaloid group [1]. Sponges in the phylum Porifera represent a diverse group of aquatic animals that have a unique composition, lacking true tissues and organs, thus relying solely on specialized cells that perform different functions, such as capturing food particles and releasing enzymes that provide structural support [2–4].
Sponges are known for their diverse secondary metabolites, which are much more abundant than in other animals. These compounds have cytotoxic, antibacterial, hemolytic, and various other biological activities [5,6]. One that has the potential to be studied further regarding its compound content and potential activity is the Aaptos sponge type. The main secondary metabolite groups of sponges of the genus Aaptos mostly contain alkaloids such as aaptamine derivatives [7,8]. Alkaloids are unique and promising compounds as potential drug candidates [9]. Derivatives of these alkaloids have inherent properties such as water solubility, acidity, and lipid solubility, which are the basis for their use as medicinal compounds. Alkaloids play an important role in both human medicine and the natural defense mechanisms of organisms. Therapeutically, alkaloids are best known for their role as anesthetics, cardioprotective agents, and anti-inflammatory agents. In addition, alkaloids are also widely known for their clinical applications, especially in treatments targeting conditions such as antibacterials and anti-malarials [10–12].
Malaria is an infectious disease that is still a health concern in several regions of Indonesia, especially in the eastern region. The number of malaria cases in Indonesia in 2021 was 304,607, a decrease compared to the number of cases in 2009 (418,439 cases). Based on these figures, the number of malaria cases expressed as an indicator of annual parasite incidence (fire) is 1.1 cases per 1,000 people. This high number of cases shows the importance of addressing the continuing prevalence of malaria in Indonesia. Malaria in Indonesia is caused by protozoan parasites from the genus Plasmodium, namely: Plasmodium falciparum, Plasmodium vivax, Plasmodium ovale, and Plasmodium malariae, which infect red blood cells in humans [13,14]. The World Health Organization recommends artemisinin-based combination therapy as the primary treatment for malaria. Artemisinin is extracted from the Artemisia annua plant and quinine is from the bark of the cinchona tree. However, inappropriate use of artemisinin in the treatment of malaria can cause resistance to the P. falciparum parasite due to gene mutations [15,16]. Artemisinin effectively inhibits the growth of malaria parasites (for 1, 3, and 7 days), but the parasites can reappear after 28 days [17,18].
The development of parasite resistance to anti-malarial drugs poses a significant challenge to their efficacy [19]. The P. falciparum parasite that causes malaria has developed resistance to antifolate drugs such as pyrimethamine, which targets the parasite enzyme dihydrofolate reductase (PfDHFR). This resistance is widespread in combinations of antifolate and sulfa drugs, so to overcome this problem, new strategies have been designed to create antifolate compounds that remain effective against PfDHFR mutations found in resistant parasites [20,21]. Therefore, this research aims to identify the content of alkaloid compounds in the sea sponge Aaptos suberitoides, originating from South Sulawesi. It is hoped that this can be used as a guide in the discovery of potential new drugs for the better treatment of malaria.
MATERIALS AND METHODS
Materials
The primary material utilized in this study was the A. suberitoides sponge from South Sulawesi, identified at the Laboratory of Biology, UGM. Chemicals employed for extraction and fractionation included dichloromethane (DCM), methanol, ethyl acetate, and n-hexane. All organic solvents utilized were of analytical grade. Silica gel with various sizes, octadecylsilane (ODS) Fuji Sylisia, thin layer chromatography plates (Merck), and ODS plates were used for purification processes. For visualization, a 10% sulfuric acid in Ethanol (EtOH) staining reagent was employed. For anti-malarial testing using the P. falciparum strain falciparum chloroquine resistant, , the following materials were utilized: Rosewell Park Memorial Institute (RPMI) 1,640 medium containing L-glutamine, 2-hydroxyethylpiperazine-N-2-ethanesulfonic acid, 5% sodium bicarbonate solution (NaHCO3), injectable gentamicin sulfate antibiotic, serum, O-group red blood cells, sorbitol (Merck), dimethyl sulfoxide (DMSO) (1/10,000 ppm DMSO), saponin (Sigma), Phosphate Buffered Saline, ammonium persulfate, citrate phosphate dextrose anticoagulant solution, Giemsa stain, and pH 7.2 phosphate buffer solution.
