INTRODUCTION
Tuberculosis (TB) disease, caused by Mycobacterium tuberculosis (Mtb), is the most frequent single infectious illness leading to global mortality [1]. Nevertheless, anti-TB drugs are more limited compared to agents existing for other bacterial infections [2]. Despite TB being declared as a global health crisis by WHO in 1993, only three new anti-TB drugs were approved and introduced to the market from 2012 to 2019: namely bedaquiline (approved by the Food and Drug Administration US in 2012), delamanid (approved by the European Medicines Agency Europe in 2014), and pretomanid (approved by the Food and Drug Administration US in 2019) [3,4]. However, the prevalence of TB drug resistance increases challenges in finding novel anti-TB drugs that are effective in shortening TB treatment [5]. Discovering new anti-TB agents involves time-intensive research due to the slow growth of Mtb. Another limitation in developing anti-TB agents is the necessity of a biosafety level 3 laboratory for conducting studies related to Mtb. Utilizing non-pathogenic and fast-growing Mycobacterium smegmatis for anti-TB investigation has exhibited several successes in past research. Mycobacterium smegmatis strains are commonly susceptible to TB medicines, such as isoniazid, rifampicin, and ethambutol [6]. It has also been well-studied that M. smegmatis is over 90% genetically identical to Mtb, shares common genes in stress response, and exhibits similarity in cell wall structure and metabolism [6,7]. Therefore, M. smegmatis serves as a suitable model for evaluating drug candidates against Mtb.
Endophytic microbes are promising sources of metabolites. These microbes live in plant tissues and play important roles in enhancing plant growth and protecting the host against pathogens [8]. Insights from ethnobotanical knowledge have proven valuable in identifying potential host plant reservoirs for endophytic microbes. Southeast Asia’s native plant, dogfruit (Archidendron pauciflorum), is locally used as a vegetable and is exploited for its therapeutic functions to cure several diseases, such as diarrhea, headaches, fevers, colds, coughs, and stomach-aches [9]. Exploiting the endophytic bacteria from this plant may provide a new alternative source of antimycobacterial agents. In vitro study supported by a genome mining approach could be an effective way to screen and evaluate endophytic bacteria as sources of secondary metabolites. The development of next-generation sequencing and powerful computational tools in the big-data era has provided researchers with a useful genomic database to easily identify diverse and novel secondary metabolite gene clusters (SMGCs) in the entire bacterial genome, which is beneficial for drug discovery [10].
In our previous studies, four endophytic Bacillus spp. isolated from A. pauciflorum exhibited antibacterial activity against antibiotic-sensitive strains (Staphylococcus aureus ATCC6538, Bacillus subtilis ATCC19659, Pseudomonas aeruginosa ATCC15442, and Escherichia coli ATCC8739), along with antibiotic-resistant strains (P. aeruginosa M19, B. subtilis M18, E. coli M4, and Klebsiella pneumoniae M19) [11,12]. However, the antimycobacterial activity of these isolates is yet to be investigated. Of note, numerous previous studies have shown that Bacillus spp. isolated from various environmental samples exhibit remarkable antimycobacterial activity. Some Bacillus strains isolated from the soil and water samples, as well as the endophyte Salicornia brachiata displayed antimycobacterial potency against Mycobacterium strains [13,14]. However, none of these studies have evaluated the effect of secondary metabolites derived from Bacillus on the inhibition of biofilm formation and eradication of Mycobacterium cell biofilms of Mycobacterium strains. This is an important insight because Mycobacterium is a biofilm-forming bacterium, and the biofilm structure promotes the persistence of the bacterium in response to antimicrobial agents. In addition, genome profiling of Bacillus is still underexplored, although it is beneficial for identifying novel SMGCs and predicting possible biosynthetic pathways related to antimycobacterial compound production. Therefore, the present study aimed to evaluate the antimycobacterial properties of four endophytic Bacillus spp. from A. pauciflorum and their inhibitory effects on M. smegmatis biofilm formation and cell biofilm eradication. Additionally, whole-genome analysis was conducted to identify secondary metabolite gene clusters (SMGCs) within the genome of the most promising endophyte isolate. The composition of volatile compounds (VOCs) in the extract was determined through analysis using gas chromatography-mass spectrometry (GC-MS). From this study, we have successfully explained the potential of endophytic Bacillus as an anti-TB agent. The data were proven from the results of in vitro analysis through antibacterial assays supported with antibiofilm tests. At the same time, we also managed to report genomic aspects by providing the whole genome sequence of the most potential Bacillus and identifying its SMGCs. The most potential Bacillus extract has also been profiled using GC-MS. These results are important reports that could incorporate the analysis of the potential of endophyte bacterial active compounds as new anti-TB agents.
MATERIALS AND METHODS
Source of endophyte isolates
Four endophytic bacteria were isolated from A. pauciflorum, namely Bacillus sp. strain DJ4, B. subtilis strain DJ7, Bacillus velezensis strain DJ9, and Bacillus megaterium strain AJ5 [12]. These isolates were stored at the Laboratory of Microbiology, Department of Biology, Faculty of Mathematics and Natural Sciences, IPB University, Indonesia. The 16S rRNA sequences of these isolates could be accessed under the NCBI accession numbers: PP178167.1, OR511995.1, OP164675.1, and OP164672.1, respectively.
Colony and cell morphology characterization
Four endophytic isolates were cultured in nutrient agar (NA) medium (Oxoid) and incubated for 24 hours. Colony characteristics, including shape, margin, elevation, optic, and pigmentation, were observed. Approximately 18-hour-old cultures were stained using the Gram staining technique to characterize their cell characteristics, such as shape, arrangement, and Gram reaction, and observed under a light microscope (Olympus CX31) with 1,000 × resolution. Additionally, the cell morphology of each isolate was also observed using a scanning electron microscope (SEM). The pellets from bacterial cultures aged 24 hours were rinsed using 100 µl of sterile distilled water. Afterward, 10 µl of the suspension was applied onto the surface of a single-polished Silicon Wafer (Sigma) and left to incubate for 18 hours. Subsequently, the SEM specimen was coated with sputter-gold particles (Hitachi®, Tokyo, Japan) and observed using an SEM-JEOL JSM-IT200 (JEOL, South Korea) at a resolution of 5,000 ×, with a working distance of 5 µm and an accelerating voltage of 10 kV.
