INTRODUCTION
Organismal metabolic processes are inextricably associated with various free radicals such as reactive oxygen species (ROS) and reactive nitrogen species. These highly reactive molecules can cause oxidative damage to cell constituents, including proteins, lipids, and nucleic acids. Oxidants are naturally neutralized by enzymatic (e.g., superoxide dismutase, catalase, and glutathione peroxidase) and non-enzymatic (e.g., ubiquinone, amino acids, and albumin) systems. However, an excess of oxidants in cells may generate oxidative stress, which is an imbalance implicated in various degenerative disorders, including cardiovascular, Alzheimer’s, and Parkinson’s diseases, as well as diabetes [1,2]. An exogenous antioxidant is required to maintain redox homeostasis and can be acquired from the bioactive compounds of plants, fungi, microalgae, and bacteria. This bioactive compound includes carotenoid, a secondary metabolite characterized by the conjugated double-bond structures, which provide attractive colors and reduce free radicals [3]. Carotenoid consists of at least six conjugated double bonds in a polyene chain that can absorb light and enhance the intensity of the colors, generating shades of yellow, orange, and red [4]. Moreover, the elongated structure of the conjugated double bonds increases the electron-rich condition, which effectively eliminates the free radicals [5].
Carotenoid is a class of isoprenoid pigments with diverse distributions and derivatives, mainly produced by plants and can be found in microalgae, fungi, yeast, and bacteria. In non-photosynthetic bacteria, carotenoid primarily acts as self-defense mechanisms, including photoprotection and radiation protection [6,7]. These self-defense mechanisms are closely related to the ability of bacteria to control oxidants, which can threaten survival. The bacterium Paracoccus is widely reported to produce carotenoid with antioxidant properties. Carotenoid extracted from Paracoccus mercusii RSPO1 and P. homiensis BKA7 have been shown to inhibit 2,2-diphenyl-1-picryhydrazyl (DPPH) and 2,2’-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) radicals [8,9]. Astaxanthin, carotenoid derived from P. carotinifaciens, has been comprehensively studied specifically for its health benefits [10–12]. However, investigations on the health-related applications of carotenoid compounds in P. haeundaesis remain limited. One study reported the antioxidant activity of cells-free supernatant-mediated gold nanoparticles (AuNPs) of P. haeundaesis BC74171T[13], but the supernatant may not represent the carotenoid compound commonly found in cells.
Paracoccus haeundaesis SAB E11 was used in this study to investigate the carotenoid compound extracted from the bacteria cells. The orange pigment extract of P. haeundaesis SAB E11 showed antioxidant activity by effectively scavenging DPPH and ABTS radicals [14]. However, further study is needed to characterize the specific compound from P. haeundaesis SAB E11 and its antioxidant effects. To investigate these effects, Schizosaccharomyces pombe ARC039 was used as the model organism. This fission yeast has been widely applied in antioxidant assays at the cellular level as a model organism because of the similarity of the mRNA splicing process to metazoans and the identical mitochondrial inheritance mechanism to mammals [15,16]. The antioxidant response of S. pombe ARC039 to carotenoid derived from P. haeundaesis SAB E11 can be used as a reference for subsequent investigation of carotenoid impact on the biological systems of multicellular organisms.
MATERIALS AND METHODS
The microorganisms and media culture
Paracoccus haeundaensis SAB E11 used in this study was the collection of Prof. Aris Tri Wahyudi isolated previously from a marine sponge Jaspis sp. (Raja Ampat Island, Southwest Papua-Indonesia) [17]. Additionally, yeast S. pombe ARC039 (h-leu-32 ura4-294) was provided by Dr. Rika Indri Astuti. Paracoccus haeundaensis SAB E11 was cultured in seawater complete (SWC) medium (5 g peptone, 1 g yeast extract, 3 ml glycerol, 750 ml seawater, and 250 ml distilled water). Meanwhile, S. pombe ARC039 was routinely cultured in yeast extract supplement (YES) medium (5 g yeast extract, 30 g glucose, 0.128 g histidine, 0.128 g leucine, 0.128 g adenine, 0.01 g uracil, 0.128 g arginine, and distilled water 1000 ml).
crtY (lycopene β-cyclase) gene analysis
The genomic DNA of P. haeundaensis SAB E11 was extracted using the PrestoTM Mini gDNA Bacteria Kit (Geneaid, Taiwan). The crtY gene of this bacteria was amplified using the specific primer pair crtYF (5′-CCA GAA ATT CGT GGG CGT CG-3′) and crtYR (5′-ATC GGA ATA GCG CGT GTC CT-3′) [18], while the target DNA fragment was ±163 bp. Polymerase chain reaction was performed under conditions, including pre-denaturation at 95oC for 5 minutes, followed by 35 cycles of denaturation, annealing, and extension at 95oC, 58oC, and 72oC for 15 seconds, 20 seconds, and 20 seconds, with a final extension at 72oC for 7 minutes. The confirmed amplicons were sequenced in FirstBase, Malaysia, then the crtY nucleotide sequence was analyzed using BioEdit and identified with the BlastX program on the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov/BLAST/). The nucleotide sequences of crtY were deduced into amino acids on the Expasy website (https://web.expasy.org/translate/) and correlated using the Clustal Omega website (https://www.ebi.ac.uk/Tools/msa/clustalo/). Additionally, a phenetic tree was constructed in the MEGA.X program using neighbor-joining with 1000x bootstrap. A three-dimensional protein structure model was built on the SWISS-MODEL website (https://swissmodel.expasy.org/interactive), while visualization and analysis of superposition were conducted using UCSF Chimera-X 1.6.1. program.