Extraction, phytochemical screening of sponges
A 300-g sample of A. suberitoides sponge was finely chopped and subjected to maceration using a solvent mixture of DCM and MeOH in a 1:1 (v/v) ratio totaling 400 ml for 2 × 1 hour at room temperature. The resulting maceration is put into a separating funnel so that it forms two phases and is separated into methanol extract and DCM extract. The DCM extract was evaporated using an evaporator below its boiling point, approximately ±35°C, resulting in a concentrated DCM extract. The concentrated DCM extract was introduced into a separating funnel, ethyl acetate, and water were added, and the mixture was vigorously shaken. After settling, it separated into two layers: the ethyl acetate layer and the water layer. The ethyl acetate layer was then evaporated at 77°C to obtain concentrated ethyl acetate extract. The concentrated ethyl acetate extract was subjected to phytochemical screening and antimalarial activity testing. Phytochemical screening was carried out using standard reagents and procedures to identify alkaloids, flavonoids, terpenoids, and steroids [22–25]. This extract underwent separation using column chromatography on silica gel (230–400 mesh), employing a gradient system. Subsequently, fractions were obtained. These fractions were further separated using column chromatography on silica gel (230–400 mesh) with a solvent system of n-hexane: ethyl acetate to yield pure compounds [20,26]. The purified fraction from the ethyl acetate extract was then tested for antimalarial activity in vitro using heme polymerization and antiplasmodial methods, and the compound content of the most potent fraction was identified using a liquid chromatography-high resolution mass spectrometry (LC-HRMS) instrument.
Identification of A. suberitoides fraction compounds using LC-HRMS
The identification of compounds contained in the A. suberitoides sponge fraction, using an LC-HRMS instrument, was done according to the compound analysis procedure described in previous research [27].
In-vitro antimalarial test
Heme polymerization inhibitory activity assay
The heme polymerization test was carried out following a method previously reported, with slight modifications [28]. The test group and positive control (chloroquine diphosphate) were prepared in various concentrations: 20, 10, 5, 2.5, and 1.25 mg/ml. 100 μl of each concentration variation was put into a microtube, and then 200 μl of hematin solution was added to each concentration series, followed by 100 μl of 100% glacial acetic acid solution (pH 2.6). The mixture was then incubated at 37°C for 24 hours. After the incubation period, each test sample in the microtube was centrifuged at 8,000 rpm for 10 minutes. The sediment and supernatant were separated, and the supernatant was discarded. The sediment was washed by adding 400 μl of 100% DMSO, centrifuged again at 8,000 rpm for 10 minutes, and the supernatant was discarded. This washing step was repeated three times. The obtained sediment was then treated with 400 μl of 0.1 M NaOH, and 100 μl was transferred to a 96-well microplate to measure the absorbance using an ELISA reader.
Antiplasmodial assay
Cultured Plasmodium was synchronized by adding 5% D-sorbitol. Plasmodium was transferred from the culture flask to a conical tube and centrifuged at 1,000 rpm for 10 minutes. After discarding the supernatant, sterile 5% sorbitol was added and incubated for 10 minutes at 37°C. The Plasmodium suspension was centrifuged again; the supernatant was discarded, and the Plasmodium was washed by adding culture media. The Plasmodium suspension was centrifuged once more, and the supernatant was discarded, resulting in Plasmodium in the form of rings only. Parasitemia was calculated from thin blood preparations. Testing was conducted using 1% parasitemia at a hematocrit of 2% in RPMI medium supplemented with 10% human serum.
The samples and chloroquine, used as a control, were dissolved in the RPMI medium. The test compounds, at various concentrations (21.25; 62.5; 125; 250; and 500 μg/ml) of 100 μl each, were added to a 96-well microplate, followed by the addition of 100 μl of Plasmodium suspension. Each concentration series was repeated three times. The microplate was then incubated at 37°C for 72 hours. At the end of the incubation period, thin blood smears were prepared using 10% Giemsa stain and observed under a microscope at 1,000× magnification. The percentage of parasitemia (counting a minimum of 1,000 erythrocytes) was calculated from thin blood smears and used to determine the percentage of growth inhibition against Plasmodium. Plasmodium culture without test compounds was considered to have 100% growth as a control [29,30]. The antiplasmodial activity was expressed as IC50, which is the concentration of a compound required to inhibit 50% of Plasmodium growth. The IC50 value was calculated using probit analysis with SPSS software.
Computation elementals
The hardware used is a computer equipped with a 13th Gen Intel i7-13700KF CPU running at 5300 GHz, 64 GB RAM with an Ubuntu 22.04.3 LTS x86_64 operating system, and an Nvidia GPU. The software utilized consists of YASARA Version 20.810 [31,32] as the primary software for molecular docking and molecular dynamics (MD) simulations. Discovery Studio by Biovia was employed for the analysis of molecular docking and MD interactions. ChemDraw by PerkinElmer was used to draw ligand structures.