Screening of antimycobacterial activity
Preliminary screening for antimycobacterial activity was performed using the streak plate method against M. smegmatis ATCC700084 (collection of the Research Center for Pharmaceutical Ingredients and Traditional Medicine, National Research, and Innovation Agency (BRIN), South Tangerang, Indonesia). Approximately 1 ml of M. smegmatis inoculum (1 × 108 CFU/ml) was added into 100 ml of Mueller Hinton Agar medium (Himedia) and poured into a petri dish. After the medium had solidified, the overnight endophyte colony was spot inoculated onto the surface of the medium and cultivated for 24 hours at ± 37°C. The diameter of the inhibition zone was expressed in millimeters [12].
Metabolites extraction
Each endophytic Bacillus strain was cultured overnight in a nutrient broth (NB) medium (Oxoid). The culture (1% v/v) was seeded to 1 l of NB medium and incubated for 3 days at ± 28°C with agitation at 120 rpm to optimize extracellular secondary metabolite production. The cultures were extracted by mixing an equal volume of ethyl acetate and shaking for 20 minutes. The upper layer was then dried at 50°C using an evaporator. The extract was re-dissolved in 1% DMSO (Merck) for antimycobacterial and antibiofilm analyses [12].
Antimycobacterial test of endophytic extracts
The antimycobacterial activity of each endophytic extract (1 mg/ml) was analyzed using the disk diffusion method as described previously. Approximately 200 µg/ml tetracycline (Sigma-Aldrich) and 1 % DMSO (Merck) were used as positive and negative controls, respectively [15].
Evaluation of the minimum inhibitory concentration (MIC) and the minimum bactericidal concentration (MBC)
The extract from each endophytic isolate underwent further assays to evaluate the MIC using a microdilution test [15]. Briefly, microdilution was performed using Mueller Hinton (MH) Broth (Himedia) containing extract (100 µl) ranging from 1,000 µg/ml to 1.95 µg/ml in 96 sterile well plates (Biologix). Subsequently, the M. smegmatis suspension (100 µl) at a density of 0.5 McFarland (1 × 108 CFU/ml, Thermo Fisher Scientific) was added into the wells (final bacterial concentration of 5 × 107 CFU/ml) and incubated overnight at 37°C. The MIC was defined as the lowest extract concentration that suppressed the visible growth of M. smegmatis. The MBC was determined by plating 100 µl suspension from the well with no growth of M. smegmatis. The lowest concentration of endophytic extract demonstrating complete killing of the target bacteria was considered the MBC. One percent of DMSO served as the negative control, while tetracycline (Sigma-Aldrich) was employed as the positive control. The MBC/MIC ratio was analyzed to classify the antimycobacterial properties of the endophytic extracts as either bacteriostatic or bactericidal. If the ratio was ≤ 4, the extract was categorized as bactericidal, but if the ratio exceeded 4, the extract was defined as bacteriostatic.
Antibiofilm analysis
The four endophytic extracts underwent antibiofilm analysis using a microtiter plate test [15]. In brief, 100 µl of M. smegmatis cells at a density of 1 × 108 CFU/ml were inoculated into brain heart infusion (BHI) medium (Merck) in the 96-well plates containing various extract concentrations (2 × MIC, 1 × MIC, ½ × MIC, ¼ × MIC) and incubated overnight. The medium was then removed from each well, and the wells were washed twice with phosphate buffer saline (PBS, Sigma-Aldrich). The formed biofilm was stained with 200 µl of 0.1% crystal violet (Merck) for 30 minutes at 37°C. The excess staining solution was removed by washing once with PBS. Subsequently, the stained biofilm was eluted in 200 µl of DMSO (99%). The optical density of the suspension was measured at 595 nm using an ELISA reader (Thermo Scientific Varioskan Flash-Thermo Fischer). Control plates without any treatment were used as reference. The percentage of inhibition of biofilm formation was determined in comparison to that of the control. The experimental data were expressed as average ± standard deviation from triplicates. The biofilm structure of each treatment was observed using an SEM. The biofilm matrix was collected and applied onto the surface of a single-polished Silicon Wafer (Sigma), and then incubated for 18 hours. Subsequently, the SEM specimens underwent coating with sputter-gold particles (Hitachi®, Tokyo, Japan), and examination was conducted using a SEM-JEOL JSM-IT200 (JEOL, South Korea) at a resolution of 5,000 ×, with a working distance of 5 µm and an accelerating voltage of 10 kV.
Biofilm cells eradication assay
To examine the biofilm cells eradication effect of endophytic extracts on preformed cell biofilm (a five-day biofilm), a methyl tetrazolium test was conducted following the procedure outlined by Wintachai et al. [16]. In the experiment, 200 µl of M. smegmatis culture (1 × 108 CFU/ml) was introduced into 96-well plates containing BHI medium (Merck) and left for 5 days at 37°C to allow for mature biofilm formation. The medium was replaced daily (every 24 hours) with fresh BHI medium supplemented with 0.25% glucose. Additionally, the endophytic extracts were added at different concentrations (2 × MIC, 1 × MIC, ½ × MIC, ¼ × MIC) and incubated for 24 hours at 37°C. The medium was removed from the well, and the cell biofilm was stained with 10 µl of 5 mg/ml methyl tetrazolium solution (Merck), and then incubated for 4 hours at 37°C. Subsequently, the formazan crystals were dissolved with 200 µl of 99% DMSO. Cell biofilms of M. smegmatis that did not undergo any treatment were utilized as controls. Optical density was measured at 595 nm using an ELISA reader (Varioskan Flash-Thermo Fischer, Thermo Scientific). The experimental data were presented as average ± standard deviation from three replications.
Analysis of VOCs composition
The composition of VOCs in the endophyte extract was analyzed using a GC-MS system (Agilent Technologies 6890N Inert C, USA) according to the manufacturer’s instructions. Mass spectra and chromatograms were processed using the MSD ChemStation Data Analysis software (G1701EA E.02.02.1431).
Whole-genome sequencing and detection of secondary metabolic gene clusters
The bacterial cells from the 24-hour-old culture underwent centrifugation at 15,000 rpm for 1 minute. Genomic DNA isolation was conducted using the Presto™ Mini gDNA Bacteria Kit (Geneaid) following the kit’s protocol. The bacterial genome sequence was acquired utilizing Oxford Nanopore Technology. De novo assembly was executed employing the Flye (v.2.8.3). Subsequently, mapping and genome polishing were iteratively performed four times polishing using Minimap2 (v.2.24–r1122) and Racon (v.1.5.0). The assembled sequence and genome completeness of closely related species were assessed using dfast-qc (v.0.4.2) and BUSCO (v.5.4.4). The quality assessment of the assembled sequence was carried out with Quast software (v5.0.2). Visualization of the bacterial genome was accomplished using Circos (v.0.69–8). AntiSMASH bacterial version v5.1.2 was employed to annotate SMGCs within the bacterial genome (https://antismash.secondarymetabolites.org) [17].
