INTRODUCTION
Research is underway on turmeric (Curcuma longa L.) for potential new drug development [1,2], but high production costs limit its application. As a result, scientists are investigating beneficial microbes found in plants and soil [3,4], particularly actinomycetes, which produce about 10,000 of the 23,000 known bioactive compounds [5], offering many potential health benefits [6]. Actinomycetes thrive in various conditions, increasing the likelihood of discovering new compounds. A study by Sapkota et al. [7] found certain soil actinomycetes to be 90% effective with minimal risk to commercial products.
Research indicates that actinomycetes from plant roots share properties with their host, including anticancer [4,8], antibacterial [9,10], antifungal, immunosuppressive, and antiparasitic effects. These benefits stem from bioactive compounds produced by biosynthetic gene clusters (BGCs), which include critical components like non-ribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) [11]. The PKS gene synthesizes polyketide (PK) chains from acyl-CoA, while the NRPS gene synthesizes peptide chains from amino acids [12]. PK and non-ribosomal peptides (NRP) are the primary secondary metabolites of the genus Streptomyces, with nearly three-quarters of its BGCs containing PKS and NRPS genes. Detecting BGCs encoding PKS1, PKS2, and NRPS provides a promising method for predicting secondary metabolites or natural products synthesized through these pathways [13].
Previous research found anticancer activity in rhizospheric actinomycetes from potential anticancer plants [4,8] but did not investigate their BGCs. Despite limitations in current research, studying the PKS-NRPS genes of turmeric’s actinomycetes could uncover valuable bioactive compounds for cancer treatment. This study focuses on isolating turmeric rhizosphere actinomycetes, detecting their NRPS, PKS1, and PKS2 gene content, and in vitro testing of the isolate’s extract on aggressive T47D cancer cells.
MATERIAL AND METHODS
Materials
Turmeric plantation soil samples were collected from Gemantar village, Jumantono, Karanganyar District, Central Java, Indonesia, at 7.6778°S, 111.0311°E, and 458 masl in May 2023. The soil was dug from 3 to 5 cm deep, about 0–10 cm away from approximately seven-month-old turmeric plants (Fig. 1) until the rhizomes were exposed [14]. About 5 g of rhizosphere soil were collected using a sterile spatula and placed into a sterile flask under sterile conditions. Before further analysis, the soil samples were stored at 4°C.
 | Figure 1. Turmeric (Curcuma longa L.) plant. [Click here to view] |
Rhizosphere actinomycetes of turmeric
Isolation and purification
Starch Casein Agar (SCA) and yeast malt agar (ISP2) were used as growth media, with their composition and preparation following the guidelines by Shirling and Gottlieb [15]. The isolation steps were adapted from Osama et al. [16]. The media were weighed according to the reference, sterilized in an autoclave at 121°C for 15 minutes, and then supplemented with 100,000 IU of nystatin (1 ml/l) to inhibit fungal growth. One gram of soil sample was mixed with 9 ml of sterile 0.9% NaCl, vortexed for 2 minutes, and filtered through filter paper. The solution was diluted from 10-¹ to 10-6. Actinomycetes were isolated using the pour plate method, where 1 ml from each dilution was added to Petri dishes containing 15 ml of SCA medium at approximately 40°C, allowed to solidify, and then incubated at 28V–30°C for 7–21 days. The streak plate method was subsequently used to transfer actinomycetes colonies with distinct characteristics to fresh SCA media, each labeled with a TC-ARCLx code. A single colony was purified on ISP2 slants (pH 8.0 and salinity 2 mg/l), and the pure actinomycetes isolates were stored in an starch casein broth (SCB) medium with 15% glycerol at −80°C for long-term preservation.
Morphology characterization
The morphology of individual actinomycetes colonies was visually examined using a stereomicroscope. Key morphological features assessed included colony form, height, edges, and consistency, focusing on the color of the aerial mycelium (in adult culture), substrate mycelium, and pigment type. The color analysis was based on the RHS Color Chart [17].