Carotenoid band detection with thin-layer chromatography (TLC) analysis
TLC analysis was performed following the modified form of the procedure described by Chekanov et al. [19]. During this process, a total of 10 µl (5% in methanol) of the carotenoid extract was applied to a silica gel G60F-254 pre-coated aluminum-backed TLC plates (10 × 5 cm) (Merck, Germany) using CAMAG Linomat 5. The plates were eluted with n-hexane:acetone (7:3), and then the chromatogram was visualized under UV light at 254 and 366 nm. The carotenoid compound was detected by comparing the band obtained from the pigment extract with a β-carotene standard (C4582, Sigma-Aldrich, Darmstadt, Germany). This comparison was based on the colors and distance of the retardation factor (Rf) presented by the bands.
Characterization of carotenoid compound
Ultraviolet-visible (UV-Vis) spectrum, FTIR, and high-performance liquid chromatography (HPLC) were used to characterize carotenoid compounds. The maximum wavelength was determined based on the procedure by Hagos et al. [20] using a UV-Vis spectrophotometer (Metertch SP-8001). For FTIR analysis, carotenoid was pelleted with potassium bromide (KBr) [21] using a Tensor II FTIR Routine Spectrometer (Bruker Optics, USA), which provided an FTIR spectrum in the range of 500–4,000 cm-1. Meanwhile, HPLC analysis was performed using a Hitachi L-2420 system to determine the retention time of the carotenoid compound peak. The sample preparation was adapted from the method described by Singh et al. [22], and each analysis result for carotenoid characterization was compared to a β-carotene standard (C4582, Sigma-Aldrich, Darmstadt, Germany).
Antioxidant activity assay
Antioxidant activity was determined using DPPH radicals according to the method described by Batubara et al. [23]. Approximately 100 µl of carotenoid compound at various concentrations was introduced into microplate wells (Costar 96) and 100 μl of DPPH radicals solution (125 μM in methanol) was added, then the mixture was incubated for 30 minutes at 25–27oC in dark conditions. The absorbance of the samples was measured using an ELISA microtiter plate reader (EPOC, USA) at 517 nm. The inhibitory concentration (IC50) was used to express antioxidant activity in the DPPH radical test.
Oxidative stress response assay (spot test)
The oxidative stress response of S. pombe ARC039 to carotenoid compound from P. haeundaensis SAB E11 was evaluated with spot assay [15]. S. pombe ARC039 was pre-cultured in YES broth medium for 24 hours at 27°C, and then transferred into 3 ml sterile YES broth medium at an initial OD600 of 0.05 as treatment culture. Carotenoid was added to the treatment culture at several concentrations based on the IC50 value of the DPPH radical assay (0.125x, 0.25x, 0.5x, 1x, and 2x). S. pombe ARC039 grown in 0.3 % (w/v) YES medium and ascorbic acid (0.1 µg.ml-1) was used as the positive control, while yeast cultured in DMSO was the negative control. A β-carotene standard was applied as the reference sample, and each treatment culture was incubated for 24 hours at 27°C. The spot assay was performed on all treatment cultures at an initial OD600 of 1, followed by serial dilutions (10-1 to 10-4). A total of 2µl of the OD600= 1 and the serial dilutions were spotted in solid YES medium containing 0, 0.5, 1, and 2 mM H2O2 as oxidative stress treatment, then cell viability was observed after incubation for 72 hours at 27°C.
Mitochondria activity assay
The mitochondrial activity assay was conducted based on the best concentration of carotenoid compound that showed a response to oxidative stress, as described by Lesmana et al. [24]. Yeast treatment cultures were prepared as previously described for the oxidative stress response assay. Furthermore, the cells were rinsed using 0.1 M phosphate buffer pH 7 and added with 300 nM Rhodamin B. The mitochondrial activity was observed under the Olympus BX51 fluorescence microscope, with S. pombe ARC039 grown in 0.3% (w/v) YES medium serving as the positive control, while DMSO was the negative control.
Anti-oxidative genes expression analysis
Antioxidant gene expression analysis was conducted following the method applied by Cahlia et al. [25]. The optimal concentration of carotenoid compound in the oxidative stress tolerance assay was selected and prepared as described in the oxidative stress response assay. DMSO was used as the negative control, while the treatment cultures were grown for 24 hours, and 1 mM H2O2 was added an hour before harvesting the cell. The total RNA of S. pombe ARC039 was extracted with a Direct-zol RNA Microprep Kit (Zymo, USA). Furthermore, cDNA was obtained by reverse transcription using the ReverTraAceTM qPCR RT Master Mix Kit (Toyobo, Japan). Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) was performed using a QuantStudio 5 instrument and Thunderbird SYBR qPCR master mix (Toyobo, Japan) as a fluorescent signal. The primer sequences of target genes were ctt1 (F5’-TCG TGA CGG CCC TAT GAA TG-3’)(R5’-AGC AAG TGG TCG GAY TGA GG-3’), sod2 (F5’-ATT TGG AGG GAG GTT GCC-3’)(R5’-GAT TGA TGT GAC CAC CGC CA-3’), and act1 (F5’-CGG TCG TGA CTT GAC TGA CT-3’)(R5’-ATT TCA CGT TCG GCG GTA GT-3’). The qRT-PCR was performed in 40 cycles of denaturation at 95°C (15 seconds), annealing at 55°C (30 seconds), and extension at 72°C (30 seconds). Moreover, the relative expression level was determined by normalizing the cycle threshold (Ct) value of ctt1 and sod2 genes to the Ct of act1 as the housekeeping gene.
Statistical analysis
The experiment in this study was performed in three independent replicates, and the results were reported as mean ± standard deviation after conducting the statistical analysis with RStudio software version 2023.09.1 ± 494. One-way analysis of variance was used to compare the mean values at 95% confidence levels. Subsequently, Duncan’s multiple range test was performed, with a p-value ≤ 0.05 being considered significant.