Statistical analysis
The experimental data obtained from the antibacterial assay, determination of MIC and MBC, antibiofilm analysis, and biofilm eradication assays were conducted in triplicate. Subsequently, the data underwent one-way analysis of variance, followed by further statistical analysis through Tukey’s test using Statistical Product and Service Solutions software version 29.0. The level of statistical significance was established at p < 0.05 to determine significance.
RESULTS
Colony and cell characteristics
The four isolates of endophytic Bacillus spp. used in this study had different colony characteristics, especially in terms of elevation and pigmentation, but the four isolates had the same shape, margins, and optics, which were circular, entire, and opaque, respectively (Table 1). These isolates also have bacilli in shape, and are stained purple using Gram staining procedure. The cell arrangement of Bacillus sp. strain DJ4 and B. subtilis strain DJ7 were monobacilli, whereas the cell arrangement of B. velezensis strain DJ9 and B. megaterium strain AJ5 were diplobacilli (Fig. 1).
 | Table 1. Colony and cell characteristics of the four endophytic Bacillus spp. isolated from A. pauciflorum. [Click here to view] |
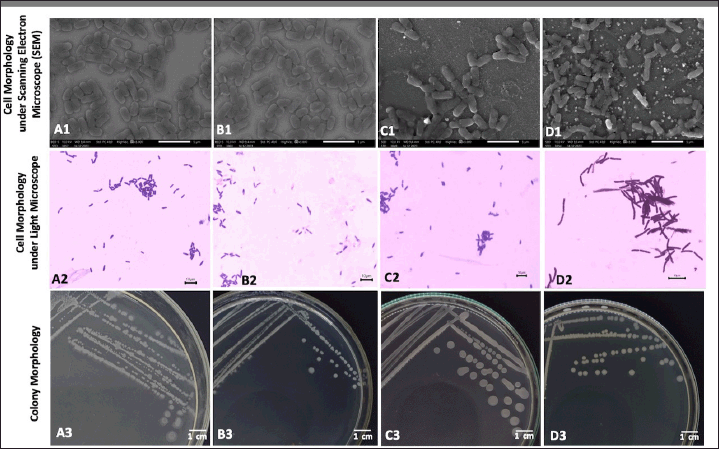 | Figure 1. Colony and cell morphology of the four endophytic Bacillus spp. isolated from A. pauciflorum. Cell morphology under SEM: (A1) B. megaterium strain AJ5, (B1) Bacillus sp. strain DJ4, (C1) B. subtilis strain DJ7, (D1) B. velezensis strain DJ9. Cell morphology under light microscope after stained using Gram technique: (A2) B. megaterium strain AJ5, (B2) Bacillus sp. strain DJ4, (C2) B. subtilis strain DJ7, (D2) B. velezensis strain DJ9. Colony morphology: (A3) B. megaterium strain AJ5, (B3) Bacillus sp. strain DJ4, (C3) B. subtilis strain DJ7, (D3) B. velezensis strain DJ9. [Click here to view] |
Antimycobacterial activity of Bacillus species isolated from A. pauciflorum
The four endophytic Bacillus spp. exhibited varied antimycobacterial activities against M. smegmatis. Inhibition zone formation was determined using endophytic bacterial colonies and their metabolite extracts (Table 2). Among the four endophytic isolates, Bacillus sp. strain DJ4 consistently displayed the strongest antimycobacterial activity, showcased by its colony and extract with inhibition zone diameters of 29.33 ± 0.47 mm and 17.7 ± 0.47 mm, respectively. Overall, the endophytic bacterial colonies demonstrated larger inhibition zone diameters against M. smegmatis compared to their metabolite extracts. The inhibition zone was also observed with tetracycline as the positive control (diameter: 15 ± 0.81 mm) and absent in 1% DMSO as the negative control (Fig. 2).
 | Table 2. Antimycobacterial activity of endophytic Bacillus spp. against M. smegmatis. [Click here to view] |
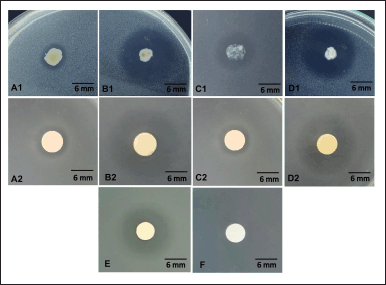 | Figure 2. Antimycobacterial activity of endophytic bacterial colony (A1) B. megaterium strain AJ5, (B1) Bacillus sp. strain DJ4, (C1) B. subtilis strain DJ7, (D1) B. velezensis strain DJ9, and endophytic bacterial extracts (A2) B. megaterium strain AJ5, (B2) Bacillus sp. strain DJ4, (C2) B. subtilis strain DJ7, (D2) B. velezensis strain DJ9, compared to (E) tetracycline, and (F) 1% DMSO. [Click here to view] |
The MIC and MBC of the four endophytic extracts were determined using a microdilution assay. The MIC ranged from 31.25 µg/ml to 125 µg/ml, while the MBC ranged from 125 µg/ml to 1,000 µg/ml (Table 3). However, the MIC and MBC of endophytic extracts are still higher than that of tetracycline, which had MIC and MBC of 7.81 µg/ml and 15.62 µg/ml, respectively. Furthermore, based on their MBC/MIC ratios, two bacterial extracts derived from B. megaterium strain AJ5 and Bacillus sp. strain DJ4 were categorized as bactericidal, whereas extracts from B. subtilis strain DJ7 and B. velezensis strain DJ9 were categorized as bacteriostatic. The remarkable antimycobacterial activity of the endophytic extract was demonstrated by Bacillus sp. strain DJ4 with the lowest MIC and MBC of 31.25 µg/ml and 125 µg/ml, respectively.
 | Table 3. The MIC, MBC, and MBC/MIC ratio of the endophytic Bacillus species against Mycobacterium smegmatis. [Click here to view] |
Mycobacterium smegmatis biofilm inhibition and biofilm cell eradication activities
The endophytic extracts reduced the biofilm-forming capability of M. smegmatis. The biofilm-forming capacity of M. smegmatis decreased in a dose-dependent manner with increasing extract concentration (Fig. 3A). The greatest inhibition of biofilm formation was observed for all extracts at 2 × MIC. At this extract concentration, biofilm formation was inhibited by 49.87 %–71.50 %. Extracts from Bacillus sp. strain DJ4 demonstrated remarkable antibiofilm activity, with the highest inhibition percentage of biofilm formation reaching up to 71.50%. The four endophytic extracts also exhibited biofilm cell eradication effects on the preformed M. smegmatis biofilms. Similar to the prevention of biofilm formation, the biofilm cell eradication effect of endophytic extracts was dose-dependent (Fig. 3B). The higher the endophytic extract applied, the greater the percentage of biofilm cell eradication effect was achieved. Consequently, 2 × MIC of endophytic extracts showed the highest cell eradication percentage, ranging from 47.99% to 60.53%. The most significant cell eradication effect was also demonstrated by Bacillus sp. strain DJ4-derived extract, with a biofilm cell eradication percentage of 60.53%.
