INTRODUCTION
Diagnostic testing forms the foundation of modern healthcare practice and provides essential clinical data about an individual’s health status by detecting disease conditions or infections [1]. These tests can range from simple blood analyses to comprehensive genetic screenings. They are crucial for ensuring diagnosis accuracy, guiding appropriate treatment plans, and tracking disease progression [2]. The validity and accuracy of these diagnostic tests must thus be guaranteed because they have a direct bearing on patient outcomes and clinical judgments. To preserve high standards and dependability in diagnostic testing, regulatory bodies, and healthcare facilities use stringent quality control and assurance procedures [3].
The utilization of reference materials (RMs) or external control materials (ECMs) in DNA-based diagnostic testing is pivotal for ensuring proper and reliable results. RMs act as benchmarks for testing procedures, enabling the detection and correction of errors and maintaining the validity of test results [4–6]. RMs and ECMs are different; RMs are a type of ECM that has been standardized and rigorously tested for specific parameters, making them vital for precise calibration and validation of diagnostic assays [7]. ECMs, which are carefully designed samples that mimic patient specimens, are used to assess the performance of diagnostic tests by providing an external standard to verify accuracy and precision [8]. Consistently incorporating both RMs and ECMs in the testing process is essential to meet quality assurance standards, particularly in healthcare and public health settings [9]. This function is critical for verifying test performance and identifying significant changes that could indicate issues with operational capabilities or equipment [10,11].
The evolution of diagnostic technology has transformed RMs from simple chemical or biological substances to more sophisticated matrices that support the immunoassay of complex substances, requiring consistent and stable RMs across various testing platforms [5,12,13]. Genetic engineering and cell culture innovations have ushered in a new era of RMs. These include synthetic DNA sequences, recombinant plasmids, genomic DNA, and cell lines with predetermined genetic compositions suited to particular diagnostic testing needs. Sanger sequencing, next-generation sequencing (NGS), Microarray and polymerase chain reaction (PCR) are examples of advanced technologies that require sensitive and specific RMs, which can improve the quality and accuracy of diagnostic tests [14–18].
Indonesia, like many other countries, faces unique challenges in its diagnostic technology sector, including limited access to high-quality RMs, variations in laboratory standards and procedures, and a need for stronger regulatory frameworks to ensure consistency and reliability in diagnostic testing [19,20]. These challenges can affect the accuracy of diagnostic results and have implications for patient care. Improving diagnostic technology in Indonesia is crucial to enhance healthcare outcomes, as accurate diagnostics are the cornerstone of effective treatment and disease management. By addressing these specific challenges and improving the quality of diagnostics, Indonesia can better manage public health, respond to disease outbreaks with greater agility, and align with international best practices. Incorporating these concerns, it is evident that there is a critical need for improvement within the diagnostic field in Indonesia to ensure precise and dependable test results, optimize patient care, and align with global healthcare standards. This review illuminates these issues and offers insights and recommendations to support Indonesian laboratories’ advancement [21].
This review focuses on summarizing current knowledge about RMs and suggests ways for laboratories to improve their diagnostic protocols in accordance with international standards. The goal is to guide laboratories in the selection of appropriate RMs to enhance the quality and accuracy of their diagnostic tests. Therefore, it covers various aspects as depicted in Figure 1. It highlights the methodologies used in the study, including the narrative review approach and the inclusion criteria for selecting relevant literature. Additionally, it encompasses the discussion on applying these RMs across various diagnostic technologies such as Sanger sequencing, NGS, microarray, and PCR (both real-time and digital). The figure concludes with recommendations for enhancing the quality and availability of RMs in Indonesia through national coordination and global collaboration initiatives to assist laboratories in making informed choices.
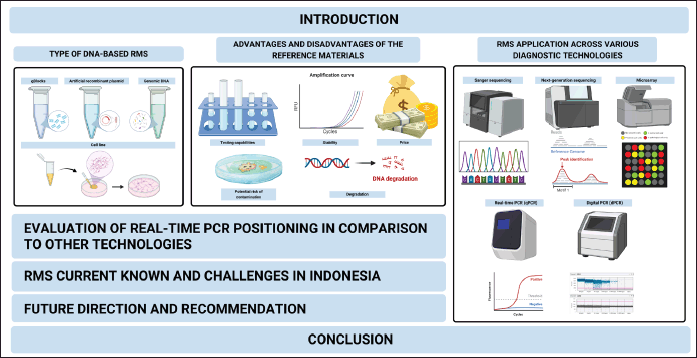 | Figure 1. A comprehensive outline for the review article illustrates the various components involved in evaluating and applying reference materials (RMs) in DNA-based diagnostic testing (Created with Biorender.com). [Click here to view] |
METHODS
This narrative review approaches the necessity and effectiveness of various control substances for precise diagnostic testing. The review focused on original research articles investigating synthetic DNA sequences, artificially constructed plasmids, genomic DNA, and cell lines as external controls and RMs. Databases such as PubMed, Scopus, and Google Scholar were utilized for the literature search. Scientific papers discussing the history, evaluation, and development of RMs and their application in diagnostic technologies, including Sanger sequencing, NGS, microarray, and real-time and digital PCR (dPCR), are covered in this review. Nonscientific publications, review articles, and sources older than ten years were excluded from this review.
TYPE OF DNA-BASED RMS
The genetic testing procedure using DNA-based PCR methods necessitates using RM to ensure the legitimacy and consistency of the results [22]. RMs play a crucial role in maintaining the integrity of the entire testing process by establishing essential performance standards [2]. Four commonly used RMs are gBlocks, artificial recombinant plasmids, genomic DNA, and cell lines.
gBlocks
gBlocks are synthetic, double-stranded DNA molecules designed to contain specific sequences [23]. In PCR-based protocols, gBlocks are primary controls to assess the assay’s effectiveness and specificity [24]. These synthetic blocks can represent various sequences, including gene variants, SNPs, or other genetic markers pertinent to the research [12]. The high fidelity and accuracy of gBlocks make them invaluable for optimizing PCR assays, allowing scientists to evaluate primer binding efficiency and the conditions necessary for precise detection and measurement [25].
Artificial recombinant plasmids
Recombinant plasmids typically contain synthetic and bacterial-amplified plasmids [26]. Synthetic plasmids are engineered in vitro to contain specific gene sequences and control elements, enhancing gene expression functions or serving as standards for molecular diagnostics [27]. Bacterial-amplified plasmids involve adding foreign DNA to a natural plasmid vector, which is then replicated in bacterial host cells [28]. These plasmids are significant in PCR methods because they can be reproduced in vitro, facilitating the production of large DNA fragment quantities and enabling accurate gene quantification through standard curves [29].