RESULTS AND DISCUSSION
The crtY (lycopene β-cyclase) gene
Paracoccus haeundaensis SAB E11 was detected to comprise the crtY gene (±163 bp), as shown in Figure 1A. The partial crtY gene sequences were deposited in the DNA Data Bank of Japan under accession number LC813284. The sequences were highly similar to those of P. mercusii lycopene β-cyclase (similarity: 100%; E-value: 3e-17; accession number: WP_282029193.1). According to the result, the bacteria produced the protein lycopene β-cyclase, a critical enzyme in carotenoid biosynthesis forming two β-ionone cyclic rings at the end of lycopene polyene chains to generate β-carotene [26]. Furthermore, β-carotene is a precursor of oxygenated carotenoid (xanthophyll) and pro-vitamin A, which are essential for health. In this study, partial nucleotide sequences were deduced into 54 amino acids, among which the crtY amino acid sequence of P. haeundaensis SAB E11 was correlated with the reference sequences of lycopene β-cyclase crtY. The reference sequences were obtained from Paracoccus PAMC 22219 (WP_042246542.1), P. mercusii (WP_282029193.1), Paracoccus sp. 228 (WP_046000944), Paracoccus sp. 08 (WP_256481049.1), Paracoccus sp. N81106 (P54974.1), Paracoccus sp. NBH48 (WP_255522031.1), P. haeundaensis BC74171 [27], MULTISPESIES: Paracoccus sp. 08 (WP_127898119.1), and Caulobacter sp. (MBP7703974.1) as an out-of-group species. Based on Figure 1B, amino acid alignment showed that most of the partial residues of crtY from P. haeundaensis SAB E11 were highly conserved in the Paracoccus spp. reference strains. However, certain different amino acid variations were observed with Caulobacter sp. The diversity of crtY among species is regulated by the evolutionary process of carotenoid biosynthesis, which leads to mutations in nucleotide sequences. Lycopene β-cyclase is divided into three groups according to the amino acid sequences, namely crtY and crtL type, heterodimeric type, and the unique CruA/CruP type. As P. haeundaensis SAB E11 belongs to the Proteobacteria phylum, it has crtY type which is also found in Pantoea agglomerans, Rhodobacter sphaeroides, and Sphingomonas spp [27–30]. The crtL type is majorly present in Cyanobacteria, but heterodimeric lycopene β-cyclases (crtYc and crtYd) are found in Brevibacterium linens and Mycobacterium aurum. Meanwhile, CruA/CruP type is characteristic of the green sulfur bacterium (Chlorobium tepidum) [31].
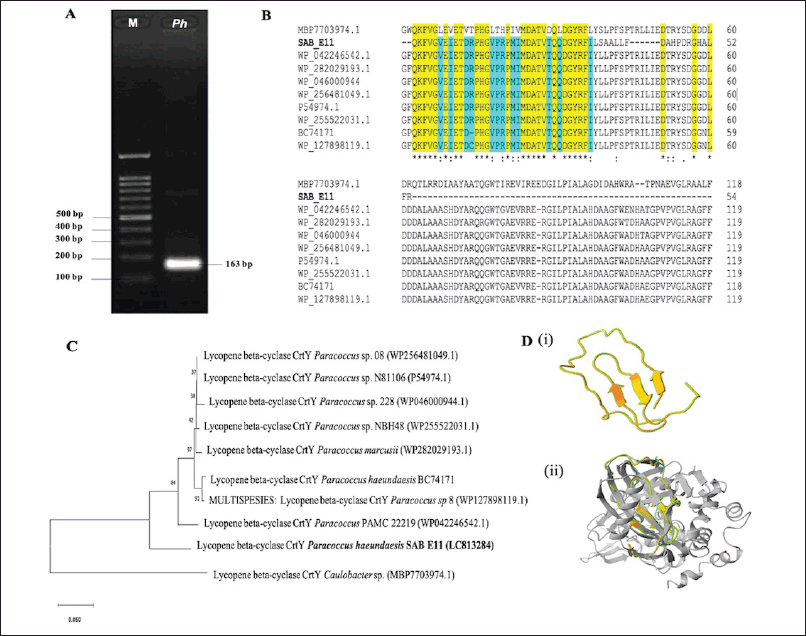 | Figure 1. (A) Amplicon visualization of crtY (lycopene β-cyclase) gene (+163 bp), Ph = P. haeundaensis SAB E11; M = 100 bp; (B) deduced amino acid alignment of partial CrtY. Yellow: conserved residues of the CrtY of Paracoccus spp. and Caulobacter sp. and Blue: conserved residues of the CrtY P. haeundaensis SABE11 among other Paracoccus strains; (C) phylogentic tree construction of lycopene β-cyclase using neighbor-joining with 1000x bootstrap; (D) 3D structure prediction and superposition analysis, (i) CrtY partial of P. haeundaensis SAB E11, (ii) superposition model CrtY P. haeundaensis SAB E11 (yellow) and CrtY Paracoccus sp. S-4012(A0A612M2J7) (grey). [Click here to view] |
A phenetic tree of lycopene β-cyclase crtY was constructed based on the deduced amino acid sequences of P. haeundaensis SAB E11 and the reference strains. According to Figure 1C, P. haeundaensis SAB E11 comprising lycopene β-cyclase crtY was grouped along with other Paracoccus strains. As shown by node numbers 84 and 97, lycopene β-cyclase crtY P. haeundaensis SAB E11 and Paracoccus PAM (WP042246542.1) were closely related to other Paracoccus strains, although clustered separately. The partial lycopene β-cyclase crtY amino acid sequences of P. haeundaensis SAB E11 in a three-dimensional structural protein were visualized using the SWISS-MODEL automated mode. Superposition analysis identified the presence of overlapping structural parts between the model and lycopene β-cyclase crtY proteins of Paracoccus sp. S-4012 shown in Figure 1D. The crtY prediction model was assumed to be acceptable and trustworthy, as evidenced by the template sequence identity percentage, global model quality estimate (GMQE) value, and prediction local distance difference test (pLDDT) score. crtY had a high sequence identity (72.55%) with the AlphaFold DB model of lycopene β-cyclase crtY Paracoccus sp. S-4012 protein template (A0A6I2M2J7.1.A). Additionally, the GMQE value of crtY was 0.86, where GMQE required a score between 0 and 1, with a higher number signifying higher reliability [32]. In this study, the protein structure of the template modeled by the AlphaFold DB method had a pLDDT score of 94.94%, denoting high confidence in the residue structure [33].