DNA isolation
The DNA isolation stages of actinomycetes were adapted from Rante et al. [10] using DNeasy PowerLyzer Microbial Extraction Kit (Qiagen) [18] with some modifications. A total of 1.8 ml of a 4-day-old liquid culture was placed in a collection tube and centrifuged at 10,000 rpm for 30 seconds. The pellet was then resuspended in 300 µl of powerbead solution and incubated in a shaker incubator at 65°C for 10 minutes to enhance DNA yield. The extraction was then continued according to the kit procedure until the genome was obtained. DNA concentration and purity were assessed using a Multiskan Sky-high microplate spectrophotometer at 260 and 280 nm wavelengths. Purity was determined by calculating the optical density (OD) ratio at these wavelengths (OD260/OD280). High DNA purity is indicated by a ratio valued between 1.8 and 2.0. A ratio below 1.8 suggests contamination from large molecular compounds, such as proteins, while a ratio above 2.0 indicates contamination from small molecular compounds, such as RNA.
Detection of PKS1, PKS2, and NRPS biosynthetic genes
Amplification of the PKS1 gene segment was performed using degenerate primers KS-F (5′-CCSCAGSAGCGCSTSYTSCTSGA-3′) and KS-R (5′-GTSCCSGTSCCGTGSGYST CSA-3′). The PKS2 gene fragment was amplified with primers KS∞ (5′-TSGCSTGCTTGGAYGCSATC-3′) and KSβ (5′-TGGAANCCGCCGAABCCTCT-3′) [19]. For NRPS, degenerate primers A3F (5′-GCSTACSYSATSTACACSTCSGG-3′) and A7R (5′-SASGTCVCCSGTSCGGTAS-3′) were used [20].
Each polymerase chain reaction (PCR) series included a negative control without a DNA template [21]. The PCR mix (25 µl) consisted of 2.5 µl DNA template, 1 µl of 10 µM forward primer, 1 µl of 10 µM reverse primer, 12.5 µl HS Taq RedMix (2x), and 8 µl nuclease free water (NFW) [22]. The PCR conditions consist of an initial denaturation at 95°C for 1 minutes, followed by 30 cycles of denaturation at 95°C for 15 seconds, annealing for 15 seconds at 55°C, 57°C, and 59°C for NRPS, PKS1, and PKS2 primers, respectively, extension at 72°C for 15 seconds, and a final extension at 72°C for 7 minutes [23].
Secondary metabolite production and extraction
The actinomycete starter cultures were grown in 100 ml of SCB medium and incubated at 28°C–30°C for 3 days at 170 rpm. The cultures were transferred to a 1-l Erlenmeyer flask containing 200 ml of SCB medium, with a 1% inoculum, and then incubated at 28°C–30°C for 10 days at 170 rpm. The actinomycete cultures were centrifuged at 6,000 rpm for 10 minutes at 4°C. The supernatant was then extracted using liquid-liquid extraction with an equal volume of ethyl acetate for 24 hours in an orbital shaker. The phases were separated with a separating funnel, and the upper layer of ethyl acetate was collected in a porcelain cup and evaporated at 40°C to obtain the extract [5].
Cytotoxicity assay
The cytotoxicity of the ethyl acetate extract from actinomycetes was evaluated on T47D and Vero cell lines using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method [24]. The cell lines were cultured in Dulbecco’s modified Eagle’s medium, supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were then seeded into 96-well plates and treated with varying concentrations of the ethyl acetate extract dissolved in Dimethyl Sulfoxide. The plates were then incubated at 37°C, 95% humidity, and 5% CO2 for 24 hours. After incubation, 1% of MTT solution was added and further incubated for 3 hours, followed by the addition of 10% SDS to stop the reaction. The absorbance of the resulting formazan crystals was measured at 595 nm using a Multiscan Sky-high microplate spectrophotometer, and the percentage of cell viability was calculated [25]. The cytotoxicity test was also conducted on hydrochloride doxorubicin (intravenous solution 2 mg/ml, Global Onkolab Farma), which is currently used as a drug, as well as on curcumin (Sigma for synthesis, Merck 20354) and a 96% ethanol extract of turmeric as the host plant of the rhizosphere actinomycete isolate.