Genomic DNA
Genomic DNA, derived from reliable sources such as cell lines or well-characterized organisms, replicates the complexity of biological samples, ensuring proper DNA extraction and amplification processes [30]. It plays a critical role in genetic testing for detecting genetic changes or assessing gene copy numbers, providing a precise comparison representing the entire human DNA complexity [31].
Cell lines
Cell lines, continuously proliferating cell cultures, offer consistent, uniform genetic material for PCR-based testing [17,32]. They can be specifically chosen or genetically altered to have relevant mutations, gene expressions, or chromosomal changes, making them ideal for validating genetic tests. Cell lines serve as relevant biological counterparts to target genes, allowing researchers to assess assay performance under conditions similar to actual patient samples, ensuring high-quality control and uniformity in routine testing [33,34].
The strategic implementation of RMs in PCR processes, including genetic material and cell lines, effectively ensures the legitimacy and reliability of test results [35]. Utilizing RMs such as gBlocks, artificial recombinant plasmids, genomic DNA, and cell lines helps laboratories identify and reduce procedural errors, optimize assay conditions, and maintain high test accuracy. The intelligent use of RMs is crucial in clinical diagnostics and genetic testing research and development, emphasizing the importance of accurate and reliable results [4,22].
ADVANTAGES AND DISADVANTAGES OF RMS
The selection of appropriate RM is crucial for ensuring the accuracy and reliability of DNA-based diagnostic testing. Different types of RMs, such as gBlocks, artificial recombinant plasmids, genomic DNA, and cell lines, each offer unique advantages and disadvantages that need to be carefully considered (Table 1). The primary goal of using RMs is to provide consistent benchmarks for testing procedures, thereby enhancing the validity and reproducibility of diagnostic results [4,22].
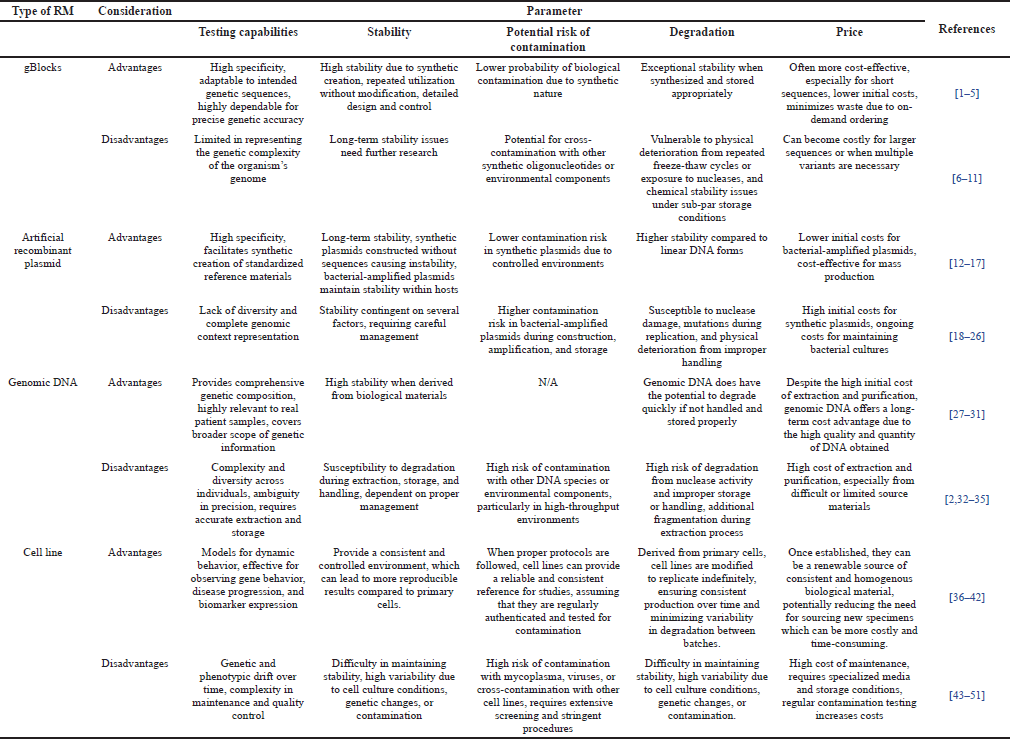 | Table 1. Advantages and disadvantages of gBlocks, recombinant plasmid, genomic DNA, and cell line RMs. [Click here to view] |
Four common types of RMs—gBlocks, artificial recombinant DNA, genomic DNA, and cell lines—each have distinct advantages and disadvantages. gBlocks are highly specific and adaptable, offering high stability and low contamination risk due to their synthetic nature, though they may not represent the entire genetic complexity of an organism and can degrade under certain conditions [23,24]. Recombinant plasmids provide high specificity and stability, particularly in controlled environments, but are susceptible to contamination and physical deterioration, with high costs associated with synthetic plasmids [26,36]. Genomic DNA offers a comprehensive genetic composition and high stability when properly managed, but it is prone to contamination and degradation, and its extraction and purification are costly [37,38]. Cell lines are effective for observing gene behavior and disease progression, yet they face challenges in maintaining stability and avoiding contamination, with high maintenance costs due to specialized requirements [39,40]. These considerations underscore the importance of selecting the appropriate RM based on the specific needs of the diagnostic application to ensure reliable and accurate results [4,22].
RMS APPLICATION ACROSS VARIOUS DIAGNOSTIC TECHNOLOGIES
Sanger sequencing
Sanger sequencing remains a fundamental method of DNA sequence analysis due to its high fidelity [37]. It is particularly effective for small-scale projects, such as identifying individual genes or regions of the genome involved in specific phenomena [41]. The principle of Sanger sequencing involves replicating a single DNA strand and incorporating fluorescently labeled nucleotides that terminate the growing chain (Fig. 2). These fragments are then separated by capillary electrophoresis, and the sequence is deduced from the labeled ends [42]. In practice, Sanger sequencing is used to validate results from other methods, confirm genetic variations like SNPs, and for forensic applications [43]. RMs, such as gBlocks or clones, ensure accuracy and repeatability by serving as benchmarks against which sequencing outputs are compared [44,45]. A notable case study demonstrated the use of Sanger sequencing in verifying gene mutations associated with various genetic disorders, underscoring its diagnostic precision [46].