Characteristics of carotenoid in P. haeundaensis SAB E11
Carotenoid compound of P. haeundaensis SAB E11 was analyzed with TLC, UV-VIS, HPLC, and FTIR. TLC was performed using 5 % crude methanolic extract of intracellular P. haeundaensis SAB E11. According to Figure 2, one orange band with an Rf value of 0.88 appeared black under UV light at both 254 nm and 566 nm. The β-carotene standard (C4582, Sigma-Aldrich) showed identical band colors and Rf values. These TLC characteristics were previously observed in Citrococcus parietis AUCs and Rhodotorula toruloides ATCC 204091 [34,35]. Additionally, the UV-visible spectrum of the carotenoid compound from P. haeundaensis SAB E11 showed a maximum peak at 449 nm. For comparison, the β-carotene standard (C4582, Sigma-Aldrich) was observed at 450 nm (Fig. 3A). A study reported that the wavelength range profile of carotenoid compounds was between 400 and 500 nm. The maximum peak wavelength of a carotenoid compound of P. haeundaensis SAB E11 at 449 nm was identified as β-carotene [36,37].
 | Figure 2. Thin layer chromatograph (TLC) of carotenoid compound P. haeundaensis SAB E11 compared with β-carotene standard (C4582 Sigma-Aldrich), (A) in visible light, (B) UV light 254 nm, and (C) 566 nm. Ph: P. haeundaensis, S: β-carotene standard. [Click here to view] |
The carotenoid compound of P. haeundaensis SAB E11 from the single band (Rf 0.88) of TLC was analyzed using FTIR and HPLC. The FTIR spectra showed a comparative pattern between the β-carotene standard and carotenoid compound at 500–4,000 cm-1. The β-carotene standard pattern was dominated by three major peaks at 2,922 cm-1 (C−H), 1,725 cm-1 (C=O), and 959 cm-1 (C−C). Meanwhile, the carotenoid compound showed four significant peaks, three of which were identical to the β-carotene standard pattern (Fig. 3B). These included three peaks at 2,861, 2,920, and 2,957 cm-1, representing asymmetric and symmetric modes of the C−H groups. The remaining one was a sharp peak at 1,052 cm-1 (CH3), which was absent in the β-carotene standard and it might be attributed to the rocking vibration of the (RH) C=C (RH) groups of the synthesized carotenoid [8]. HPLC analysis confirmed the dominant peaks of the carotenoid compound and β-carotene standard at 10.607 and 10.547 minutes, respectively [31]. These peaks had comparable values, including the area and height with high similarity values of 95.26 % and 85.86 %, respectively (Fig. 3C). The characteristic data obtained were insufficient for the definitive identification of carotenoid compounds. Therefore, additional investigation is needed to purify carotenoids and determine their structure.
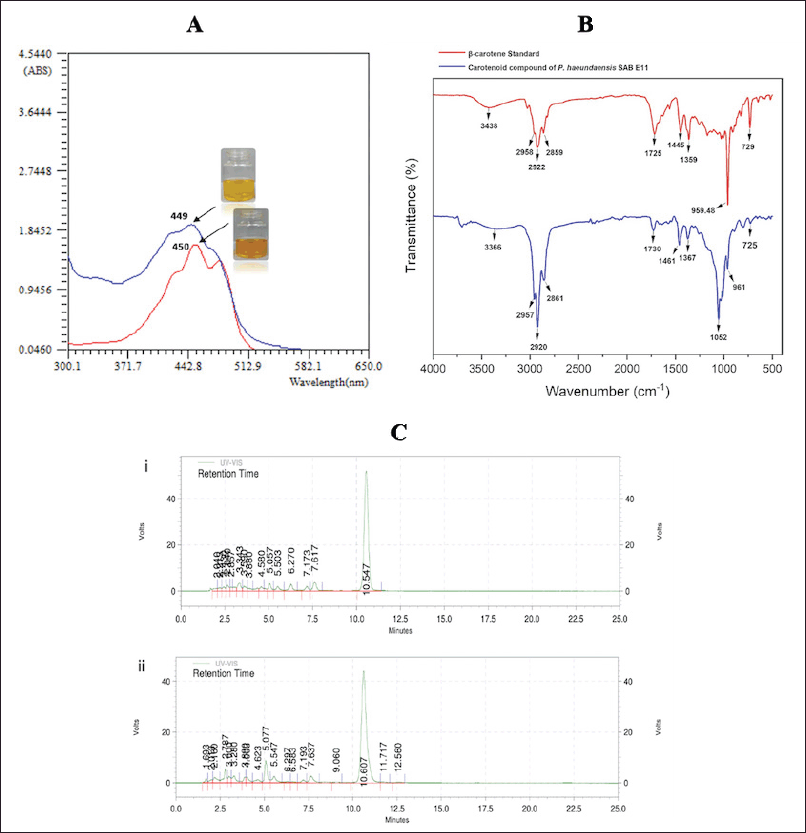 | Figure 3. Characterzation of carotenoid compound of P. haeundaensis SAB E11, (A) Maximum wavelength measurement of β-carotene standard (C4582, Sigma-Aldrich, Darmstadt, Germany) (red line) and the carotenoid compound (blue line). (B) FTIR pattern of β-carotene standard (red line) and carotenoid compound P. haeundaensis SAB E11 (blue line). (C) HPLC chromatogram of (i) β-carotene standard and (ii) carotenoid compound P. haeundaensis SAB E11. [Click here to view] |
Antioxidant activity of carotenoid in P. haeundaensis SAB E11
The DPPH radical assay was used to assess the antioxidant activity of the carotenoid compound. The scavenging activity against DPPH radicals increased proportionally with carotenoid concentration. According to Figure 4, the IC50 value required to reduce 50 % of DPPH radicals was 203.85 ± 3.12 μg.ml−1, while the IC50 of the β-carotene standard was 43.53 ± 0.59 µg.ml−1. These results corresponded with previous studies showing that the carotenoid compound derived from Kocuria sp. RAM1, Virgibacillus sp., and Citricoccus parietis AUC had antioxidant properties [35,38,39]. Carotenoid extract generated from Paracoccus marcusii RSPO1 reduced 65% ± 0.06% DPPH and 42% ± 0.7% ABTS radicals at 20 µg.ml-1 concentration. Meanwhile, 10 mg/ml β-carotene derived from P. homiensis BKA7 reduced DPPH radicals by 72% [8,9]. Carotenoid effectively eliminates free radicals due to the electron-rich conditions, which arise from the elongated structure with conjugated double bonds [5]. The presence of a cyclic structure including two β-ionic rings along with a lipophilic microenvironment provides an opportunity for carotenoids to reduce oxidants through mechanisms such as electron transfer, hydrogen atom transfer, or inhibition of peroxidase on lipids [40,41].