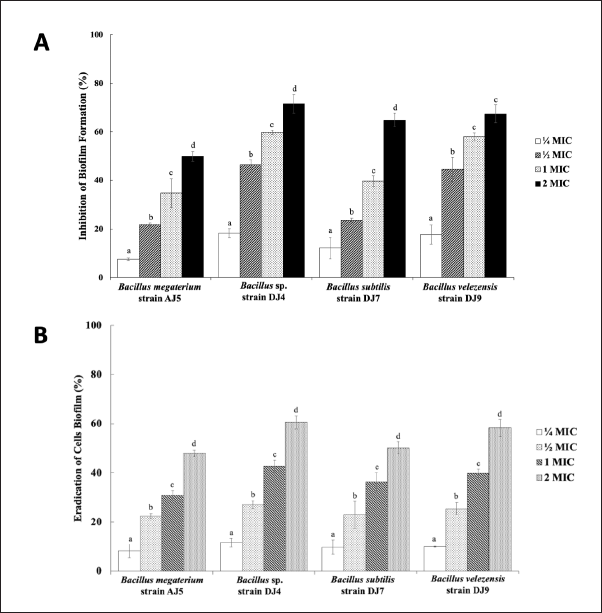 | Figure 3. The effect of endophytic Bacillus spp. extracts on (A) the biofilm-forming capacity of M. smegmatis, and (B) in eradicating M. smegmatis cell biofilm. Dissimilar letters following the bars in each treatment mean statistically significant difference (p-value of each treatment group is 0.000). [Click here to view] |
Furthermore, SEM images revealed a pronounced accumulation of the exopolysaccharide matrix in both the DMSO (Fig. 4A), and medium-only treatment (Fig. 4B). Conversely, a notable decrease in the exopolysaccharide matrix was observed in the treatments with endophytic extracts derived from Bacillus sp. strain DJ4 (Fig. 4C), B. megaterium strain AJ5 (Fig. 4D), B. subtilis strain DJ7 (Fig. 4E), and B. velezensis strain DJ9 (Fig. 4F). Following endophytic extract treatments, the exopolysaccharide layer of M. smegmatis was thinner, less compact and separated into small pieces, indicating that the endophytic extract treatments inhibited the production of exopolysaccharide and the formation of the biofilm matrix. Notably, the sample treated with extract derived from Bacillus sp. strain DJ4 exhibited the thinnest biofilm structure compared to untreated control, DMSO, and other endophytic extract treatments.
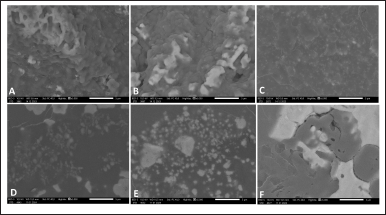 | Figure 4. Biofilm structure of M. smegmatis after treated with (A) DMSO, (B) medium only, and extract derived from (C) Bacillus sp. strain DJ4, (D) B. megaterium strain AJ5, (E) B. subtilis strain DJ7, (F) B. velezensis strain DJ9, at the concentration of 2 × MIC. [Click here to view] |
VOCs composition of extract from Bacillus sp. strain DJ4
GC-MS analysis identified 58 VOCs peaks in the extract from Bacillus sp. strain DJ4. Eighteen compounds were found in the bacterial extract by matching with the spectra of the recognized compounds (Table 4). These compounds include 2,4-di-tert-butylphenol (4.05%), 1-octadecene (0.44%), cyclo (L-prolyl-L-valine) (3.40%), 9-di-tert-butyl-1-oxaspiro(4,5)deca-6,9-diene-2,8-dione (0.41%), dibutyl phthalate (2.60%), cycloeicosane (0.96%), palmitic acid (0.32%), 9,12-octadecadienoic acid (Z,Z)-methyl ester (0.29%), methyl stearate (0.25%), tributyl acetyl citrate (1.73%), phthalic acid (0.46%), pyrrolo [1,2-a]pyrazine-1,4-dione, hexahydro-3-(phenylmethyl) (1.84%), oxacyclopentadecan-2-one (0.36%), lauric acid (6.66%), 1-hexacosene (0.71%), squalene (0.73%), dodecanamide (0.34%), dl-alpha-tocopherol (0.92%), and other unknown compounds.
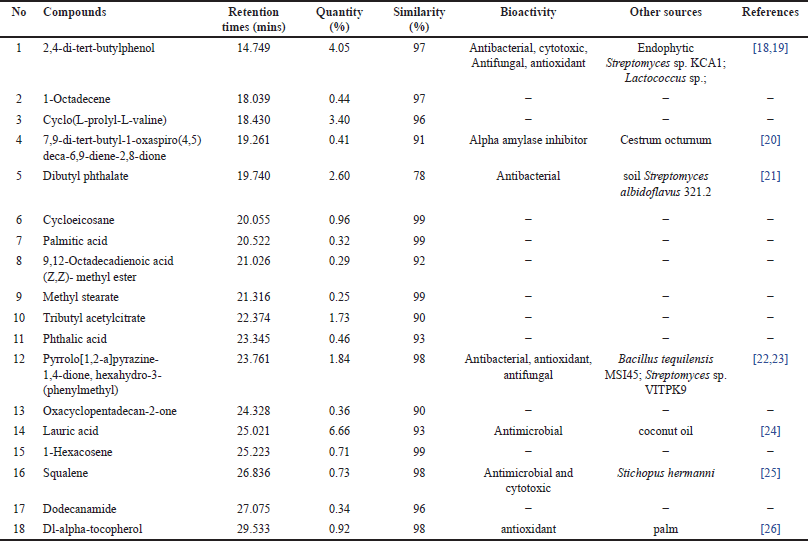 | Table 4. VOCs profile of the metabolite extract from Bacillus sp. strain DJ4. [Click here to view] |
Whole genome sequence and SMGCs of the most potential isolate
Based on the complete genome sequence, Bacillus sp. strain DJ4 exhibited a close relation (97.55%) to B. velezensis strain KCTC 13012 (accession no: GCA_001267695.1). The genome assembly size of this bacterium was 4,016,867 bp, having a 46.3% GC content (Fig. 5A). The genome assembly reached a completion rate of 99.1%, with the largest contig length at 1,945,594 bp and a read length N50 of 10,774 bp. Genome annotation via NCBI PGAP identified 3,909 coding sequences within the bacterial genome (Table 5). The complete genome sequence of this bacterium is accessible in the NCBI GenBank database under accession number CP144358.1.