Molecular identification with the 16S rRNA gene
Isolates that demonstrated cytotoxic activity against T47D cancer cells were identified by amplifying the 16S rRNA Gene using the PCR method. The procedure employed HS Taq RedMix (2x) according to the specified protocol, utilizing primers 27F (5′-AGAGTTTGATCC TGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) [10]. The reaction mixture was prepared to a total volume of 50 μl, comprising 1 μl of DNA template (approximately > 10 ng), 2 μl of 10 nM forward primer, 2 μl of 10 nM reverse primer, 25 μl of HS Taq RedMix (2x), and 20 μl of NFW. The PCR conditions consist of an initial denaturation at 98°C for 2 minutes, followed by 29 cycles of denaturation at 98°C for 15 seconds, annealing at 55°C for 15 seconds, extension at 72°C for 15 seconds, and a final extension at 72°C for 7 minutes.
Following amplification, the PCR products were visualized by electrophoresis on a 1% agarose gel dissolved in Tris/Borate/EDTA Buffer and 2 µl of SYBR Safe stain, running for 50 minutes at 70V. The results were observed using the Gel Documentation System Alliance™ Q9 Manual™. The completed PCR products were then sent to 1st BASE Malaysia for sequencing via Sanger DNA Sequencing with Capillary Electrophoresis.
Data analysis
A descriptive analysis of the morphological data was conducted by Bergey’s Manual of Systematic Bacteriology [26]. The IC50 value was calculated using Microsoft Excel with a linear regression formula. Sequencing results were analyzed with the Bioedit program, and comparisons were made against the NCBI GenBank database using BLAST (http://www.ncbi.nih.gov/Blast).
RESULTS AND DISCUSSION
Rizhosphere actinomycetes of turmeric
Actinomycetes were initially isolated on SCA and subsequently purified using an ISP2 medium. Twelve isolates were obtained from the rhizosphere soil of turmeric, each exhibiting unique morphological characteristics. However, effective purification was successful for only seven of these isolates, as the other five remained tightly associated with other bacteria during the growth phase. The seven isolates were then subjected to morphological characterization (Table 1 and Fig. 2).
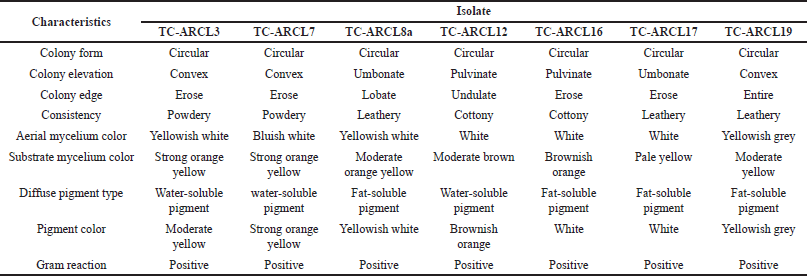 | Table 1. Morphological characteristics of rhizosphere turmeric actinomycetes isolates in ISP2 media. [Click here to view] |
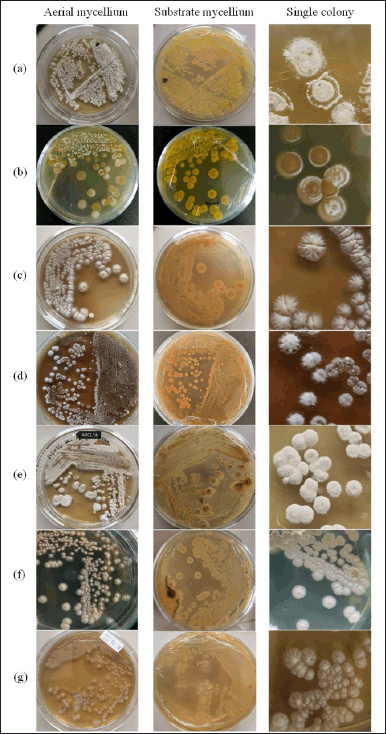 | Figure 2. Actinomycetes colony isolated with ISP2 medium. (a) TC-ARCL3, (b) TC-ARCL7, (c) TC-ARCL8a, (d) TC-ARCL12, (e) TC-ARCL16, (f) TC-ARCL17, and (g) TC-ARCL19. [Click here to view] |
The actinomycetes colonies from the turmeric rhizosphere displayed a circular morphology. In ISP2 medium, the colors of the aerial mycelium varied from white to grey, while the substrate mycelium ranged from yellow to brown. As illustrated in Table 1 and Figure 2, these actinomycetes produced pigments in a spectrum from light yellow to brownish orange. Rante et al. [10] reference a “colour wheel” that demonstrates the range of colors that can be produced by substrate and aerial mycelium. The pigments and colors of the mycelium are substrate-dependent, reflecting their essential role in nutrient absorption [27]. Furthermore, the concentration of hydrogen ions in the medium may influence the color of these pigments [28].