Redocking (validation method)
The process of redocking natural ligands into proteins was carried out using YASARA software. Fourfold mutant P. falciparum dyhydrofolate reductase protein and double mutant P. falciparum dyhydrofolate reductase protein with PDB codes 1J3K and 1J3J. Each was made by separating proteins and their natural ligands from other molecules, such as water molecules, Ca atoms, and other ligands. Next, the protein’s pH was adjusted using the default setting of the YASARA software. Then, the protein was “Clean All” to display the hydrogen atoms, and energy minimization was performed to correct the angles and distances of the protein molecules. After preparation was carried out, the protein was set to become rigid and the redocking process was carried out using additional methods from molmod.id [33,34].
Re-docking of the natural ligand was carried out on the active site of the protein using the VINA autodock method with the AMBER03 force field for 100 repetitions, with each repetition producing the 25 best conformations of the natural ligand to form a bond with the active site. The results of re-docking using additional methods from molmod.id will automatically produce protein files that have been separated from their ligands, ligands before and after re-docking, and root mean square deviation (RMSD) data. Re-docking is said to be accurate if it produces an RMSD value of <2 Å and forms a ligand-receptor interaction that is similar to the ligand-receptor bond in the protein crystal structure [13,14].
Molecular docking
The molecular docking study began with energy minimization for each ligand, both natural compounds and acarbose as a positive control using the default settings of the YASARA software. Molecular docking parameters were carried out using re-docking parameters that had been done previously. The results of molecular docking were analyzed by paying attention to the bonds formed between the ligand and the receptor. Molecular docking is expected to form interactions with the main catalytic residues, where the main catalytic residues in the 1J3K protein are residues Ile14, Cys15, Asp54, Met55, Phe58, Leu119, Ile164, and Tyr170. In the 1J3J protein, namely le14, Cys15, Asp54, Phe58, Pro113, and Ile164. Apart from the interaction of amino acid residues with various specific amino acids, the natural ligands of 1J3K and 1J3J have a more specific interaction, namely Asp54 [17,35].
RESULT AND DISCUSSION
Aaptos suberitoides sponge extract yield and phytochemical analysis
The sponge used in this research came from the waters of South Sulawesi and was the result of sample determination carried out by an animal systematist from the animal systematics Laboratory of the Biology Faculty. The sample was named A. suberitoides in Latin (number: 008/B1/SH/111/2024). The extract obtained was partitioned using ethyl acetate solvent, and 0.2 g (0.067%) was obtained in the form of a paste and greenish-brown in color, which was then referred to as ethyl acetate extract. The extract obtained was further investigated to determine the content of phytochemical compounds using standard reagents. This information is very important to reflect success during the extraction process of secondary metabolites contained in the sample. The extraction yield is closely related to the active compound content in the sample, implying that a higher extraction yield is correlated with an increased active compound content [36,37]. The results of the phytochemical screening of the partitioned ethyl acetate extract were positive for alkaloid, terpenoid, and steroid compounds.
The results of hematin polymerization inhibition
Antimalarial activity testing was carried out on all fractions resulting from column chromatography. In its life cycle, P. falciparum multiplies and breaks down hemoglobin in food vacuoles to obtain nutrients. This hemoglobin breakdown process produces free globin and heme molecules, which are toxic to Plasmodium and are known as ferriprotoporphyrin IX (FPIX). Plasmodium utilizes globin to survive in host cells because it can be broken down into amino acids for protein synthesis. The toxin-free heme, FPIX, then breaks down into a harmless substance known as hemozoin (malarial pigment) [38,39]. The ability of an antiplasmodial agent to inhibit hematin polymerization is directly related to its effectiveness as an antimalarial agent, although it is known that the mechanism of action of antiplasmodial agents is not limited to inhibiting hematin polymerization. The activity of inhibiting hematin polymerization actually involves one or two mechanisms. First, there is an interaction between terpenoid compounds, phenols, or sterols and the hematin electron system. Second, the extract contains compounds that have hydroxyl groups that are able to bind to iron ions in hematin.
The B-hematin compound is a heme crystal that has a chemical structure identical to hemozoin. The use of fourier transform infrared spectroscopy spectrophotometry can show the bond between the iron-carboxylate ion of two heme molecules in hemozoin, which has the same analog, namely B-hematin [38,40,41]. Therefore, in in vitro assays, inhibition of B-hematin formation is considered an ideal target for antimalarial drugs. The results of heme polymerization inhibition are presented in Table 1, which shows that fraction number 12 has an antimalarial activity with an IC50 value of 0.423± 0.243 µg/ml. The activity of fraction 12 was the best against antimalarials tested by the heme polymerization inhibition method compared to other fractions. Furthermore, antiplasmodium activity testing was carried out using chloroquine as a positive control. The results of antiplasmodium activity testing are presented in Table 2.