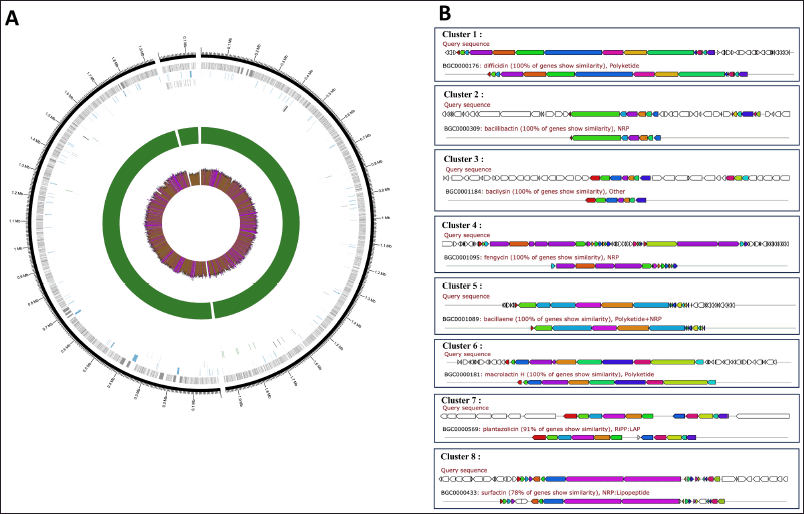 | Figure 5. (A) Annotated genome of Bacillus sp. strain DJ4. Figure Legend (from outer): contig (blue), genes (grey), pseudogenes (blue), coding sequences (CDS; black), rRNA (green), tRNA (purple), depth (depth >50 = green; depth <50 = red), gc content (gc content >50% = purple; gc content <50% = brown); (B) Putative SMGCs from the Bacillus sp. strain DJ4 genome with the antiSMASH database. [Click here to view] |
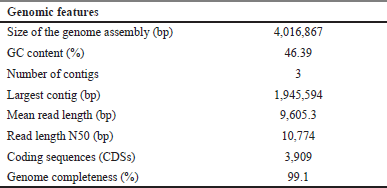 | Table 5. Genomic features of Bacillus sp. strain DJ4. [Click here to view] |
The presence of SMGCs within the genome of Bacillus sp. strain DJ4 was predicted using antiSMASH. Annotation results revealed eight SMGCs, encompassing gene clusters involved in the production of non-ribosomal peptides, polyketides, linear azole (in)e-containing peptides (LAP), hybrid non-ribosomal peptides and polyketides, hybrid non-ribosomal peptides and lipopeptides, as well as an unknown compound. Some genes shared their similarity with biosynthesis clusters of known compounds, such as difficidin, bacillibactin, bacilysin, fengycin, bacillaene, macrolactin H, plantazolicin, and surfactin, with similarities ranging from 78% to 100% (Fig. 5B). Interestingly, all these gene clusters were identified across the Bacillus group, encompassing B. velezensis and B. subtilis (Table 6).
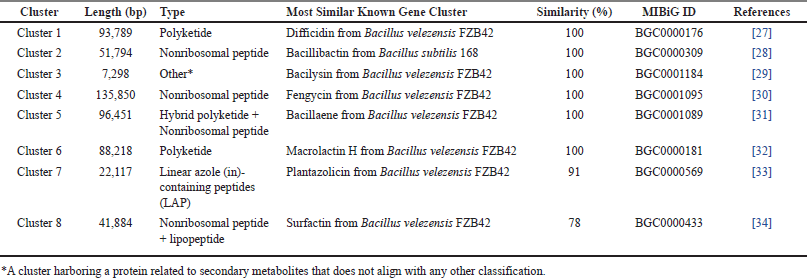 | Table 6. Putative SMGCs present in the genome of Bacillus sp. strain DJ4. [Click here to view] |
DISCUSSION
Bacillus species isolated from the internal tissues of medicinal plants are considered promising sources of compounds with valuable pharmacological functions, particularly as antimicrobial agents [35]. Various chemical substances produced by endophytic bacteria may contribute to their antimicrobial activity. In this study, antimycobacterial properties of four endophytic Bacillus from A. pauciflorum, namely Bacillus sp. strain DJ4, B. subtilis strain DJ7, B. megaterium strain AJ5, and B. velezensis strain DJ9, have been evaluated against M. smegmatis, a surrogate bacterium commonly used in preliminary study of antituberculosis candidates.
The four isolates had different colony and cell characteristics, particularly different in colony pigmentation, and elevation, as well as their cell arrangement. All isolates also belonged to Bacilli Gram-positive bacteria according to Gram staining. Interestingly, all endophytic colonies showed antagonistic activity against M. smegmatis, as indicated by the formation of a clear zone around the endophytic colony. Endophytic bacteria produce antibacterial compounds that are excreted extracellularly near the colony. Consequently, M. smegmatis growth in this area was inhibited. Secondary metabolites from the four endophytic bacteria were extracted and assessed using a disc diffusion test. The results showed that their metabolites also showed antimycobacterial activities with different degrees of significance. Extract from Bacillus sp. strain DJ4 displayed the strongest antimycobacterial activity with the largest inhibition zone of 17.7 ± 0.47 mm, followed by B. velezensis strain DJ9, B. megaterium strain AJ5, and B. subtilis strain DJ7. The diverse antimycobacterial effects of the endophytic isolates indicate the diverse chemical constituents produced by these endophytic bacteria.