Detection of NRPS/PKS genes
This study identified seven actinomycete isolates with at least one BGC gene marker (Table 2 and Fig. 3). The BGC gene markers detected were PKS1, PKS2, and NRPS, measuring 700–800, 554, and 1,200–1,400 bp in size, respectively. These findings align with those of Peng et al. [29], who successfully isolated actinomycetes from the rhizosphere of Panax notoginseng [29].
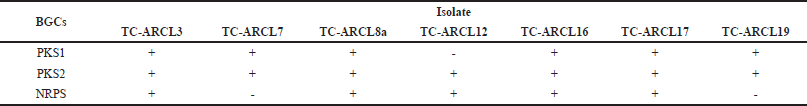 | Table 2. BGC (NRPS/PKS1/PKS2) of rhizosphere turmeric actinomycetes. [Click here to view] |
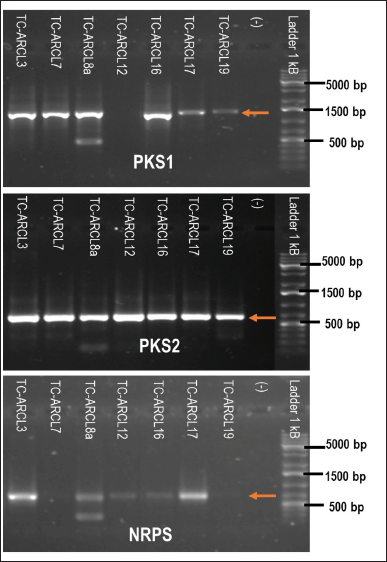 | Figure 3. Electropherogram of amplified PKS1, PKS2, and NRPS genes from actinomycetes isolates. [Click here to view] |
Among the seven isolates, three combinations of BGC content were observed: TC-ARCL3, TC-ARCL8a, TC-ARCL16, and TC-ARCL17 contained PKS1, PKS2, and NRPS genes; TC-ARCL7 and TC-ARCL19 had only PKS1 and PKS2; and TC-ARCL12 contained PKS2 and NRPS. According to Safari et al. [30], these isolates have the potential to produce various bioactive compounds from the non-ribosomal peptide and PK families due to the presence of KS and A domains in the PKS and NRPS gene clusters [30]. The core domain determines the addition of peptide units, while optional domains modify the peptide or ketide backbone [31]. Doxorubicin, an anthracycline chemotherapy agent currently used in the medical field, is produced from the PKS2 gene cluster of Streptomyces peucetius varietas caesius [6,13,32]. These results show that data from genome sequences pointing to secondary metabolite BGCs is essential for discovering new compounds and designing them using synthetic biology. Specifically, PK and NRP can be re-engineered with modular enzymes that interact with specific CoAs or amino acids [33].