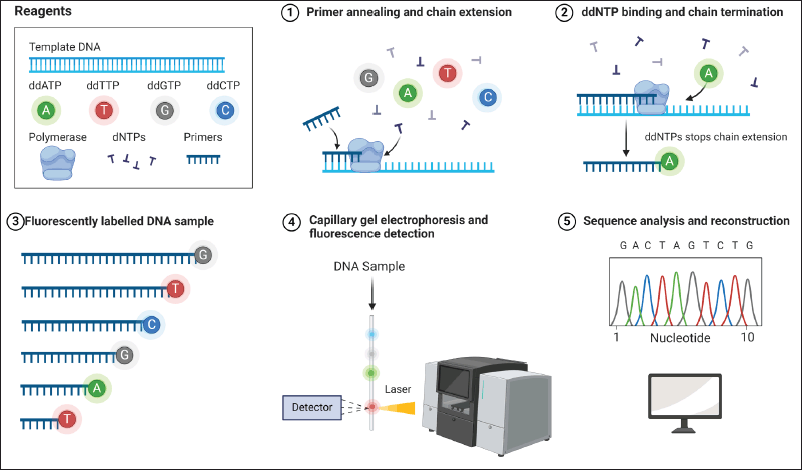 | Figure 2. Sanger sequencing workflow. In Sanger sequencing, essential reagents such as ddATP, ddTTP, ddGTP, ddCTP, polymerase, dNTPs, and primers are used. Initially, primers are anneal to the DNA template, and polymerase is extended using dNTPs. Subsequently, ddNTPs are incorporated, causing chain termination. The DNA fragments produced are then labeled with specific fluorescent dyes. These fragments undergo separation by size via capillary gel electrophoresis, followed by fluorescence detection. Ultimately, the fluorescence data is analyzed to decode the DNA sequence [52]. [Click here to view] |
Next-generation sequencing
NGS has revolutionized genetic analysis by enabling the simultaneous sequencing of multiple DNA strands [47]. This technology supports a variety of genomic analyses, including whole-genome sequencing, targeted re-sequencing, metagenomics, epigenomics, and transcriptome analysis [48]. NGS is characterized by its high throughput and cost-effectiveness, making it ideal for applications in personalized medicine, cancer genomics, and microbiome research [49]. The NGS workflow involves library creation, sequencing, image analysis, and data processing (Fig. 3).
 | Figure 3. Next-generation sequencing workflow. The process begins with library preparation, involving DNA fragmentation, adapter attachment, and the creation of a DNA fragment library. These fragments are amplified on a flow cell during library bridge amplification, forming clusters of identical sequences. In the DNA library sequencing step, fluorescently labeled nucleotides are used to sequence these fragments, generating raw data. Finally, the data undergoes alignment and analysis to reconstruct the final sequence, including contigs and the assembled sequence [53]. [Click here to view] |
RM are critical in NGS-based diagnosis because they rely on germline variant detection using high-throughput DNA sequencing for rare diseases. RMs such as standardized cell line genomes or synthetic DNA fragments ensure the quality and reliability of the data by serving as controls to validate the sequencing process and assess assay sensitivity and specificity [11]. For human whole genome sequencing, the NA12878 genome from the Coriell cell line GM12878 with a European ancestry background has been known as the sole leading human genome reference standard for various NGS applications. Despite the availability of Ashkenazi Jewish Asian and Han Chinese ancestry as genome references, as listed in the NIST-hosted Genome in a Bottle consortium, work continues to create reference genomes from various ancestries that cover all types of human genetic variation. The Genetic Testing Reference Materials Coordination Programme (GeT-RM) also provides various cell lines harboring specific mutations representing inherited diseases in ans. Additionally, for microbial reference standards, the US Food and Drug Administration (FDA) has released numerous microbial reference genomes for testing related to infectious microorganisms in the FDA-ARGOS database. Additionally, the Human Microbiome Project Consortium offers RMs for microbiome studies, assembling mock microbial communities from extracted gDNA samples.
Microarray
Microarray technology, also known as DNA chips, is a powerful tool for simultaneously analyzing multiple genes or DNA regions with high precision and efficiency [50]. Microarrays are solid supports, typically glass or silicon, onto densely packed DNA molecules. The principle of microarray technology, also known as DNA chips, involves the high-throughput analysis of gene expression or genetic variation. Microarrays consist of solid support, typically glass or silicon, onto which thousands of DNA probes are immobilized in a precise grid pattern. Each probe is designed to hybridize with a specific DNA or RNA sequence from the sample, allowing for the simultaneous analysis of many genes or genomic regions. During the assay, labeled nucleic acids from the sample are hybridized to the probes on the microarray. The hybridization signals are then detected and quantified using fluorescence or other detection methods, providing a comprehensive profile of gene expression or genetic variations (Fig. 4) [51]. Applications of microarrays include expression profiling, comparative genomic hybridization, and SNP detection [54].
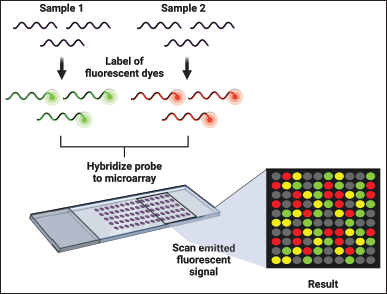 | Figure 4. Microarray workflow. In microarray, samples are tagged with fluorescent dyes and matched with the microarray. Subsequently, the emitted fluorescent signals are scanned and examined to gauge gene expression levels. Different colors signify various outcomes: gray indicates absent genes, green represents genes expressed solely in normal cells, yellow denotes genes expressed in both normal and pathological cells, and red indicates genes expressed only in pathological cells. This robust technique facilitates the identification of genes implicated in disease processes and contributes to developing diagnostic and therapeutic approaches [58]. [Click here to view] |
RM like gBlocks and recombinant plasmids are used to calibrate microarrays, ensuring data accuracy and consistency across different arrays and experiments [53]. A case study on the use of microarrays for detecting chromosomal abnormalities in prenatal diagnostics illustrated the technology’s utility in identifying genetic disorders early [56].