 | Table 1. Heme polymerization inhibition by fractions from EtOAc extract. [Click here to view] |
 | Table 2. Antiplasmodial activity of fractions obtained by column chromatography of the EtOAc extract. [Click here to view] |
Chloroquine, an antimalaria drug that was used as positive control, had an IC50 of 11.13 ± 4.23 µg/ml (Table 2). On the other hand, the EtOAc extract of A. suberitoides sponge showed an IC50 of 11.65 ± 4.76 µg/ml (fraction 8), 10.21 ± 3.63 µg/ml (fraction 12). Remarkably, fraction 12 showed inhibitory potential comparable to chloroquine. These results indicate the potential for new alternatives to be developed as promising antimalarial agents. Given that the active compounds are sourced from natural materials and common antimalarial drugs cause side effects of resistance. Therefore, further investigation is needed on the potential and side effects of using these bioactive compounds as antimalarials. Based on the antimalarial activity value (IC50), it is categorized as if IC50 ≤10 µg/ml is in the very good category, 10< IC50 <50 µg/ml is in the good category, and IC50 ≥50 µg/ml is in the poor category [42], then fractions 12, 8, and 7 are included in the good category.
The composition of sponge extract by LC-HRMS
The identification of bioactive compounds in sponge extracts was revealed using LC-HRMS, and the results are presented in Table 3. There were four compounds identified, which may be responsible for the antimalarial activity. These compounds included aaptamine and its derivatives, such as aaptanone, which is included in the alkaloid group. These findings are consistent with Herlt et al. [7]’s research in 2004, which highlighted alkaloids as typical compounds in sponges, especially the genus Aaptos, which mostly produces aaptamine derivatives alkaloids. LC-HRMS analysis produced a total of 5 compounds, including 3 previously identified compounds and 2 new potential compounds. However, the new compounds are coded NP-011220 and NP-013736 and have structures predicted using LC-HRMS, which are also presented in Figure 1 and Table 3. Based on the identification results, all compounds belong to the alkaloid group. Apart from that, two new potential compounds were identified in the extract, namely NP-011220 with a retention time of 4.941 minute, and NP-001501 with a retention time of 3.600 minute. A thorough literature review revealed that these two compounds had not been previously reported. Thus, it can be concluded that the ethyl acetate extract of the A. suberitoides sponge contains secondary metabolite alkaloid compounds.
 | Table 3. Analysis of compounds in fraction number 12 by the LC-HRMS instrument. [Click here to view] |
 | Figure 1. Compound identified in fraction number 12 by the LC-HRMS instrument. [Click here to view] |
Redocking and molecular docking
The validation method for molecular docking involves the process of redocking native ligands that are known to bind to the enzyme crystal structure. The validation is considered successful if the conformation of the native ligand after redocking closely matches its conformation when bound to the enzyme crystal. This match is assessed by comparing the redocked position of the native ligand to its crystallographic coordinates, using the RMSD value as a measure. A lower RMSD value indicates a higher degree of similarity between the redocked conformation and the original conformation of the native ligand on the enzyme crystal [38,40].
The redocking process to validate the method for the 1J3K and 1J3J (Fig. 2) proteins was carried out by setting the grid box size to 19.52 × 19.52 × 19.52 Å. The RMSD value of redocking was chosen to be the lowest among all conformations so that an RMSD value of 0.5469 Å for 1J3J and 0.7014 Å for 1J3K was obtained with a binding energy value of −8.9340 kcal/mol for 1J3J and −8.7470 kcal/mol for 1J3K. The RMSD value obtained meets the RMSD requirements for redocking, namely below 2.0 Å. So, the docking results have met the validity criteria and can be continued with docking test compounds obtained from the extract and EtOAc fraction of the sponge, A. suberitoides.
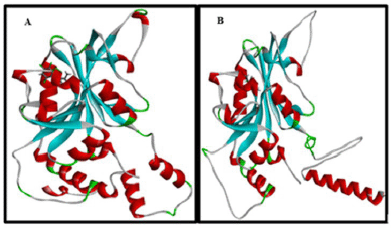 | Figure 2. Redocking of natural ligand, (A) 1J3K, and (B) 1J3J. [Click here to view] |
A visualization of the similarity of natural ligands after re-docking with natural ligands is shown in Figure 3. 2D visualization of the 1J3K Figure 4 shows the presence of hydrogen bonds that occur between the natural ligand and the amino acid residues Cys15, Asp54, Ile14, and Leu164, which are conventional hydrogen bond interactions. Meanwhile, the results of 2D visualization of the 1J3J Figure 5 redocking show that hydrogen bonds occur between the natural ligand and the amino acids Ile164, Ile14, Cys15, and Asp54. This interaction occurs between the H atom of the secondary amine group on the natural ligand and the O atom on the amino acid residue, which acts as a proton acceptor. Another interaction is in the form of a salt bridge between the O atom in the amino acid residue and the H atom from the tertiary amine group, which has a positive charge. This salt bridge interaction occurs when there are two opposite charges at a fairly close distance; this distance is usually below 4 Å [16,31]. This interaction with the natural ligand is in accordance with the journal [43]. The next study investigated how three compounds identified from fractions of 12 A. suberitoides sponge extracts analyzed by LC-HRMS interacted with the same proteins. The results of the molecular docking analysis are presented in Table 4, and images of the interactions between the three compounds on each protein are shown in Figure 4.