Cytotoxicity assay
A widely used cell line in cancer research is T47D, a human breast cancer cell line. T47D cells are a great model for studying the specific effects of progesterone in luminal A subtype breast cancer [34]. This study successfully extracted secondary metabolites from five of the seven isolates using ethyl acetate as the solvent, followed by cytotoxicity tests. The IC50 values for T47D cancer cells and Vero cells are detailed in Table 3. Based on Nordin et al. [35], the cytotoxicity analysis of the T47D cell line categorized the ethyl acetate extracts into two groups: high (TC-ARCL7) and weak (TC-ARCL12, TC-ARCL16, TC-ARCL17, and TC-ARCL19). In contrast, all isolates showed weak inhibition of Vero cell proliferation.
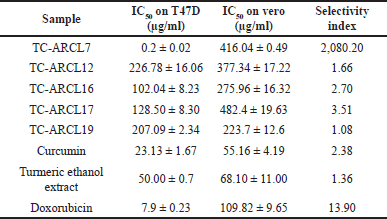 | Table 3. IC50 value of ethyl acetate extract of turmeric rhizosphere actinomycetes on T47D cancer cells and vero cells. [Click here to view] |
Among the extracts, only TC-ARCL7 demonstrated a high cytotoxic effect against the T47D cell line, with an IC50 value of 0.2 ± 0.02 µg/ml, while showing weak toxicity toward Vero cells, which had an IC50 value of 416.04 ± 0.49 µg/ml, so resulted in a selectivity index 2,080.20. A selectivity index of ≥10 is considered promising and indicates strong potential for further research [36,37]. Notably, the cytotoxic efficacy of the TC-ARCL7 extract against T47D cells was 39.5 times lower than hydrochloride doxorubicin (IC50 = 7.9 µg/ml), 115.65 times lower than curcumin (IC50 = 23.13 µg/ml), and 250 times lower than turmeric ethanol extract (IC50 = 50 µg/ml). The cytotoxic activity of ARCL7 ethyl acetate extract against T47D cells is comparable to Doxorubicin from Ebewe Pharma, Unterach, Austria (IC50 of 1.845 µg/ml) [32], and Doxorubicin from Sigma, Germany (IC50 = 0.2 ± 0.04 µg/ml) [38]. The IC50 value of Doxorubicin can vary significantly in different literature due to factors such as cell line conditions, formulation type, and brand.
Considering that Doxorubicin causes various side effects [32,39], these results suggest that TC-ARCL7 could serve as a valuable source for developing new anticancer drugs, given its high toxicity to T47D cancer cells coupled with low toxicity to Vero cells in vitro.
Molecular identification of potential isolate
The TC-ARCL7 isolate was identified through molecular amplification of the 16S rRNA biomarker gene. The 16S rRNA gene is favoured for this purpose due to its multicopy nature (150–300 copies in the bacterial genome) and high level of conservation. Its variable regions allow for effective differentiation among species, while its lack of horizontal gene transfer contributes to a slower evolutionary rate [40].
Nucleotide sequences from the forward and reverse reads were assembled into contigs and were compared to the GenBank database using BLAST. The TC-ARCL7 isolate exhibited a sequence similarity of 99.08% with Kitasatospora misakiensis (NBRC 12891, NR 112321.1) and Kitasatospora purpeofusca (NBRC 12905, NR 1123330). A similarity percentage between 98.7% and 99% indicates species delineation [41]. Therefore, this finding implies that the actinomycetes isolate belongs to the same species as these identified organisms. The GenBank accession number for TC-ARCL7 is PP212015.
The genus Kitasatospora is notable for its ability to produce a wide range of bioactive compounds, with over 50 known secondary metabolites exhibiting various biological activities, including antitumor, immunomodulatory, antiviral, herbicidal, and antiprotozoal effects [42]. Research by Li et al. [43] further indicates that Kitasatospora has a high diversity of secondary metabolite biosynthetic gene clusters distinguishing it from the closely related genus Streptomyces [43].