Polymerase chain reaction
Real-time PCR (qPCR) is widely used in molecular diagnostics to amplify and quantify specific DNA segments. This method employs fluorescent dyes or probes that emit fluorescence upon binding to DNA, with the fluorescence intensity being proportional to the amount of DNA amplified [57]. The principle of qPCR is based on the detection and quantification of a fluorescent signal that increases proportionally with the amount of DNA amplified during the PCR cycles. The process involves using fluorescent dyes, such as SYBR Green, which binds to double-stranded DNA, or specific fluorescently labeled probes, like TaqMan probes, which emit fluorescence upon hybridization with the target DNA sequence. As the PCR progresses, the fluorescent signal is measured in real-time at each cycle, allowing for the monitoring of DNA amplification throughout the reaction (Fig. 5).
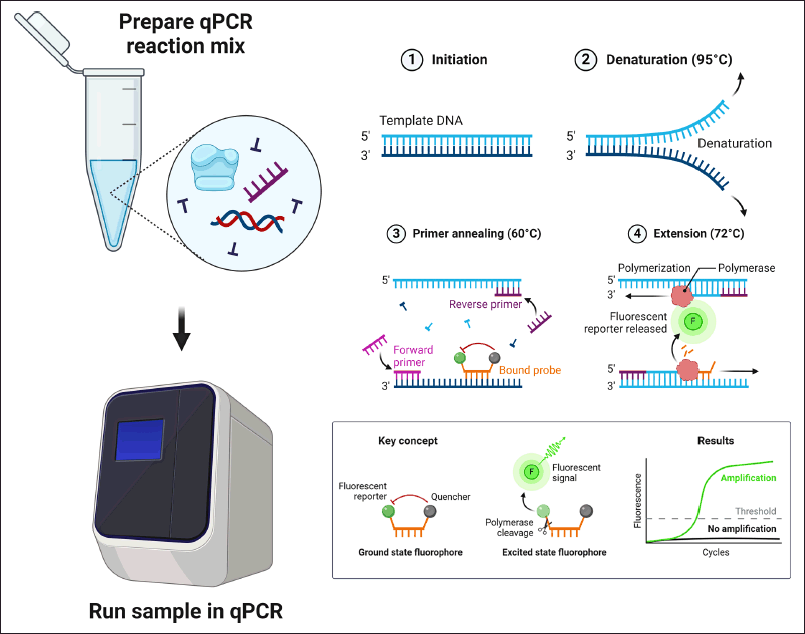 | Figure 5. Real-time PCR workflow. Initially, a reaction mix is prepared with template DNA (sample), master mix (enzymes and buffers), forward and reverse primers, a fluorescent probe (reporter), and nuclease-free water (NFW). This mix is then loaded into a qPCR machine. The qPCR process involves cycles including initiation, denaturation (DNA strand separation at 90°C), annealing (primers binding to specific DNA sequences at 60°C), and extension (polymerase extending the primers and cleaving the probe at 72°C). The behavior of the fluorescent probe is crucial; in the ground state, reporter and quencher dyes emit no signal, while in the excited state, polymerase cleaves the probe, generating a fluorescent signal. The measured signal is shown as an amplification curve, where a signal below a threshold level indicates undetected target DNA (Created with Biorender.com). [Click here to view] |
RMs with known DNA or RNA concentrations are crucial for creating standard curves to measure unknown samples accurately [56]. qPCR is employed in various diagnostic tests, including the detection of SARS-CoV-2, malaria, and Chagas disease, and genetic variant detection crucial for personalized medicine [60–62]. A case study on qPCR’s role in the rapid detection of SARS-CoV-2 during the COVID-19 pandemic highlighted its importance in managing public health emergencies [60].
On the other hand, dPCR offers exceptional precision and sensitivity in nucleic acid measurement by partitioning the DNA sample into numerous small-volume reactions, some containing the target molecule [63]. Each partition is then assessed for amplification, providing an absolute count of the target DNA without external references [64]. RMs ensure accuracy and reproducibility by validating the partitioning system and PCR efficiency [65]. dPCR is used in clinical applications such as analyzing tumor DNA, detecting drug resistance in viruses, early detection of infections, and monitoring transplant patients by measuring donor DNA levels [13,66,67]. A case study demonstrated the effectiveness of dPCR in detecting low-abundance tumor DNA, proving its value in early cancer diagnosis and monitoring [66]. Figure 6 illustrates the workflow of dPCR.
 | Figure 6. Digital PCR workflow. This diagram outlines the key steps in a dPCR workflow, allowing for precise quantification of target molecules. Initially, a reaction mixture is prepared with all necessary components for PCR amplification. Following this, the sample is carefully divided into numerous nanoliter-sized droplets using specialized equipment, ensuring each droplet encapsulates individual target molecules. Subsequent to droplet generation, the PCR cycling process occurs, amplifying target DNA within each droplet alongside any background DNA present. After amplification, droplets undergo analysis to differentiate positive droplets containing amplified targets from negative ones lacking. This analysis facilitates the accurate determination of the absolute abundance of target molecules in the original sample. dPCR offers researchers a robust tool with heightened sensitivity and the capability for absolute quantification, eliminating the need for standard curves (Created with Biorender.com). [Click here to view] |
EVALUATION OF REAL-TIME PCR POSITIONING IN COMPARISON TO OTHER TECHNOLOGIES
PCR technology is a cornerstone of DNA-based genetic testing, enabling the amplification of specific gene sequences from minute DNA samples [68,69]. qPCR, an advancement of traditional PCR, is a routinely used tool in molecular diagnostic laboratories [70]. qPCR’s adoption as a commonly found technology in diagnostic testing laboratories underlines its significance and thus provides a solid foundation for why this particular technology is the benchmark against which other methodologies are compared. qPCR is widely used due to its ability to quantify nucleic acids in real time, which is crucial for various applications, including disease diagnostics, genetic research, and pathogen detection [71]. This real-time quantification aspect offers a significant advantage over traditional PCR, enabling more detailed analyses of gene expression patterns, viral load determination, and genotyping [72]. Consequently, qPCR stands out as a highly sensitive, specific, and efficient diagnostic tool, making it a primary choice for comparing with other emerging genetic technologies [65].
PCR amplifies target DNA sequences through repeated cycles of temperature changes, which denature DNA, anneal primers, and extend new DNA strands [73]. This process results in an exponential increase in the target DNA region [68]. The advent of quantitative PCR (qPCR) introduced real-time DNA amplification and quantification using fluorescent dyes, enhancing genetic testing’s complexity and capabilities [65]. The essential stages of DNA-based PCR diagnostic testing are illustrated in Figure 7, which details sample collection, DNA extraction, reaction mixture preparation, and PCR amplification.