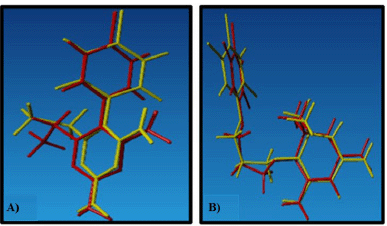 | Figure 3. Visualization of the similarity of natural ligands, (A) 1J3K and (B) 1J3J. [Click here to view] |
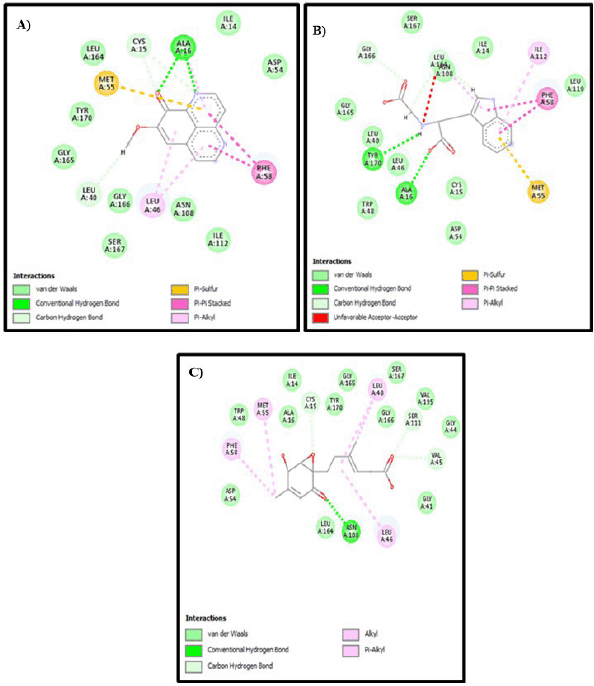 | Figure 4. 2D visualization of each compound with 1J3K protein: (A) Demethylaaptamine; (B) Conbamidine; and (C) Penicylone D. [Click here to view] |
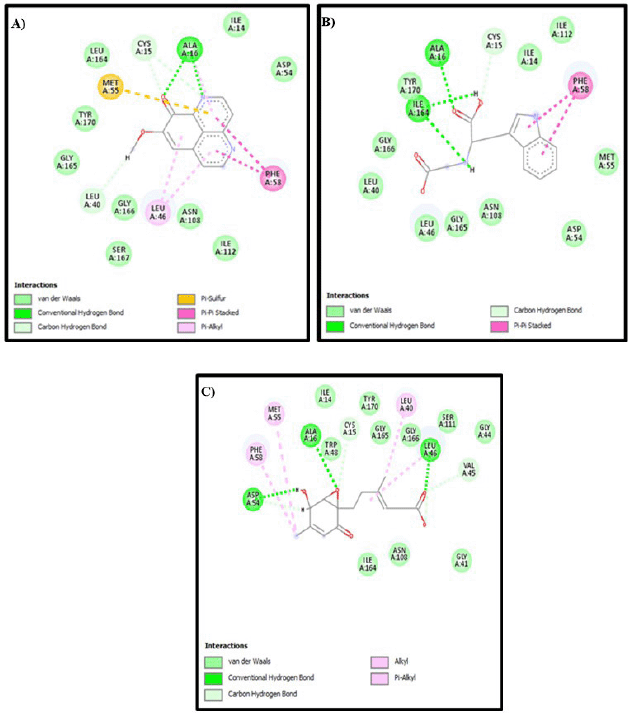 | Figure 5. 2D visualization of each compound with 1J3J protein: (A) Demethylaaptamine; (B) Conbamidine; and (C) Penicylone D. [Click here to view] |
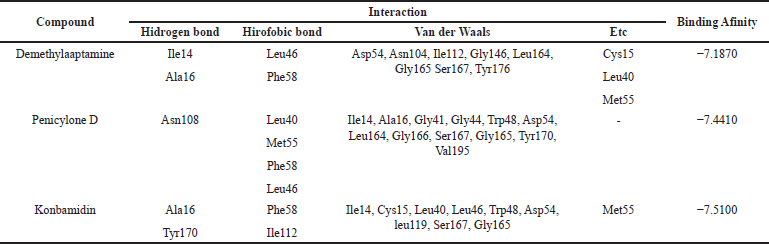 | Table 4. The results of molecular docking of each compound with 1J3K protein. [Click here to view] |
Based on the interaction of three compounds identified by LC-HRMS analysis, Fraction 12 contains metabolites with the ability to inhibit the growth of Plasmodium. This inhibitory effect is due to a specific hydrogen bond interaction between the bioactive compound (demethylaaptamine) and a specific amino acid (Ile14) from the Plasmodium protein code IJ3K, which is shown in Table 4 and Figure 4. In addition, each bioactive compound shows a specific hydrogen bond interaction with Plasmodium amino acids (protein code 1J3J), as shown in Table 5 and Figure 5. However, looking at the number of hydrogen bond interactions formed, Penicylone D has more potential to show antimalarial activity because it has three hydrogen bonds (Ala16, Leu46, and Asp54), while the others have two hydrogen bonds each. Molecular docking studies of the two compounds coded NP-011220 and NP-013736 are not reported, considering that further structural analysis is needed to confirm their molecular structure.