CONCLUSION
This study reveals that various microbes, including actinomycetes, thrive in the rhizosphere of turmeric, with seven isolates identified that feature three combinations of BGCs. The presence of specific BGCs does not necessarily correlate with anticancer activity. Notably, only one extract—the TC-ARCL7 ethyl acetate extract—exhibited significant anticancer effects against T47D cancer cells. This isolate was closely related to K. misakiensis NBRC 12891 (NR 112321.1) and K. purpeofusca NBRC 12905 (NR 1123330). Further research, such as optimization of growth conditions to obtain various extracts and purifications of bioactive metabolites, is needed to gather comprehensive data that can assist in the discovery of new cancer treatments.
ACKNOWLEDGMENT
The authors are grateful for funding the Research Organization for Life Sciences and Environmental of the National Research and Innovation Agency (BRIN), Indonesia.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Hu S, Xu Y, Meng L, Huang L, Sun H. Curcumin inhibits proliferation and promotes apoptosis of breast cancer cells. Exp Ther Med. 2018;16(2):1266–72.
2. Liu ST, Zheng SW, Hou AJ, Zhang JX, Wang S, Wang XJ, et al. A review: the phytochemistry, pharmacology, and pharmacokinetics of Curcumae Longae Rhizoma (Turmeric). World J Tradit Chinese Med. 2022;8(4):463–90.
3. Mandale SD, Dagar V, Dagar V. Antimicrobial activity of rhizospheric bacteria of Curcuma longa (Turmeric) producing metabolites against human pathogens. IOSR J Pharm Biol Sci. 2017;12(01):37–42.
4. Romero-Arguelles R, Romo-Sáenz CI, Morán-Santibáñez K, Tamez-Guerra P, Quintanilla-Licea R, Orozco-Flores AA, et al. In vitro antitumor activity of endophytic and rhizosphere gram-positive bacteria from Ibervillea sonorae (S. Watson) Greene against L5178Y-R lymphoma cells. Int J Environ Res Public Health. 2022;19(2):1–11.
5. Bahri A, Moazamian E, Azarpira N. Molecular identification, isolation and evaluation of Persian gulf actinomycetes as candidates of cytotoxic metabolites against breast cancer. Multidiscip Cancer Investig. 2017;1(3):10–4.
6. Abdallah OM, Tolba STM, Mohamed MR. Antimicrobial, antioxidant and anticancer potential of Streptomyces species isolated from the rhizosphere of Avicennia marina (Forssk.) Vierh. Indian J Exp Biol. 2022;60(February):81–90.
7. Sapkota A, Thapa A, Budhathoki A, Sainju M, Shrestha P, Aryal S. Isolation, Characterization, and Screening of Antimicrobial-Producing Actinomycetes from Soil Samples. Int J Microbiol. 2020 Mar 26;2020:2716584. doi: http://doi.org/10.1155/2020/2716584
8. Taechowisan T, Chaisaeng S, Phutdhawong WS. Antibacterial, antioxidant and anticancer activities of biphenyls from Streptomyces sp. BO-07: an endophyte in Boesenbergia rotunda (L.) Mansf A. Food Agric Immunol [Internet]. 2017;28(6):1330–46. doi: https://doi.org/10.1080/09540105.2017.1339669
9. Nova MR, Handayani N. Antibacterial activity of the sambiloto leaf extracts (Andrographis paniculata) to Bacillus cereus and Pseudomonas aeruginosa. Asian J Nat Prod Biochem. 2017;14(1):19–24.
10. Rante H, Alam G, Pakki E, Usmar U, Ali A. Identification and antibacterial activity of actinomycetes isolated from medicinal plant Andrographis paniculata rhizosphere soil. Crescent J Med Biol Sci. 2020;7(4):467–73.
11. Marfil-Santana MD, Martínez-Cárdenas A, Ruíz-Hernández A, Vidal-Torres M, Márquez-Velázquez NA, Figueroa M, et al. A meta-omics analysis unveils the shift in microbial community structures and metabolomics profiles in mangrove sediments treated with a selective actinobacterial isolation procedure. Molecules. 2021;26:1–24.