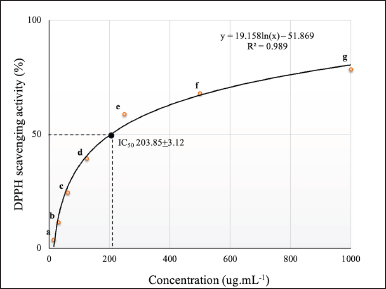 | Figure 4. IC50 of P. haeundaensis SAB E11 carotenoids was presented by the mean ± SD (n = 3). The mean value of each percentage inhibition in different letters indicators a significant distinction (p < 0.05) based on the DMR test. [Click here to view] |
Effect of the carotenoid on S. pombe ARC039 cell viability
The antioxidant activity of the carotenoid compound was assessed at the cellular level in S. pombe ARC039. Furthermore, yeast viability was determined by colony spot density after 72 hours of incubation in YES medium supplemented with H2O2. The antioxidant assay identified that the carotenoid compound showed lower DPPH radical scavenging activity than the β-carotene standard. However, in the cellular level assay, treatment with carotenoid compound maintained yeast viability against H2O2-induced oxidative stress until a concentration of 2 mM (Fig. 5). Treatment with carotenoid compound concentrations of 56 µg.ml-1 and 28 µg.ml-1 generated the same colony density as the positive control (CR treatment) and reference sample (ascorbic acid and β-carotene standard). Meanwhile, the negative control prepared with 3% glucose in the YES medium generated a lower colony density compared to the carotenoid compound treatment. Ascorbic acid was applied as the reference sample because previous studies showed the capacity to maintain yeast cell viability under oxidative stress conditions induced by H2O2 [24,25]. Additionally, the β-carotene standard was used to support the data obtained for the carotenoid compound. CR treatment was used as a positive control to investigate the oxidative response in yeast and has been widely reported to maintain yeast cell adaptability under low-glucose conditions. This induced a metabolic shift in yeast cells from glucose fermentation to mitochondrial respiration to maximize energy usage in response to limited glucose availability. Consequently, the shift enhanced mitochondrial activity, leading to increased intracellular ROS levels at mild concentrations, which induced an adaptive oxidative response in yeast cells [42]. Under the CR condition, yeast cells inhibited nutrient signaling kinases, such as Sck2 and the Git/PKA1 pathways in S. pombe [43,44]. In correspondence with the CR condition, carotenoids tended to trigger the adaptive oxidative response of yeast cells, which showed a colony density reaching 10-4 dilutions without low-glucose treatment. Previous studies found that S. pombe responded to oxidative stress through key mediators of mitogen-activated protein kinase signaling. The mediators stimulated the core transcriptional response to diverse stress conditions, including the core environmental stress response (CESR) [45,46]. This prediction needs to be supported by another cellular analysis, such as the analysis of mitochondrial activity and the oxidative response of antioxidant genes in S. pombe cells ARC039.
 | Figure 5. The effect of caretenoid compound (406 µg.ml-1, 203 µg ml-1, 102 µg ml-1, 56 µg ml-1 and 28 µg ml-1) on the S. pombe ARC093 cell viability with H2O2 induced S. pombe ARC093 in YES medium (0.3 % glucose) was assigned as positive control, while negative control was prepared in YES medium (3 % glucose Ascorbic acid (0.1µg ml-1) and β-carotene standard (C4582 Sigma-Aldrich ) (43µg ml-1) were used as a reference. [Click here to view] |
Mitochondrial activity of S. pombe ARC039
The adaptive oxidative response of S. pombe ARC039 induced by carotenoid compound was confirmed through mitochondrial activity. Carotenoid from P. haeundaensis SAB E11 increased mitochondrial activity, as shown by bright fluorescence (Fig. 6). This compound produced the highest fluorescence intensity in cells at a concentration of 28 µg.ml-1. The CR treatment and β-carotene standard presented the same fluorescence intensity as the value obtained from carotenoid. However, yeast cells treated with 3 % glucose in YES medium and DMSO showed no fluorescence. In this study, the observed fluorescence originated from the reactions between Rhodamine B and cations produced during respiration in the mitochondria [47]. As previously predicted, S. pombe ARC039 cells withstand oxidative stress at 3 % glucose in the medium due to the influence of carotenoid. The mitochondrial activity of yeast is probably enhanced to produce ROS, thereby allowing S. pombe ARC039 to tolerate ROS level. Exposure to ROS in mitochondria generated an adaptive response to oxidants, as evidenced by the viability of yeast cells that remained alive after 72 hours of incubation. Induction of mitochondrial activity can develop mitochondrial adaptive responses to ROS and induce the CESR system through the transcription factors (TF) Sty1-Atf, Pap1, and Prr1 [16,46]. Sty1-Atf1 modulates the transcriptional response to severe oxidative stress conditions, while Pap1 and Prr1 respond to weak oxidative stress [43,48].