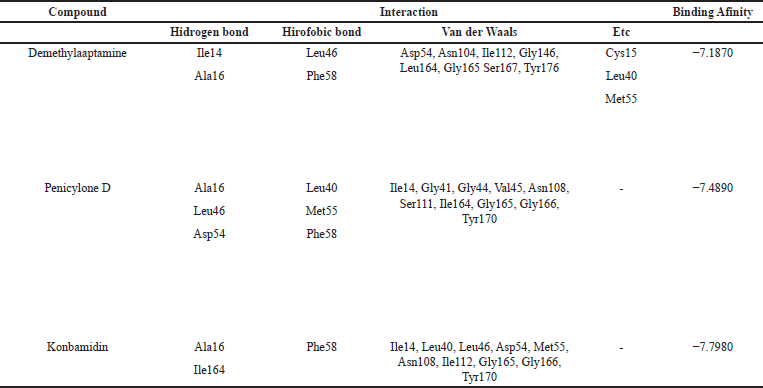 | Table 5. The results of molecular docking of each compound with 1J3J protein. [Click here to view] |
CONCLUSION
Based on the research that has been carried out, it can be concluded that the extraction of A. suberitoides sponge samples from the waters of Podang-Podang Island, South Sulawesi, shows antimalarial activity based on in vitro test analysis using the HEME polymerization method (IC50 of 0.4223 μg/ml) and the antiplasmodium method (IC50 10.21 ± 3.63 μg/ml), all of which are categorized as active antimalarials. The results of analysis using molecular docking studies show a positive correlation where the Demethylaaptamine compound has specific hydrogen interactions, and the Penicylone D compound has specific and more hydrogen interactions with the amino acids of the Plasmodium protein (code IJ3J). The two suspected new compounds with the codes NP-011220 and NP-013736 require further structural analysis and testing both in vitro, in silico, and in vivo.
ACKNOWLEDGMENTS
Wahyuni acknowledges LPDP (Lembaga Pengelola Dana Pendidikan), Ministry of Finance, the Republic of Indonesia, for the Postgraduate Indonesia Education Scholarship Program and thank you to Universitas Gadjah Mada for funding this research through the 2024 RTA program (number: 5286/UN1.P1/PT.01.03/2024).
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Abdelmohsen UR, Bayer K, Hentschel U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat Prod Rep. 2014;31:381–99. CrossRef
2. Engelberts JP, Robbins SJ, de Goeij JM, Aranda M, Bell SC, Webster NS. Characterization of a sponge microbiome using an integrative genome-centric approach. ISME J. 2020;14:1100–10. CrossRef
3. Imhoff JF, Truper HG, Pfennig N. Rearrangement of the species and genera of the phototrophic “purple nonsulfur bacteria.” Int J Syst Bacteriol. 1984;34:340–3. CrossRef
4. Moeller FU, Webster NS, Herbold CW, Behnam F, Domman D, Albertsen M, et al. Characterization of a thaumarchaeal symbiont that drives incomplete nitrification in the tropical sponge Ianthella basta. Environ Microbiol. 2019;21:3831–54. CrossRef
5. Orani AM, Barats A, Vassileva E, Thomas OP. Marine sponges as a powerful tool for trace elements biomonitoring studies in coastal environment. Mar Pollut Bull. 2018;131:633–45. CrossRef
6. Skropeta D, Wei L. Recent advances in deep-sea natural products. Nat Prod Rep. 2014;31:999–1025. CrossRef
7. Herlt A, Mander L, Rombang W, Rumampuk R, Soemitro S, Steglich W, et al. Alkaloids from marine organisms. Part 8: isolation of bisdemethylaaptamine and bisdemethylaaptamine-9-O-sulfate from an Indonesian Aaptos sp. marine sponge. Tetrahedron. 2004;60:6101–4. CrossRef
8. Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev. 2007;71:295–347. CrossRef
9. Sevindik M, Mohammed FS, Uysal I. Autism: plants with neuro-psychopharmacotherapeutic potential. Prospects Pharm Sci. 2023;21:38–48. CrossRef
10. Christopher R, Msonga A, Hoppe HC, Boyom FF. Ethanol extracts from selected Tanzanian medicinal plants selectively inhibit Plasmodium falciparum growth in vitro. Tanzania J Sci. 2023;49:41–7. CrossRef
11. Mohammed FS, Uysal ?, Sevindik M. A review on antiviral plants effective against different virus types. Prospects Pharm Sci. 2023;21:1–21. CrossRef
12. Uysal I, Koçer O, Mohammed FS, Lekesiz Ö, Do?an M, ?abik AE, et al. Pharmacological and nutritional properties: genus Salvia. Adv Pharmacol Pharm. 2023;11:140–55. CrossRef