12. Komaki H, Tamura T. Profile of PKS and NRPS gene clusters in the genome of Streptomyces cellostaticus NBRC 12849T. Fermentation. 2023;9(11):924.
13. Ye JJ, Zou RJ, Zhou DD, Deng XL, Wu NL, Chen DD, et al. Insights into the phylogenetic diversity, biological activities, and biosynthetic potential of mangrove rhizosphere Actinobacteria from Hainan Island. Front Microbiol. 2023;14(1157601):1–17.
14. Osaro-Matthew RC, Ire FS, Frank-Peterside N. Screening of actinomycetes from turmeric (Curcuma longa L.) and ginger (Zingiber officinale) rhizosphere for antifungal activity. J Adv Microbiol. 2020;20(2):18–28.
15. Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16(3):313–40.
16. Osama N, Bakeer W, Raslan M, Soliman HA, Abdelmohsen UR, Sebak M. Anticancer and antimicrobial potential of five soil Streptomycetes: a metabolomics-based study. R Soc Open Sci. 2022;9(2):1–17.
17. Media RHS. RHS colour chart. 6th ed. London, UK: RHS Media; 2019.
18. Qiagen. DNeasy powerlyzer microbial kit handbook [Internet]. Hilden, Germany: Qiagen; 2017. Available from: www.qiagen.com
19. Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol. 2005;49:10–24.
20. Farida Y, Widada J, Meiyanto E. Combination methods for screening marine actinomycetes producing potential compounds as anticancer. Indones J Biotechnol [Internet]. 2007 Dec 9 [cited 2022 Jun 14];12(2):988–97. Available from: https://jurnal.ugm.ac.id/ijbiotech/article/view/7772
21. Shan W, Zhou Y, Liu H, Yu X. Endophytic actinomycetes from tea plants (Camellia sinensis): isolation, abundance, antimicrobial, and plant-growth-promoting activities. Biomed Res Int. 2018;2018:1–12.
22. Bioline. MyTaq TM HS Red Mix Kit. Meridian Life Science; 2017. p. 1–2.
23. Anggelina AC, Pringgenies D, Setyati WA. Presence of biosynthetic gene clusters (NRPS/PKS) in actinomycetes of mangrove sediment in Semarang and Karimunjawa, Indonesia. Environ Nat Resour J. 2021;19(5):391–401.
24. Haryanti S, Widiyastuti Y. Aktivitas Sitotoksik pada Sel MCF-7 dari Tumbuhan Indonesia untuk Pengobatan Tradisional Kanker Payudara. Media Penelit dan Pengemb Kesehat. 2017;27(4):247–54.
25. Andayani DGS, Sukandar U, Sukandar EY, Adnyana IK. Antibacterial, antifungal and anticancer, activity of five strains of soil microorganisms, isolated from Tangkuban Perahu Mountain by Fermentation. HAYATI J Biosci [Internet]. 2015;22(4):186–90. Available from: http://dx.doi.org/10.1016/j.hjb.2016.01.003
26. Goodfellow M, Kampfer P, Busse HJ, Trujillo ME, Suzuki KI, Ludwig W, et al. The actinobacteria. In: Whitman WB, editor. Bergey’s manual of systematic bacteriology. 2nd ed. Athens, Georgia: Department of Microbiology, University of Georgia; 2012. pp. 1446–804.
27. Li Q, Chen X, Jiang Y, Jiang C. Morphological identification of actinobacteria. In: Actinobacteria—basics and biotechnological applications. London, UK: Intech Open Science; 2016. pp. 59–86.
28. Akilandeswari P, Pradeep BV. Microbial pigments: potential functions and prospects. In: Singh OV, editor. Bio-pigmentation and biotechnological implementations. 1st ed. Hoboken, NJ: John Wiley and Sons, Inc.; 2017. 241–61 pp.
29. Peng F, Zhang MY, Hou SY, Chen J, Wu YY, Zhang YX. Insights into Streptomyces spp. Isolated from the rhizospheric soil of Panax notoginseng: isolation, antimicrobial activity and biosynthetic potential for polyketides and non-ribosomal peptides. BMC Microbiol. 2020;20:1–16.