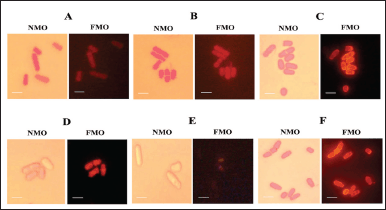 | Figure 6. The effect of the treatments on mitochondrial activity of S. pombe ARC039:carotenoid compound of P. haeundaensis SAB E11 of (A) 102 µg ml-1; (B) 56 µg ml-1; (D) positive control (0.3 % glucose of YES medium ); (E) negative control % glucose of YES medium); (F) reference sample β-carotene standard (43 µg ml-1). Nonfluorescent mitochondrial obderved (NMO), Fluorescent mitochondrial observed (FMO); bars 5 µm. [Click here to view] |
Effect of the carotenoid on anti-oxidative genes expression
Carotenoid compound was used to assess the potential to induce CESR genes in S. pombe ARC039. The ctt1 and sod2 genes present in the CESR system encode the enzymes catalase (Ctt1) and superoxide dismutase (Sod2). These genes are critical components of the detoxification system, functioning as ROS detoxifiers to enhance the oxidative response and promote reduction-oxidation homeostasis in S. pombe [46]. At a carotenoid concentration of 28 µg.ml-1, a significant increase was observed in the mRNA level of the ctt1 and sod2 genes of S. pombe ARC039. One hour after exposure to 1 mM H2O2, the relative expression of ctt1 and sod2 increased by 3.18 and 2.55 folds, respectively (Fig. 7). These data suggested that carotenoid compound induced the CESR system in S. pombe ARC039 by targeting ctt1 and sod2. The ctt1 gene in S. pombe is downstream of the TF Sty1-Atf1 and Pap1, which regulate gene expression differently depending on the H2O2 level [49]. Meanwhile, sod2 encodes mitochondrial Sod2, which Pap1 modulates in mild oxidative stress [16]. These genes may be regulated simultaneously by Pap1 TF to protect yeast cells from H2O2 and the ROS produced. The regulation is probably caused by carotenoid compound-induced increases in mitochondrial activity, leading to an adaptive response to ROS signaling. This result showed a mechanism similar to the type observed in the previous study of antioxidant compounds oxyphyllacinol, valine, and sugiol extracted from the water fraction of the Asteraceae plant. The three compounds induced an adaptive response to oxidative stress and promoted longevity in S. pombe ARC039 by modulating Pap1-ctt1 and Pap1-sod2 [16].
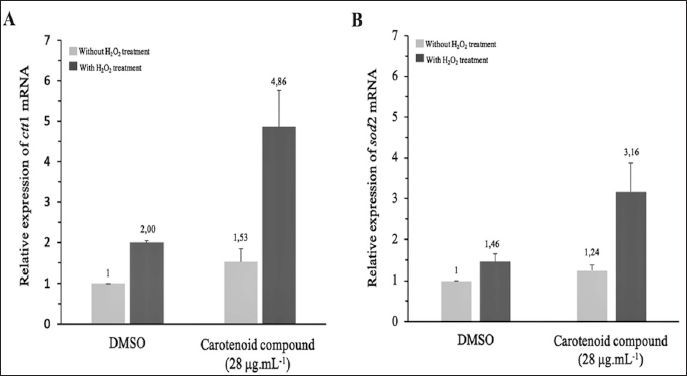 | Figure 7. Carotenoid compound P. haeundaensis SAB E11 affected the expression of ctt1 and sod2 genes. (A), The carotenoid compound (28 µg ml-1) induced relative expression of ctt1 and (B) sod2 genes. DMSO treatment is used as the negative control. The estimation of relative expression value was shown by the mean ± SD (n = 3). [Click here to view] |
CONCLUSION
In conclusion, this study showed the capabilities of P. haeundaensis SAB E11 to produce carotenoid compound, as evidenced by the presence of the crtY gene encoding lycopene β-cyclase. TLC, UV-Vis, HPLC, and FTIR confirmed carotenoid in P. haeundaensis SAB E11 requiring more purification through structural analysis, while DPPH radicals assay detected antioxidant activity. The compound effectively induced the adaptive response of S. pombe ARC039 by increasing mitochondrial activity and the relative expression of ctt1 and sod2 genes. Due to the results, carotenoids can be developed as functional dietary additives or nutraceuticals. However, this preliminary study requires further investigations, particularly regarding the impact on oxidative responses in multicellular organisms.
ACKNOWLEDGMENT
The authors are grateful to The Education Fund Management Institute (Lembaga Pengelola Dana Pendidikan/LPDP) from the Ministry of Finance, Indonesia, for the funding assistance provided under LoG number: SKPB3720/LPDP/LPDP.3/2023.