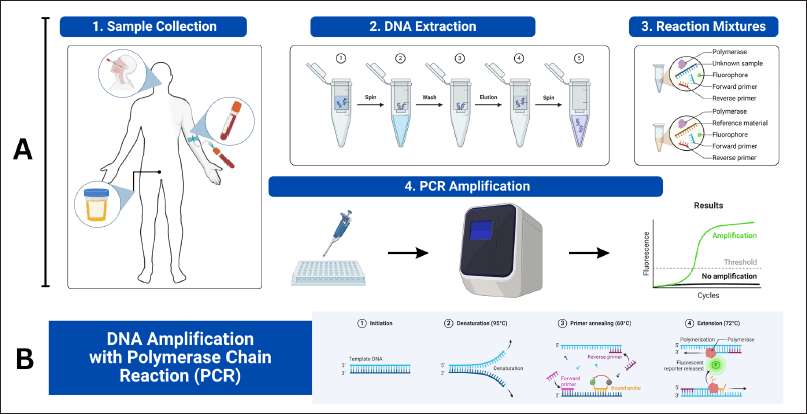 | Figure 7. A) The essential stages of DNA-based PCR diagnostic testing. Firstly, sample collection involves gathering biological material like blood or urine from the patient. Next, DNA extraction isolates DNA from other cellular components. Then, reaction mixture preparation mixes the extracted DNA with primers, DNA polymerase, and a fluorophore, divided into tubes for testing (unknown samples) and controls (reference material). Lastly, PCR amplification uses a PCR machine to amplify the DNA. B) The DNA amplification process through PCR. It begins with initiation, where double-stranded DNA is denatured into single strands. Denaturation heats the mixture to separate the DNA. Annealing follows, lowering the temperature for primers to bind to the target DNA. Extension occurs as DNA polymerase adds nucleotides, amplifying the target DNA exponentially (Created with Biorender.com). [Click here to view] |
The exceptional specificity, sensitivity, and quantitative capabilities of qPCR serve as the basis for molecular diagnostics. This efficacy comes from pre-designed, optimized primers and real-time fluorescence monitoring, which ensures excellent specificity and low background noise [68,74]. In addition to primer design, exact control over reaction parameters like temperature profiles and salt concentrations contributes to qPCR’s specificity, which makes it possible to identify single base pair variances and clinically meaningful genetic alterations [75]. The method’s superior sensitivity is achieved through efficient amplification and real-time fluorescence-based monitoring, providing accurate and reproducible quantitative data essential for applications like viral load measurement [72,76].
qPCR also offers operational simplicity, with master mixtures containing all necessary components, reducing the need for extensive optimization and minimizing potential errors. This setup simplicity, combined with user-friendly software for data interpretation, makes qPCR well-suited for rapid diagnostic environments [77,78]. This efficiency underscores qPCR’s popularity for rapid and accurate diagnostics [79]. Most laboratories are equipped with qPCR technology, making it a common choice for routine molecular testing.
While qPCR is a powerful tool in diagnostic testing, it is not without competition. Table 2 compares the cost and scope of various diagnostic technologies, including Sanger sequencing, NGS, Micro Arrays, and dPCR. Each technology has unique benefits and limitations that make it suitable for specific applications. For example, Sanger sequencing excels in sequencing small DNA fragments but is costly, while NGS offers large-scale sequencing capabilities but requires substantial initial investment [48,77]. Microarrays allow simultaneous analysis of multiple genes but are expensive and complex [50]. dPCR provides high precision and sensitivity but is slightly more costly than qPCR [63,81].
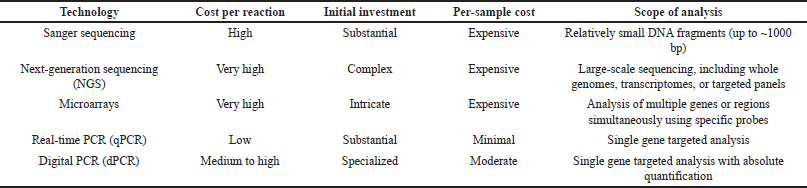 | Table 2. Cost comparison of diagnostic testing technologies. [Click here to view] |
The selection of molecular diagnostic technology depends on cost, accuracy, and scalability. Although qPCR offers significant advantages in certain areas, a comprehensive evaluation is essential to determine the most appropriate technology for specific laboratory needs [82,83].
RMS CURRENT KNOWN AND CHALLENGES IN INDONESIA
In the field of medical diagnostics, RMs play a crucial role in establishing standards and ensuring the legitimacy of tests for various diseases. RMs are essential for the validation of methods, the calibration of equipment, and the verification of diagnostic results [6,84]. Their diverse array of uses ensures consistency and reproducibility in diagnostic testing, which are fundamental to the accurate diagnosis and subsequent treatment of patients [5,10,84]. Studies employing RMs in the diagnosis of diseases have been conducted globally, covering a wide range of conditions, as summarized in Table 3.
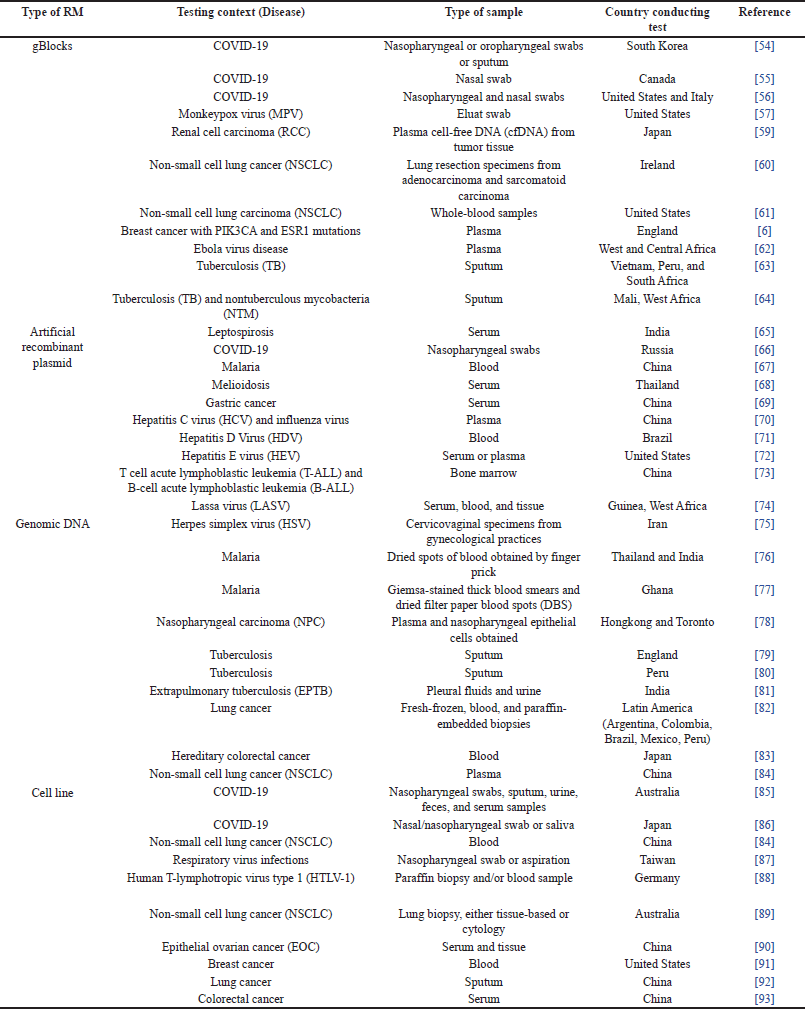 | Table 3. Current RMs used in diagnostic testing. [Click here to view] |
This worldwide research promotes increased quality control in healthcare and facilitates the development of genuine and pertinent diagnostic methods. The utilization of RM’s in this context is significant because of their importance in the progression of medical diagnosis and therapeutic strategies [10].