The MIC of the four endophyte extracts was investigated to define the lowest extract concentration capable of inhibiting the visible growth of M. smegmatis, along with the MBC, as the lowest concentration that could kill M. smegmatis cells completely. The MIC of the extracts varied from 31.25 µg/ml to 125 µg/ml, and MBC from 125 µg/ml to 1,000 µg/ml, indicating that M. smegmatis has different sensitivity to each endophytic extract tested. However, the extracts with MIC less than 1,000 µg/ml are considered to have noteworthy antibacterial properties [36]. In this case, extract from Bacillus sp. strain DJ4 demonstrated the strongest antimycobacterial activity because the extract had the lowest MIC and MBC (MIC: 31.25 µg/ml; MBC: 125 µg/ml) compared to that of other extracts. Extracts from Bacillus sp. strain DJ4 and B. megaterium strain AJ5 were categorized as bactericidal (MBC/MIC ratio: 4), whereas extracts from B. subtilis strain DJ7 and B. velezensis strain DJ9 were categorized as bacteriostatic (MBC/MIC ratio: 8). These results indicate that endophytic Bacillus spp. isolated from A. pauciflorum have potential as antimycobacterial agents. Several studies have investigated the antimycobacterial mechanisms of natural compounds against Mycobacterium. Well-known mechanisms include destructing bacterial cell membranes [37], blocking specific metabolic pathways [38], inhibiting cell wall synthesis [39], inhibiting ribosome function [40], inhibiting efflux pumps [41], and inhibiting DNA replication [42].
Biofilm formation stands as the primary virulence factor in Mycobacteria. TB treatment spans an extended period (around 6–9 months) due to Mtb cell’s ability to survive within human lungs, forming biofilms for shielding against the host’s immune response and various antibiotics [43]. Consequently, the infection persists, showing high persistence and tolerance to available anti-TB drugs. Hence, targeting the inhibition and destruction of biofilms holds promise for enhancing TB treatment success. Encouragingly, the four endophytic extracts exhibited inhibitory effects on M. smegmatis biofilm formation in vitro. Notably, metabolites extracted from Bacillus sp. strain DJ4 displayed a significant effect. At a concentration of 2 × MIC, the extract inhibited biofilm formation by up to 71.50% after 24 hours of treatment. The effectiveness increased with higher extract concentrations, suggesting a need for a higher concentration than that required to suppress the bacterium’s growth. However, the same bacterial extract showed lower effectiveness in eradicating M. smegmatis cells within the biofilm, evidenced by a lower percentage (60.53%) of cell eradication. This indicates increased resistance of the target bacterium to the endophytic extract after biofilm formation. Hence, a combination of antibacterial agents might be necessary to eliminate biofilms and bolster TB chemotherapy efficacy.
VOCs produced by the most potent bacteria (Bacillus sp. strain DJ4) were identified using GC-MS analysis. Surprisingly, some substances with a high similarity to the spectra database (≥90%) found in the extract are known to possess pharmaceutical properties (e.g., antimicrobial, antioxidant, alpha-amylase inhibitor, and cytotoxic) (Table 5). Other compounds not listed in Table 5 showed low similarity (<90%), indicating that VOCs might be unidentified or new compounds. The VOCs identified in the extract are also be produced by other microbial sources, such as endophytic Streptomyces sp. KCA1, Lactococcus sp., soil Streptomyces albidoflavus 321.2, Bacillus tequilensis MSI45, and Streptomyces sp. VITPK9, by plants (such as Cestrum octurnum, and coconut oil), and by animals (such as Stichopus hermanni) (Table 4). VOCs derived from Bacillus and other natural sources exert antimicrobial action, possibly by destroying the bacterial cell membrane, damaging DNA, interfering with protein structure and function, and downregulating the expression of genes involved in pathogen virulence [44,45]. Additionally, particular VOCs, such as eugenol inhibit biofilm formation by interfering with quorum-sensing signals and controlling transmembrane transport [46]. VOCs also have an eradicating effect on established biofilms due to their ability to diffuse and destabilize the polysaccharide matrix in mature biofilms [47]. Therefore, VOCs found in the extract may contribute to the antimycobacterial and antibiofilm activities of the Bacillus sp. DJ4 derived extract.
Bacillus sp. strain DJ4 was deemed the most potent isolate based on its antibacterial and antibiofilm activities. In a prior study, the bacterium showed a close relation (similarity 100%) to Bacillus amyloliquefaciens strain B29 through the 16S rRNA sequence [12]. However, the 16S rRNA-based identification has limitations as the sequence is not sufficient for species-level bacterial identification and intra-strain differentiation due to its short size (~1,300 bp) compared to the general 4–6 million bp size of complete bacterial genomes [48]. Therefore, a more comprehensive bacterial identification requires sequencing the complete bacterial genome. In this study, the complete genome of this bacterium was sequenced. According to the average nucleotide identity from dfast_qc, the bacterium was closely related (97.55% similarity) to B. velezensis strain KCTC 13012. Bacillus velezensis strain KCTC 13012 was suggested as a relative of B. amyloliquefaciens strain KCTC 13012, a well-known producer of broad-spectrum antibiotics [49].
Secondary metabolic gene clusters within the genome of Bacillus sp. strain DJ4 were annotated by AntiSMASH. Polyketide, non-ribosomal peptide, LAP, and lipopeptide-coding genes were found in the bacterial genome. The clusters shared high similarity (similarity: 78%–100%) with known products, such as difficidin (100%), bacillibactin (100%), bacilysin (100%), fengycin (100%), bacillaene (100%), macrolactin H (100%), plantazolicin (91%), and surfactin (78%), which were synthesized by B. velezensis and B. subtilis (Table 6). Interestingly, bacillibactin produced by endophytic B. subtilis NPROOT3 has previously been reported as having antimycobacterial activity against M. smegmatis MTCC6 [14]. Additionally, all similar gene products showed antibacterial properties against a broad spectrum of bacteria (Table 5). Moreover, the bacterium may also carry genes contributed in the biosynthesis of VOCs because the products of these genes have been detected in bacterial extracts. However, these genes could not be identified through this methodology because of the limited genome database of VOCs-related genes in bacteria. Bacillus VOCs are mostly derived from amino acid degradation, sulfur reduction, terpene synthesis, glucose oxidation, and lactate fermentation [50]. Further studies are necessary to investigate whether the secondary metabolic gene clusters found in the bacterium are expressed under controlled laboratory conditions.
CONCLUSION
The current study showcases the potential of the four endophytic Bacillus species, especially Bacillus sp. strain DJ4 isolated from A. pauciflorum, as promising sources of novel antimycobacterial substances. Extract from this bacterium showed significant inhibition of growth, biofilm formation, and eradication of M. smegmatis cell biofilm. The identification of eight secondary metabolic gene clusters within the bacterial genome suggests their potential contribution to the production of antimycobacterial compounds. These findings offer compelling evidence for the antimycobacterial capacity of endophytic Bacillus and highlight its genomic features, which hold promise for drug discovery. This opens avenues for uncovering new antimycobacterial agents from endophytic bacteria, fostering opportunities within the pharmaceutical industry. Future investigations should prioritize isolating active constituents and optimizing metabolite production using diverse biological, chemical, and molecular techniques. Additionally, in-depth exploration of the functionality of the identified secondary metabolic gene clusters in Bacillus sp. strain DJ4 through mutagenesis or heterologous overexpression, coupled with understanding the biosynthesis pathways of antimycobacterial compounds, will enhance the exploration and harnessing of novel compounds from this endophytic bacterium.