AUTHOR CONTRIBUTION
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analized are included in this research article.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Nandi A, Yan LJ, Jana CK, Das N. Role of catalase in oxidative stress- and age-associated degenerative diseases. Oxid Med Cell Longev. 2019;9613090:1–19. CrossRef
2. Dubois-Deruy E, Peugnet V, Turkieh A, Pinet F. Oxidative stress in cardiovascular diseases. Antioxidants. 2020;9(864):1–15. CrossRef
3. Rodriguez-Concepcion M, Avalos J, Bonet ML, Boronat A, Gomez-Gomez L, Hornero-Mendez D, et al. A global perspective on carotenoids: metabolism, biotechnology, and benefits for nutrition and health. Prog Lipid Res. 2018;70:62–93. CrossRef
4. Meléndez-Martínez AJ, Britton G, Vicario IM, Heredia FJ. Relationship between the colour and the chemical structure of carotenoid pigments. Food Chem. 2007;101:1145–50. CrossRef
5. Mordi RC, Ademosun OT, Ajanaku CO, Olanrewaju IO, Walton JC. Free radical mediated oxidative degradation of carotenes and xanthophylls. Molecules. 2020;25:1–13. CrossRef
6. Asker D, Beppu T, Ueda K. Unique diversity of carotenoid-producing bacteria isolated from Misasa, a radioactive site in Japan. Appl Microbiol Biotechnol. 2007;77(2):383–92. CrossRef
7. Licht MK, Nuss AM, Volk M, Konzer A, Beckstette M, Berghoff BA, et al. Adaptation to photooxidative stress: common and special strategies of the alphaproteobacteria Rhodobacter sphaeroides and Rhodobacter capsulatus. Microorganisms. 2020;8(2):1–22. CrossRef
8. Naik R, Gupte S. Characterization of pigment produced by high carotenoid yielding bacteria Paracoccus marcusii RSPO1 and evaluation of its anti-diabetic, anti-microbial and antioxidant properties. Nat Prod Res. 2024;38(6):968–77. CrossRef
9. Basim I, Jaber Kithar A, Majeed Alaa Gazi ALhashimi R. Antioxidant and antibacterial activity of β-carotene pigment extracted from Paracoccus homiensis strain BKA7 isolated from air Basrah, Iraq. Ann Rom Soc Cell Biol. 2021;25(4):14006–28.
10. Mussagy CU, Duffose L. A review of natural astaxanthin production in a circular bioeconomy context using Paracoccus carotinifaciens. Bioresour Technol. 2023;369:1–9. CrossRef
11. Hirakida H, Nakamura S, Inagaki S, Tsuji S, Hayashi M, Shimazawa M, et al. Anti-diabetic effects of astaxanthin-rich extract derived from Paracoccus carotinifaciens on pancreatic β cells. J Funct Foods. 2022;97:105252.
12. Hayashi M, Kawamura M, Kawashima Y, Uemura T, Maoka T. Effect of astaxanthin-rich extract derived from Paracoccus carotinifaciens on the status of stress and sleep in adults. J Clin Biochem Nutr. 2020;66(2):92–102. CrossRef
13. Patil MP, Kang M jae, Niyonizigiye I, Singh A, Kim JO, Seo YB, et al. Extracellular synthesis of gold nanoparticles using the marine bacterium Paracoccus haeundaensis BC74171T and evaluation of their antioxidant activity and antiproliferative effect on normal and cancer cell lines. Colloids Surf B Biointerfaces. 2019;183(110455):1–7. CrossRef
14. Abubakar H, Astuti RI, Listyowati S, Batubara I, Wahyudi AT. An orange pigment from the marine bacterium Paracoccus haeundaensis SAB E11 as a prospective source of natural antioxidants. Biodiversitas. 2022;23(9):4730–7. CrossRef
15. Prastya ME, Astuti RI, Batubara I, Takagi H, Wahyudi AT. Natural extract and its fractions isolated from the marine bacterium Pseudoalteromonas flavipulchra STILL-33 have antioxidant and antiaging activities in Schizosaccharomyces pombe. FEMS Yeast Res. 2020;20(3):1–14. CrossRef
16. Astuti RI, Prastya ME, Batubara I, Budiarti E, Ilmiyawati A. Antiaging and antioxidant bioactivities of Asteraceae plant fractions on the cellular functions of the yeast Schizosaccharomyces pombe. Adv Pharmacol Pharm Sci. 2021;2021:1–12. CrossRef
17. Abubakar H, Wahyudi AT, Yuhana M. Skrining bakteri yang berasosiasi dengan spons Jaspis sp. sebagai penghasil senyawa antimikroba. IJMS. 2012;16(1):35–40. CrossRef
18. Choi SS, Seo YB, Nam SW, Kim G Do. Enhanced production of astaxanthin by co-culture of Paracoccus haeundaensis and lactic acid bacteria. Front Mar Sci. 2021;7:1–12. CrossRef
19. Chekanov K, Litvinov D, Fedorenko T, Chivkunova O, Lobakova E. Combined production of astaxanthin and β-carotene in a new strain of the microalga Bracteacoccus aggregatus BM5/15 (IPPAS C-2045) cultivated in photobioreactor. Biology (Basel). 2021;10(7):1–17. CrossRef
20. Hagos M, Redi-Abshiro M, Chandravanshi BS, Yaya EE. Development of analytical methods for determination of β-carotene in pumpkin (Cucurbita maxima) flesh, peel, and seed powder samples. Int J Anal Chem. 2022;2022:1–11. CrossRef
21. Allahkarami S, Akhavan Sepahi A, Hosseini H, Razavi MR. Isolation and identification of carotenoid-producing Rhodotorula sp. from Pinaceae forest ecosystems and optimization of in vitro carotenoid production. Biotechnol Rep. 2021;32:1–12. CrossRef
22. Singh DP, Khattar JS, Rajput A, Chaudhary R, Singh R. High production of carotenoids by the green microalga Asterarcys quadricellulare PUMCC 5.1.1 under optimized culture conditions. PLoS One. 2019;14(9):1–19. CrossRef
23. Batubara I, Komariah K, Sandrawati A, Nurcholis W. Genotype selection for phytochemical content and pharmacological activities in ethanol extracts of fifteen types of Orthosiphon aristatus (Blume) Miq. leaves using chemometric analysis. Sci Rep. 2020;10(1):1–11. CrossRef