30. Safari WF, Chasanah E, Wahyudi AT. Antibacterial and anticancer activities of marine bacterial extracts and detection of genes for bioactive compounds synthesis. Int J Pharm Pharm Sci [Internet]. 2016;8(2):55–9. Available from: https://innovareacademics.in/journals/index.php/ijpps/article/viewFile/9778/3815.pdf
31. Paguirigan JAG, Kim JA, Hur JS, Kim W. Identification of a biosynthetic gene cluster for a red pigment cristazarin produced by a lichen-forming fungus Cladonia metacorallifera. PLoS One [Internet]. 2023;18(6 June):1–15. doi: http://dx.doi.org/10.1371/journal.pone.0287559
32. Yusuf H, Satria D, Suryawati S, Fahriani M. Combination therapy of eurycomanone and doxorubicin as anticancer on T47D and MCF-7 cell lines. Syst Rev Pharm. 2020;11(10):335–41.
33. Lee N, Hwang S, Lee Y, Cho S, Palsson B, Cho BK. Synthetic biology tools for novel secondary metabolite discovery in Streptomyces. J Microbiol Biotechnol. 2019;29(5):667–86.
34. Yu S, Kim T, Yoo KH, Kang K. The T47D cell line is an ideal experimental model to elucidate the progesterone-specific effects of a luminal A subtype of breast cancer. Biochem Biophys Res Commun [Internet]. 2017;486(3):752–8. doi: http://dx.doi.org/10.1016/j.bbrc.2017.03.114
35. Nordin ML, Kadir AA, Zakaria ZA, Abdullah R, Nazrul M, Abdullah H. In vitro investigation of cytotoxic and antioxidative activities of Ardisia crispa against breast cancer cell lines, MCF-7 and. BMC Complement Altern Med. 2018;18(87):1–10.
36. Indrayanto G, Putra GS, Suhud F. Validation of in-vitro bioassay methods: application in herbal drug research [Internet]. Profiles Drug Subst Excip Relat Methodol. 2021;46:273–307. doi: http://dx.doi.org/10.1016/bs.podrm.2020.07.005
37. Peña-Morán OA, Villarreal ML, Álvarez-Berber L, Meneses-Acosta A, Rodríguez-López V. Cytotoxicity, post-treatment recovery, and selectivity analysis of naturally occurring podophyllotoxins from Bursera fagaroides var. fagaroides on breast cancer cell lines. Molecules. 2016;21(8):1–15.
38. Putri H, Jenie RI, Handayani S, Kastian RF, Meiyanto E. Combination of potassium pentagamavunon-0 and Doxorubicin induces apoptosis and cell cycle arrest and inhibits metastasis in breast cancer cells. Asian Pacific J Cancer Prev. 2016;17(5):2683–8.
39. Gao Q, Deng S, Jiang T. Recent developments in the identification and biosynthesis of antitumor drugs derived from microorganisms. Eng Microbiol [Internet]. 2022;2(4):1–23. doi: https://doi.org/10.1016/j.engmic.2022.100047
40. Nurkanto A, Agusta A. Molecular identification and morpho—physiological characterization of actinomycetes with antimicrobial properties. J Biol Indones. 2015;11(2):195–203.
41. Barco RA, Garrity GM, Scott JJ, Amend JP, Nealson KH, Emerson D. A genus definition for bacteria and archaea based on a standard genome relatedness index. MBio. 2020;11(1):1–20.
42. Takahashi Y. Genus Kitasatospora, taxonomic features and diversity of secondary metabolites. J Antibiot (Tokyo). 2017;70(5):506–13.
43. Li Y, Wang M, Sun ZZ, Xie BB. Comparative genomic insights into the taxonomic classification, diversity, and secondary metabolic potentials of Kitasatospora, a genus closely related to Streptomyces. Front Microbiol. 2021;12(June):1–13.