Multiple studies have concentrated on devising novel techniques to produce DNA RMs with certain attributes, which may be utilized as benchmarks for diverse purposes [85–88]. Seo et al. [82] reported a new bioprinting technique that can create RMs with a precise amount of target DNA, referred to as “cell number-based DNA reference material” [85]. This development offers a rigorous and replicable technique for producing DNA RMs with specified quantities, which is essential for the standardization and quality assurance of DNA analysis. In addition, the literature has emphasized the creation of RMs for certain DNA sequences or genetic markers [89–92]. In the research they conducted, [92] detailed the development of DNA molecules that functioned as model Standard RMs for the purpose of determining DNA sequences. Their findings highlighted the significance of well-characterized RMs in genetic testing. The mentioned references highlight the crucial importance of DNA RMs in upholding the quality and precision of genetic testing processes. In addition, DNA RMs have a wide range of uses, including identifying the mechanisms of antibiotic resistance in foodborne pathogens [87], creating genomic RMs for detecting meat adulteration [93], and constructing DNA hydrogels for biomedical applications [94]. Niu et al. [87] presented the creation of plasmid DNA RMs to identify antibiotic resistance mechanisms. They highlighted the usefulness of these materials as universal calibrators in qPCR analysis [87]. Furthermore, [93] emphasized the significance of quality control procedures in food analysis and the usefulness of DNA-based RMs in identifying adulterated meat [93]. NGS-based diagnosis heavily relies on RMs for identifying the presence of germline variants in rare diseases through the use of high-throughput DNA sequencing [95]. RMs, such as standardized cell line genomes or synthetic DNA fragments, are used to validate the sequencing procedure and evaluate the sensitivity and specificity of the assay. This ensures the data’s quality and reliability [11]. The NA12878 genome from the Coriell cell line GM12878, which has a European ancestry background, is widely recognized as the primary reference standard for different NGS applications in human whole genome sequencing [95,96]. Although the NIST-hosted Genome in a Bottle consortium provides genome references for Ashkenazi Asian and Han Chinese ancestry, efforts are still ongoing to develop reference genomes from diverse ancestries that encompass all forms of human genetic variation [96]. The Genetic Testing Reference Materials Coordination Programme (GeT-RM) offers a range of cell lines that contain specific mutations associated with inherited disorders in humans [97]. In addition, the US Food and Drug Administration (FDA) has made available a large number of microbial reference genomes in the FDA-ARGOS database for the purpose of testing infectious microorganisms [95]. In addition, the Human Microbiome Project Consortium provides RMs for microbiome studies by creating artificial microbial communities using extracted gDNA samples [95]. Organizations such as the Reference Materials and Measurements of the European Commission (IRMM-EU) and the American Oil Chemists’ Society (AOCS) are the main producers of commercially accessible certified RMs for matrix-based and genomic DNA. This information is supported by Li et al. [98]. These well-established organizations play a crucial role in standardizing and ensuring the quality of genetic testing methods by providing highly defined RMs. Collaborative ring trials have been carried out to evaluate the suitability of reference plasmid DNA calibrants in quantitative analyses of genetically modified (GM) contents. These trials emphasize the significance of certified reference materials (CRMs) in guaranteeing the precision and dependability of GM quantification methods [99]. In addition, the meticulous processes involved in certifying DNA reference RM materials to meet stringent quality standards are demonstrated through the development and characterization of quantitative RMs for specific applications, such as Legionella detection and quantification by qPCR [83].
The development of RMs has challenges, particularly in achieving interlaboratory repeatability. This issue has been emphasized in research that specifically investigates the use of new DNA RM formats in droplet digital PCR [100]. Ensuring the ability to replicate results in other laboratories is essential for the widespread acceptance of DNA RMs in diverse scientific fields. In Indonesia, the progress in developing DNA-based RMs is still in its early stages compared to international benchmarks. Although there is agreement with worldwide patterns in DNA nanotechnology and biomaterials research [101,102], there are still notable obstacles to overcome. These factors comprise restricted infrastructure, unavailability of new technological equipment, and inadequate money for comprehensive research and development projects.
To put it simply, the current situation in Indonesia regarding the establishment of DNA-based RMs encompasses the process of developing, characterizing, and certifying RMs for different purposes, such as genetic testing, and clinical diagnostics. The challenges involved in developing RMs encompass the need to guarantee consistent results across different laboratories. To address these difficulties, a significant investment in infrastructure, technology, and human resources is needed.
FUTURE DIRECTION AND RECOMMENDATION
Future research should focus on developing and standardizing RMs to improve the quality and reliability of DNA-based diagnostics. This includes the establishment of a national coordination body in Indonesia to oversee the production, validation, and distribution of RMs. Collaboration between research institutions, standardization bodies, and regulatory agencies is essential to create a robust framework for RM utilization. These efforts should prioritize the creation of standardized reference controls that align with international standards, ensuring consistent and reproducible diagnostic results [103]. By incorporating locally relevant pathogen strains and epidemiological data, the selected RMs will enhance the accuracy and validity of diagnostic tests within the Indonesian context [104].