ACKNOWLEDGMENTS
All authors acknowledge Dr. Tjandrawati Mozef (BRIN, Indonesia) for providing Mycobacterium smegmatis ATCC700084 used in this study.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
Funding for this research was provided by the Directorate of Research and Innovation at IPB University, through the Young Lecturer Research Program (Penelitian Dosen Muda) 2023 (grant number: 11425/IT3/PT.01.03/P/B/2023) awarded to Jepri Agung Priyanto.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study did not involve human or animal participants.
DATA AVAILABILITY
All experimental data collected in this research are provided in this paper.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. NIAID (National Institute of Allergy and Infectious Diseases). Tuberculosis. [cited 2024 June 4]. [Internet]. Available from: https://www.niaid.nih.gov/diseases-conditions/tuberculosis-tb
2. WHO. Treatment of tuberculosis: guidelines. 4th ed. Geneva, Switzerland: WHO; 2010. [cited 2024 June 4]. Available from: http://apps.who.int/iris/bitstream/10665/44165/1/9789241547833_eng.pdf?ua=1&ua=1
3. Mabhula A, Singh V. Drug-resistance in Mycobacterium tuberculosis: where we stand. Medchemcomm. 2019;10(8):1342−60. CrossRef
4. Food and Drug Administration (FDA) approves new drug for treatment-resistant forms of tuberculosis that affects the lungs. 2019 [cited 2024 June 4]. [Internet]. Available from: https://www.fda.gov/news-events/press-announcements/fda-approves-new-drug-treatment-resistant-forms-tuberculosis-affects-lungs
5. Perveen S, Kumari D, Singh K, Sharma R. Tuberculosis drug discovery: progression and future interventions in the wake of emerging resistance. Europ J Med Chem. 2022;229:114066−86. CrossRef
6. Lelovic N, Mitachi K, Yang J, Lemieux MR, Ji Y, Kuurosu MM. Application of Mycobacterium smegmatis as a surrogate to evaluate drug leads against Mycobacterium tuberculosis. J Antibiot (Tokyo). 2020;73(11):780−9. CrossRef
7. Tensingh JAS, Ranjitha J, Rajan A, Shankar V. Features of the biochemistry of Mycobacterium smegmatis, as a possible model for Mycobacterium tuberculosis. J Infect Publ Health. 2020;13(9):1255−64. CrossRef
8. Fouda A, Eid AM, Elsaied A, El-Belely EF, Barghoth MG, Azab E, et al. Plant growth promoting endophytic bacterial community inhabiting the leaves of Pulicaria incisa (Lam.) DC inherent to arid regions. Plants. 2021;10(76):2−22. CrossRef
9. Bunawan H, Dusik L, Bunawan SN, Amin NM. Botany, traditional uses, phytochemistry and pharmacology of Archidendron jiringa: a review. Global J Pharmacol. 2013;7(4):474−8. CrossRef
10. Hemmerling F, Pie J. Strategies to access biosynthetic novelty in bacterial genomes for drug discovery. Nat Rev Drug Discov. 2022; 21:359–78. CrossRef
11. Priyanto JA, Prastya ME, Astuti RI, Minarti, Mozef T. Endophytic Bacillus spp. isolated from Archidendron pauciflorum: pharmacological property and their phytochemical constituents. J Res Pharm. 2023;27(6):2511–21. CrossRef
12. Priyanto JA, Prastya ME, Astuti ME, Kristiana R. The antibacterial and antibiofilm activities of the endophytic bacteria associated with Archidendron pauciflorum against multidrug-resistant strains. Appl Biochem Biotechnol. 2023;195(11):6653–74. CrossRef
13. Guendouzi SE, Suzanna D, Hassi M, Haggoud A, Souda SI, Houari A, et al. Isolation and identification of Bacillus strains with antimycobacterial activity. Afr J Microbiol Res. 2011;5(18):2786–92. CrossRef
14. Nalli Y, Singh S, Gajjar A, Mahizhaveni B, Dusthackeer VNA, Shinde PB. Bacillibactin class siderophores produced by the endophyte Bacillus subtilis NPROOT3 as antimycobacterial agents. Lett Appl Microbiol. 2023;76(2):ovac026. CrossRef
15. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. CLSI Supplement M100. 31st edition, Wayne, PA: Clinical and Laboratory Standards Institute; 2023.
16. Wintachai P, Paosen S, Yupanqui CT, Voravuthikunchai SP. Silver nanoparticles synthesized with Eucalyptus critriodora ethanol leaf extract stimulate antibacterial activity against clinically multidrug-resistant Acinetobacter baumannii isolated from pneumonia patients. Microb Pathogen. 2019;126:245–57. CrossRef
17. Dat TTH, Oanh PTT, Cuong LCV, Anh LT, Minh LTH, Ha H, et al. Pharmacological properties, volatile organic compounds, and genome sequences of bacterial endophytes from the mangrove plant Rhizophora apiculata Blume. Antibiotics. 2021;10(1491):1–23. CrossRef
18. Seenivasan A, Manikkam R, Kaari M, Sahu AK, Said M, Dastager SG. 2,4-Di-tert-butylphenol (2,4-DTBP) purified from Streptomyces sp. KCA1 from Phyllanthus niruri: isolation, characterization, antibacterial and anticancer properties. J King Saudi Univ Sci. 2022;34:1–7. CrossRef
19. Varsha KK, Devendra L, Shilpa G, Priya S, Pandey A, Nampoothiri KM. 2,4-Di-tert-butyl phenol as the antifungal, antioxidant bioactive purified from a newly isolated Lactococcus sp. Int J Food Microbiol. 2015;211:44–50. CrossRef
20. Ahmad S, Alrouji M, Alhajlah S, Alomeir O, Pandey RP, Ashraf MS, et al. Secondary metabolite profiling, antioxidant, antidiabetic and neuroprotective activity of Cestrum octurnum (Night Scented-Jasmine): use of in vitro and in silico approach in determining the potential bioactive compound. Plants. 2023;12:1–22. CrossRef
21. Roy RN, Laskar S, Sen SK. Dibutyl phthalate, the bioactive compound produced by Streptomyces albidoflavus 321.2. Microbiol Res. 2006;161(13):121–6. CrossRef
22. Kiran GS, Priyadharsini S, Sajayan A, Ravindran A, Selvin J. An antibiotic agent pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro isolated from a marine bacteria Bacillus tequilensis MSI45 effectively controls multi-drug resistant Staphylococcus aureus. RSC Adv. 2018;8:17837–46. CrossRef