24. Lesmana D, Andrianto D, Astuti RI. Antiaging properties of the ethanol fractions of clove (Syzygium aromaticum l.) bud and leaf at the cellular levels: study in yeast Schizosaccharomyces pombe. Sci Pharm. 2021;89(4):1–13. CrossRef
25. Cahlia U, Astuti RI, Nomura J, Wahyudi AT. Antioxidant properties of active fraction extract derived from yellow-red pigment produced by the marine sponge-associated bacterium Bacillus haikouensis AGS112 and identification of related compounds. Hayati. 2023;30(5):874–84. CrossRef
26. Foong LC, Loh CWL, Ng HS, Lan JCW. Recent development in the production strategies of microbial carotenoids. World J Microbiol Biotechnol. 2021;37(1):1–11. CrossRef
27. Lee JH, Kim YT. Cloning and characterization of the astaxanthin biosynthesis gene cluster from the marine bacterium Paracoccus haeundaensis. Gene. 2006;370:86–95. CrossRef
28. Siddaramappa S, Viswanathan V, Thiyagarajan S, Narjala A. Genomewide characterization of the genetic diversity of carotenogenesis in bacteria of the order Sphingomonadales. Microb Genom. 2018;4(4):1–15. CrossRef
29. Qiang S, Su AP, Li Y, Chen Z, Hu CY, Meng YH. Elevated β-carotene synthesis by the engineered Rhodobacter sphaeroides with enhanced CrtY expression. J Agric Food Chem. 2019;67(34):9560–68. CrossRef
30. Guevarra RB, Magez S, Peeters E, Chung MS, Kim KH, Radwanska M. Comprehensive genomic analysis reveals virulence factors and antibiotic resistance genes in Pantoea agglomerans KM1, a potential opportunistic pathogen. PLoS One. 2021;16:1–27. CrossRef
31. Zhao Z, Liu Z, Mao X. Biotechnological advances in lycopene β-Cyclases. J Agric Food Chem. 2020;68(43):11895–907. CrossRef
32. Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–8. CrossRef
33. Guo HB, Perminov A, Bekele S, Kedziora G, Farajollahi S, Varaljay V, et al. AlphaFold2 models indicate that protein sequence determines both structure and dynamics. Sci Rep. 2022;12(1):1–15. CrossRef
34. Sinha S, Das S, Saha B, Paul D, Basu B. Anti-microbial, anti-oxidant, and anti-breast cancer properties unraveled in yeast carotenoids produced via cost-effective fermentation technique utilizing waste hydrolysate. Front Microbiol. 2023;13:1–13. CrossRef
35. Hagaggi NSA, Abdul-Raouf UM. Production of bioactive β-carotene by the endophytic bacterium Citricoccus parietis AUCs with multiple in vitro biological potentials. Microb Cell Fact. 2023;22(1):1–9. CrossRef
36. Vila E, Hornero-Méndez D, Azziz G, Lareo C, Saravia V. Carotenoids from heterotrophic bacteria isolated from Fildes Peninsula, King George Island, Antarctica. Biotechnol Rep. 2019;21:1–7. CrossRef
37. Kaur P, Ghoshal G, Jain A. Bio-utilization of fruits and vegetables waste to produce β-carotene in solid-state fermentation: characterization and antioxidant activity. Process Biochem. 2019;76(1):155–64. CrossRef
38. Kusmita L, Nur Prasetyo Edi A, Dwi Franyoto Y, Mutmainah, Haryanti S, Dwi Retno Nurcahyanti A. Sun protection and antibacterial activities of carotenoids from the soft coral Sinularia sp. symbiotic bacteria from Panjang Island, North Java Sea. Saudi Pharm J. 2023;31(8):1–10. CrossRef
39. Metwally RA, El-Sersy NA, El Sikaily A, Sabry SA, Ghozlan HA. Optimization and multiple in vitro activity potentials of carotenoids from marine Kocuria sp. RAM1. Sci Rep. 2022;12(1):1–19. CrossRef
40. Sandhiya L, Zipse H. Conformation-dependent antioxidant properties of β-carotene. Org Biomol Chem. 2022;20(1):152–62. CrossRef
41. Stafsnes MH, Bruheim P. Pigmented marine heterotrophic bacteria: occurrence, diversity, and characterization of pigmentation. In: Kim S-K, editor. Marine biomaterials: characterization, isolation and applications. First, London, UK: Taylor & Francis; 2013, p. 117–47. CrossRef
42. Skinner C, Lin SJ. Effects of calorie restriction on life span of microorganisms. Appl Microbiol Biotechnol. 2010;88(4):817–28. CrossRef
43. Chen D, Wilkinson CR, Watt S, Penkett CJ, Mark Toone W, Jones N, et al. Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol Biol Cell. 2008;19(1):308–17. CrossRef
44. Ohtsuka H, Shimasaki T, Aiba H. Genes affecting the extension of chronological lifespan in Schizosaccharomyces pombe (fission yeast). Mol Microbiol. 2021;115(4):623–42. CrossRef
45. González-Rubio G, Fernández-Acero T, Martín H, Molina M. Mitogen-activated protein kinase phosphatases (MKPs) in fungal signaling: conservation, function, and regulation. Int J Mol Sci. 2019;20(7):1–16. CrossRef
46. Papadakis MA, Workman CT. Oxidative stress response pathways: fission yeast as archetype. Crit Rev Microbiol. 2015;41(4):520–35. CrossRef
47. Reungpatthanaphong P, Dechsupa S, Meesungnoen J, Loetchutinat C, Mankhetkorn S. Rhodamine B as a mitochondrial probe for measurement and monitoring of mitochondrial membrane potential in drug-sensitive and -resistant cells. J Biochem Biophys Methods. 2003;57(1):1–16. CrossRef
48. Quinn J, Findlay VJ, Dawson K, Millar JBA, Jones N, Morgan BA, et al. Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in fission yeast Schizosaccharomyces pombe. Mol Biol Cell. 2002;13(2):805–16. CrossRef
49. Vivancos AP, Jara M, Zuin A, Sansó M, Hidalgo E. Oxidative stress in Schizosaccharomyces pombe: different H2O2 levels, different response pathways. Mol Geneti Genomics. 2006;276(6):495–502. CrossRef