The selection and implementation of RMs must balance cost, capability, stability, and contamination risks. Laboratories with limited budgets can utilize cost-effective synthetic targets like gBlocks for high-volume applications, while more complex diagnostics may require genomic DNA or cell lines despite their higher costs. Continuous proficiency testing and external quality assessment are vital to validate laboratory performance against common RMs, identifying areas for improvement and ensuring high diagnostic standards [104]. Investment in infrastructure, personnel training, and policy enforcement is critical for maintaining high-quality diagnostic testing. Robust infrastructure supports effective testing, while trained personnel ensure reliable results, and policy enforcement promotes a culture of excellence [97].
Innovation and adaptability are crucial in the evolving field of diagnostic testing. Embracing new technologies and staying updated with advancements will keep Indonesia at the forefront of global diagnostic standards. This commitment to innovation will improve diagnostic accuracy, enhance patient care, and strengthen public health. By focusing on continuous improvement and aligning with international best practices, Indonesia can ensure its diagnostic testing capabilities remain advanced and effective, ultimately benefiting patient outcomes and the healthcare system as a whole.
CONCLUSION
The comprehensive review highlights the pivotal role of RMs in ensuring the accuracy and reliability of DNA-based diagnostic testing. This study identifies their unique characteristics, advantages, and limitations by examining various types of RMs—gBlocks, recombinant plasmids, genomic DNA, and cell lines. gBlocks, for example, offer high specificity and stability but may lack genomic complexity. Recombinant plasmids are flexible and stable but costly and prone to contamination. Genomic DNA provides a comprehensive genetic composition and high stability, yet it is susceptible to degradation and high contamination risk. While effective for observing gene behavior and disease progression, cell lines face challenges in maintaining stability and avoiding contamination, with high maintenance costs. The strategic selection of RMs, considering specificity, complexity, stability, and cost factors, is essential for optimizing diagnostic testing. The establishment of a national coordination body in Indonesia to standardize RM utilization and collaboration with international organizations will enhance the quality and availability of RMs, ensuring diagnostic accuracy and reliability that meets international standards.
LIST OF ABBREVIATIONS
dPCR: digital PCR; DNA: deoxyribonucleic acid; ECM: external control material; NGS: next-generation sequencing; PCR: polymerase chain reaction; RM: reference material; qPCR: real-time PCR/quantitative PCR.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FUNDING
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
DATA ACCESS AND RETENTION
All data analyzed during this study is included in this published article in the references section.
REFERENCES
1. Balogh EP, Miller BT, Ball JR, editors. Improving diagnosis in health care. Washington, DC: National Academies Press; 2015.
2. DiGangi BA. Diagnostic testing. In: Miller L, Hurley KF, editors. Infectious disease management in animal shelters. Hoboken, NJ: Wiley; 2021. pp. 60–93.
3. Land KJ, Boeras DI, Chen XS, Ramsay AR, Peeling RW. Reassured diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat Microbiol. 2018;4:46–54. CrossRef
4. Ahmad-Nejad P, Ashavaid T, Vacaflores Salinas A, Huggett J, Harris K, Linder MW, et al. Current and future challenges in quality assurance in molecular diagnostics. Clinica Chimica Acta. 2021;519:239–246. CrossRef
5. Devonshire AS, O’Sullivan DM, Honeyborne I, Jones G, Karczmarczyk M, Pavši? J, et al. The use of digital PCR to improve the application of quantitative molecular diagnostic methods for tuberculosis. BMC Infect Dis. 2016;16:1–10. CrossRef
6. van Pelt-Verkuil E, van Leeuwen WB. Quality assurance, management and control in molecular diagnostics. In: van Pelt-Verkuil E, van Leeuwen W, te Witt R, editors. Molecular Diagnostics. Singapore, Singapore: Springer; 2019.
7. International Organization for Standardization. Reference materials—Selected terms and definitions (ISO Guide 30:2015). Geneva, Switzerland: ISO; 2015. p. 8.
8. De la Salle B, Meijer P, Thomas A, Simundic AM. Special issue on external quality assessment in laboratory medicine—current challenges and future trends. Biochem Med (Zagreb). 2017;27:19–22. CrossRef
9. Borysiak MD, Thompson MJ, Posner JD. Translating diagnostic assays from the laboratory to the clinic: analytical and clinical metrics for device development and evaluation. Lab Chip. 2016;16:1293–313. CrossRef
10. Arnold JE, Camus MS, Freeman KP, Giori L, Hooijberg EH, Jeffery U, et al. ASVCP guidelines: principles of quality assurance and standards for veterinary clinical pathology (version 3.0): developed by the American Society for Veterinary Clinical Pathology’s (ASVCP) Quality Assurance and Laboratory Standards (QALS) Committee. Vet Clin Pathol. 2019;48:542–618. CrossRef
11. Gargis AS, Kalman L, Lubin IM. Assuring the quality of next-generation sequencing in clinical microbiology and public health laboratories. J Clin Microbiol. 2016;54:2857–65. CrossRef
12. Bhadra S, Paik I, Torres JA, Fadanka S, Gandini C, Akligoh H, et al. Preparation and use of cellular reagents: a low-resource molecular biology reagent platform. Curr Protoc. 2022;2:1–54. CrossRef
13. Whale AS, Bushell CA, Grant PR, Cowen S, Gutierrez Aguirre I, O’Sullivan DM, et al. Detection of rare drug resistance mutations by digital PCR in a human influenza a virus model system and clinical samples. J Clin Microbiol. 2016;54:392–400. CrossRef
14. Abdi G, Tarighat MA, Jain M, Tendulkar R, Tendulkar M, Barwant M. Revolutionizing genomics: exploring the potential of next-generation sequencing. In: Singh V, Kumar A, editors. Advances in Bioinformatics. Singapore, Singapore: Springer Nature;2024. pp. 1–33.
15. Ghasemi M, Nabipour I, Omrani A, Alipour Z, Assadi M. Precision medicine and molecular imaging: new targeted approaches toward cancer therapeutic and diagnosis. Am J Nucl Med Mol Imaging. 2016;6(6):310–27.
16. Huang F, Spangler JR, Huang AY. In vivo cloning of up to 16 kb plasmids in E. coli is as simple as PCR. PLoS One. 2017;12:1–21. CrossRef
17. Wilding JL, Bodmer WF. Cancer cell lines for drug discovery and development. Cancer Res. 2014;74:2377–84. CrossRef
18. Xu L, Chen H, Canales M, Ciric L. Use of synthesized double-stranded gene fragments as qPCR standards for the quantification of antibiotic resistance genes. J Microbiol Methods. 2019;164:105670. CrossRef
19. ARISE+ Indonesia. Amplifying the Importance of Reference Materials to Foster RMP Growth in Indonesia [Internet]. 2023 [cited 2024 May 30]. Available from: https://ariseplus-indonesia.org/en/activities/amplifying-the-importance-of-reference-materials-to-foster-rmp-growth-in-indonesia.html.