13. Wicht KJ, Mok S, Fidock DA. Molecular mechanisms of drug resistance in Plasmodium falciparum Malaria. Annu Rev Microbiol. 2020;74:431–54. CrossRef
14. Mensah BA, Aydemir O, Myers-Hansen JL, Opoku M, Hathaway NJ, Marsh PW, et al. Antimalarial drug resistance profiling of Plasmodium falciparum infections in Ghana using molecular inversion probes and next-generation sequencing. Antimicrob Agents Chemother. 2020;64:1–17. CrossRef
15. Astuti E, Raharjo TJ, Boangmanalu PM, Putra ISR, Waskitha SSW, Solin J. Synthesis, molecular docking, and evaluation of some new curcumin analogs as antimalarial agents. Indones J Chem. 2021;21:452–61. CrossRef
16. Nisa SA, Jumina J, Mardjan M, Kurniawan YS. View of synthesis, activity test and molecular docking of novel nitrophenylcalix[4]-2-methylresorcinarene derivatives as antimalarial agent. Molekul. 2023;18:404–13.
17. Wood BR, Langford SJ, Cooke BM, Glenister FK, Lim J, McNaughton D. Raman imaging of hemozoin within the food vacuole of Plasmodium falciparum trophozoites. FEBS Lett. 2003;554:247–52. CrossRef
18. Turkiewicz A, Manko E, Sutherland CJ, Benavente ED, Campino S, Clark TG. Genetic diversity of the Plasmodium falciparum GTP-cyclohydrolase 1, dihydrofolate reductase and dihydropteroate synthetase genes reveals new insights into sulfadoxine-pyrimethamine antimalarial drug resistance. PLoS Genet. 2020;16:e1009268. CrossRef
19. Fadel Diagana M, Bira Gueye S. Analysis, design, and test of CDMA LFSR with offset mask using standard ICs. Engineering. 2016;8:226–31. CrossRef
20. Gurning K, Haryadi W. Potential antioxidants of secondary metabolite isolates ethyl acetate fraction Coleus amboinicus Lour. Leaves. ScienceRise: Pharm Sci. 2022;39:100–5. CrossRef
21. Sinaga SP, Lumbangaol DA, Situmorang R, Gurning K. Determination of phenolic, flavonoid content, antioxidant and antibacterial activities of seri (Muntingia calabura L.) leaves ethanol extract from North Sumatera, Indonesia. Rasayan J Chem. 2022;15:2022. CrossRef
22. Kibungu WC, Clarke A, Fri J, Njom HA. Antimicrobial potential and phytochemical screening of Clathria sp . 1 and Tedania (Tedania ) Stylonychaeta sponge crude extracts obtained from the South East coast of South Africa. Biomed Res Int. 2021;2021:1–10. CrossRef
23. Situmorang RFR, Gurning K, Kaban VE, Butar-Butar MJ, Perangin-Angin SAB. Determination of total phenolic content, analysis of bioactive compound components, and antioxidant activity of ethyl acetate seri (Muntingia calabura L.) leaves from North Sumatera province, Indonesia. Open Access Maced J Med Sci. 2022;10:240–4. CrossRef
24. Lubis MF, Kaban VE, Gurning K, Parhan P, Syahputra H, Aira Juwita N, et al. Phytochemicals and biological activities of ethanolic extract of Garcinia atroviridis leaf grown in Indonesia. J Med Chem Sci. 2023;6:2456–69. CrossRef
25. Sinaga SP, Situmorang RFR, Singarimbun N, Lestari W, Gurning K. Determination of total phytochemical compounds from ethanol extract nangka (Artocarpus heterophyllus Lam.) leaves and antioxidant activity from North Sumatera, Indonesia. Rasayan J Chem. 2023;16:19–23. CrossRef
26. Gurning K, Haryadi W, Sastrohamidjojo H. Isolation and characterization of antioxidant compounds of bangun-bangun (Coleus amboinicus, L.) leaves from North Sumatera, Indonesia. Rasayan J Chem. 2021;14:248–53. CrossRef