23. Sanjenbam P, Kannabiran K. Bioactivity of pyrrolo [1,2-a]pyrazine-1,4-dione,hexahydro-3-(phenylmethyl)- extracted from Streptomyces sp. VITPK9 isolated from the salt spring habitat of Manipur, India. Asian J Pharmaceut. 2016;10(4):265–70. CrossRef
24. Anzaku AA, Akyala JI, Juliet A, Obianuju, EC. Antibacterial activity of lauric acid on some selected clinical isolates. Annals Clin Lab Res. 2017;5(2):1–5. CrossRef
25. Nazemi M, Motallebi A, Abbasi E, Kheladi M, Zare M. Antibacterial, antifungal, and cytotoxic activity of the fraction contains squalene in the acetone extract of a sea cucumber, Stichopus hermanni. Iranian J Fish Sci. 2022;21(6):1495–507. CrossRef
26. Selamat SN, Muhamad II, Idham Z, Pae. Retention of alpha tocopherol and antioxidant activity of encapsulated palm mixed vitamin E in formulated blends. MOJ Food Process Technol. 2018;6(3):272?8. CrossRef
27. Chen XH, Vater J, Piel J, Franke P, Scholz R, Schneider K, et al. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J Bacteriol. 2006;188(11):4024–36. CrossRef
28. Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, et al. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology (Reading). 2009;155(6):1758–75. CrossRef
29. Wu L, Wu H, Chen L, Yu X, Borriss R, Gao X. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci Rep. 2015;5(12975):1–9. CrossRef
30. Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, et al. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol. 2007;25(9):1007–14. CrossRef
31. Moldenhauer J, Chen XH, Borriss R, Piel J. Biosynthesis of the antibiotic bacillaene, the product of a giant polyketide synthase complex of the trans-AT family. Angewandte Chemie Int Ed. 2007;46(43):8195–7. CrossRef
32. Schneider K, Chen XH, Vater J, Franke P, Nicholson G, Borriss R, et al. Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J Nat Prod. 2007;70(9):1417–23. CrossRef
33. Liu Z, Budiharjo A, Wang P, Shi H, Fang J, Borriss R, et al. The highly modified microcin peptide plantazolicin is associated with nematicidal activity of Bacillus amyloliquefaciens FZB42. Appl Microbiol Biotechnol. 2013;97(23):10081–90. CrossRef
34. Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, et al. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol. 2004;186(4):1084–96. CrossRef
35. Nisa S, Shoukat M, Bibi Y, Al Ayoubi S, Shah W, Masood S, et al. Therapeutic prospects of endophytic Bacillus species from Berberis lycium against oxidative stress and microbial pathogens. Saudi J Biol Sci. 2022;29(1):287–95. CrossRef
36. Nxumalo CI, Ngidi LS, Shandu JSE, Maliehe TS. Isolation of endophytic bacteria from the leaves of Anredera cordifolia CIX1 for metabolites and their biological activities. BMC Compl Med Ther. 2020;20(1):1–11. CrossRef
37. Riaz MS, Kaur A, Shwayat SN, Behboudi S, Kishore U, Pathan AA. Dissecting the mechanism of intracellular Mycobacterium smegmatis growth inhibition by platelet activating factor C-16. Front Microbiol. 2020;11:1–14. CrossRef
38. Jeffrey LN, Ardrey A, Hafiz TA, Dyer LA, Warman AJ, Mosallam N, et al. Identification of 2-aryl-quinolone inhibitors of cytochrome bd and chemical validation of combination strategies for respiratory inhibitors against Mycobacterium tuberculosis. ACS Infect Diseases. 2023;9(2):221–38. CrossRef
39. Shetty A, Dick T. Mycobacterial cell wall synthesis inhibitors cause lethal ATP burst. Front Microbiol. 2018;9:1–9. CrossRef
40. Lin Y, Li Y, Zhu N, Han Y, Jiang W, Wang Y, et al. The antituberculosis antibiotic capreomycin inhibits protein synthesis by disrupting interaction between ribosomal proteins L12 and L10. Antimicrob Agents Chemother. 2014;58(4):2038–44. CrossRef
41. Šimunovi? K, Solnier J, Alperth F, Kunert O, Možina S, Bucar F. Efflux pump inhibition and resistance modulation in Mycobacterium smegmatis by Peucedanum ostruthium and its coumarins. Antibiotics. 2021;10:1–17. CrossRef
42. Szafran MJ, Ko?odziej M, Skut P, Medapi B, Domaga?a A, Trojanowski D, et al. Amsacrine derivatives selectively inhibit mycobacterial topoisomerase I (topa), impair M. smegmatis growth and disturb chromosome replication. Front Microbiol. 2018;17(9):1–13. CrossRef
43. Richards JP, Cai W, Zill NA, Zhang W, Ojha AK. Adaptation of Mycobacterium tuberculosis to biofilm growth is genetically linked to drug tolerance. Antimicrobact Agents Chemoter. 2019;63(11):1–17. CrossRef
44. Grahovac J, Paj?in I, Vlajkov V. Bacillus VOCs in the context of biological control. Antibiotics. 2023;12:1–40. CrossRef
45. Naksang P, Tongchitpakdee S, Thumanu K, Oruna-Concha MJ, Niranjan K, Rachtanapun C. Assessment of antimicrobial activity, mode of action and volatile compounds of Etlingera pavieana essential oil. Molecules. 2020;25(14):1–14. CrossRef
46. Wang WQ, Feng XC, Shi HT, Wang YM, Jiang CY, Xiao ZJ, et al. Biofilm inhibition based on controlling the transmembrane transport and extracellular accumulation of quorum sensing signals. Environ Res. 2023;221:115218. CrossRef
47. Abdallah FB, Lagha R, Gaber A. Biofilm inhibition and eradication properties of medicinal plant essential oils against methicillin-resistant Staphylococcus aureus clinical isolates. Pharmaceut (Basel). 2020;13(11):1–15. CrossRef
48. Land M, Hauser L, Jun SR, Nookaew I, Leuze MR, Ahn TH, et al. Insights from 20 years of bacterial genome sequencing. Functional Integr Genomics. 2015;15(2):141–61. CrossRef
49. Jeong H, Park SH, Choi SK. Genome sequence of antibiotic-producing Bacillus amyloliquefaciens strain KCTC 13012. Genome Announc. 2015;3(5):1–2. CrossRef
50. Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front Microbiol. 2019;10:1–19. CrossRef