27. Gurning K, Suratno S, Astuti E, Haryadi W. Untargeted LC/HRMS metabolomics analysis and anticancer activity assay on MCF-7 and A549 cells from Coleus amboinicus Lour leaf extract. Iran J Pharm Res. 2024;23(1):e143494. CrossRef
28. Basilico N, Pagani E, Monti D, Olliaro P, Taramelli D. A microtitre-based method for measuring the haem polymerization inhibitory activity (HPIA) of antimalarial drugs. J Antimicrob Chemother. 1998;42:55–60. CrossRef
29. Quadros HC, Silva MCB, Moreira DRM. The role of the iron Protoporphyrins Heme and Hematin in the antimalarial activity of Endoperoxide drugs. Pharmaceuticals. 2022;15:60. CrossRef
30. Zakiah M, Syarif RA, Mustofa M, Jumina J, Fatmasari N, Sholikhah EN. In vitro antiplasmodial, heme polymerization, and cytotoxicity of hydroxyxanthone derivatives. J Trop Med. 2021;2021:1–11. CrossRef
31. Krieger E, Vriend G. YASARA view—molecular graphics for all devices—from smartphones to workstations. Bioinformatics. 2014;30:2981–2. CrossRef
32. Kurniawan YS, Yudha E, Nugraha G, Fatmasari N, Pranowo HD, Jumina J, et al. Molecular docking and molecular dynamic investigations of xanthone-xhalcone derivatives against epidermal growth factor receptor for preliminary discovery of novel anticancer agent. Indones J Chem. 2024;24:250–66. CrossRef
33. Haryadi W, Gurning K, Astuti E. Molecular target identification of two Coleus amboinicus leaf isolates toward lung cancer using a bioinformatic approach and molecular docking-based assessment. J Appl Pharm Sci. 2024;14:203–10. CrossRef
34. Moustakas DT, Lang PT, Pegg S, Pettersen E, Kuntz ID, Brooijmans N, et al. Development and validation of a modular, extensible docking program: DOCK 5. J Comput Aided Mol Des. 2006;20:601–19. CrossRef
35. Haryadi W, Pranowo HD. Molecular docking and dynamics analysis of halogenated imidazole chalcone as anticancer compounds. Pharmacia. 2023;70:323–9. CrossRef
36. Begum S, Munissi JJE, Buriyo AS, Makangara JJ, Lucantoni L, Avery VM, et al. Antiplasmodial, antimicrobial and cytotoxic activities of extracts from selected medicinal plants growing in Tanzania. J Biol Act Prod Nat. 2020;10:165–76. CrossRef
37. Nuzul MI, Jong VYM, Koo LF, Chan TH, Ang CH, Idris J, et al. Effects of extraction methods on phenolic content in the young bamboo culm extracts of Bambusa beecheyana Munro. Molecules. 2022;27:2359. CrossRef
38. Rajguru T, Bora D, Modi MK. Identification of promising inhibitors for Plasmodium haemoglobinase falcipain-2, using virtual screening, molecular docking, and MD simulation. J Mol Struct. 2022;1248:131427. CrossRef
39. Arnold MSJ, Macdonald JR, Quinn RJ, Skinner-Adams TS, Andrews KT, Fisher GM. Antiplasmodial activity of the natural product compounds alstonine and himbeline. Int J Parasitol Drugs Drug Resist. 2021;16:17–22. CrossRef
40. Zhou X, Ling M, Lin Q, Tang S, Wu J, Hu H. Effectiveness analysis of multiple initial states simulated annealing algorithm, a case study on the molecular docking tool AutoDock Vina. IEEE/ACM Trans Comput Biol Bioinform. 2023;20:3830–41. CrossRef
41. Tang S, Chen R, Lin M, Lin Q, Zhu Y, Ding J, et al. Accelerating AutoDock Vina with GPUs. Molecules. 2022;27:3041. CrossRef
42. Gessler MC, Nkunya MH, Mwasumbi LB, Heinrich M, Tanner M. Screening Tanzanian medicinal plants for antimalarial activity. Acta Trop. 1994;56(1):65–77.
43. Yuvaniyama J, Chitnumsub P, Kamchonwongpaisan S, Vanichtanankul J, Sirawaraporn W, Taylor P, et al. Insights into antifolate resistance from malarial DHFR-TS structures. Nat Struct Biol. 2003;10:357–65. CrossRef