20. Prasetya B, Restu Wahono D, Yopi, Prasetya C. Innovation opportunity and challenge of standardization in response to COVID-19 pandemic and the socio-economic impact: a case study in Indonesia. Standards. 2022;2:66–82. CrossRef
21. Cree IA, Deans Z, Ligtenberg MJL, Groenen P, Van Krieken JH, Normanno N, et al. Guidance for laboratories performing molecular pathology for cancer patients. J Clin Pathol. 2014;67:923–31. CrossRef
22. Chan M, Jiang B, Tan TY. Using pooled recombinant plasmids as control materials for diagnostic real-time PCR. Clin Lab. 2016;62:1893–901. CrossRef
23. Bogožalec Košir A, Cvelbar T, Kammel M, Grunert HP, Zeichhardt H, Milavec M. Digital PCR method for detection and quantification of specific antimicrobial drug-resistance mutations in human cytomegalovirus. J Virol Methods. 2020;281:113864. CrossRef
24. Rowlands V, Rutkowski AJ, Meuser E, Carr TH, Harrington EA, Barrett JC. Optimisation of robust singleplex and multiplex droplet digital PCR assays for high confidence mutation detection in circulating tumour DNA. Sci Rep. 2019;9:1–13. CrossRef
25. Sikkema AP, Tabatabaei SK, Lee Y, Lund S, Lohman GJS. High-complexity one-pot golden gate assembly. Curr Protoc. 2023;3:e882. CrossRef
26. Martínez-García E, Benedetti I, Hueso A, de Lorenzo V. Mining environmental plasmids for synthetic biology parts and devices. In: Marcelo E. Tolmasky, Juan C. Alonso, editors. Plasmids: Biology and Impact in Biotechnology and Discovery. Hoboken, NJ: John Wiley & Sons; 2015. pp. 633–49. CrossRef
27. Roquet N, Soleimany AP, Ferris AC, Aaronson S, Lu TK. Synthetic recombinase-based state machines in living cells. Science. 2016 Jul 22;353(6297):aad8559. CrossRef
28. Green MR, Sambrook J. Cloning in plasmid vectors: directional cloning. Cold Spring Harb Protoc. 2020;2020:485–8. CrossRef
29. Kit M. Development of recombinant positive control for African Swine fever virus pcr detection. Biotechnol Acta. 2020;13:58–63. CrossRef
30. Fang LT, Zhu B, Zhao Y, Chen W, Yang Z, Kerrigan L, et al. Establishing community reference samples, data and call sets for benchmarking cancer mutation detection using whole-genome sequencing. Nat Biotechnol. 2021;39:1151–60. CrossRef
31. Garcia EP, Minkovsky A, Jia Y, Ducar MD, Shivdasani P, Gong X, et al. Validation of oncopanel a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med. 2017;141:751–8. CrossRef
32. Segeritz CP, Vallier L. Cell culture: growing cells as model systems in vitro. In: Jalali M, Saldanha FYL, Jalali M, editors. Basic Science Methods for Clinical Researchers. London,UK: Elsevier Inc.; 2017.
33. El Harane S, Zidi B, El Harane N, Krause KH, Matthes T, Preynat-Seauve O. Cancer spheroids and organoids as novel tools for research precision medicine. Cells. 2023;12:1001
34. Lin G, Zhang K, Han Y, Li J. Quality control materials for pharmacogenomic testing in the clinic. Clin Chem Lab Med. 2017;55:926–33. CrossRef
35. Cirmena G, Dameri M, Ravera F, Fregatti P, Ballestrero A, Zoppoli G. Assessment of circulating nucleic acids in cancer: from current status to future perspectives and potential clinical applications. Cancers (Basel). 2021;13:1–24. CrossRef
36. Flagg MP, Kao A, Hampton RY. Integrating after CEN Excision (ICE) Plasmids: Combining the ease of yeast recombination cloning with the stability of genomic integration. Yeast. 2019;36:593–605. CrossRef
37. Volckmar A, Sültmann H, Riediger A, Fioretos T, Schirmacher P, Endris V, et al. A field guide for cancer diagnostics using cell-free DNA: from principles to practice and clinical applications. Genes Chromosomes Cancer. 2018;57:123–39. CrossRef
38. Kasu M, Shires K. The validation of forensic DNA extraction systems to utilize soil contaminated biological evidence. Leg Med. 2015;17:232–8. CrossRef
39. Mirabelli P, Coppola L, Salvatore M. Cancer cell lines are useful model systems for medical research. Cancers (Basel). 2019;11:1098. CrossRef
40. Geraghty RJ, Capes-Davis A, Davis JM, Downward J, Freshney RI, Knezevic I, et al. Guidelines for the use of cell lines in biomedical research. Br J Cancer. 2014;111:1021–46. CrossRef
41. El-Metwally S. Next Generation Sequencing Technologies and Challenges in Sequence Assembly. Volume 7 of SpringerBriefs in Systems Biology. New York, NY: Springer; 2014.
42. Marma MS. Terminator to reversible terminator: generational shift in DNA sequencing technologies. In: Islam MM, Hossain MM, editors. Science and Technology Innovation for a Sustainable Economy. Cham, Switzerland: Springer International Publishing; 2020. pp. 43–58.
43. Sturk Andreaggi K, Peck MA, Boysen C, Dekker P, McMahon TP, Marshall CK. AQME: A forensic mitochondrial DNA analysis tool for next-generation sequencing data. Forensic Sci Int Genet. 2017;31:189–97. CrossRef
44. Li Z, Sinha A, Zhang Y, Tanner N, Cheng HT, Premsrirut P, et al. Extraction-free LAMP assays for generic detection of Old World Orthopoxviruses and specific detection of Mpox virus. Sci Rep. 2023;13:1–12. CrossRef
45. Head SR, Kiyomi Komori H, LaMere SA, Whisenant T, Van Nieuwerburgh F, Salomon DR, et al. Library construction for next-generation sequencing: overviews and challenges. Biotechniques. 2014;56:61–77. CrossRef
46. Choochuen P, Laochareonsuk W, Tanaanantarak P, Kanjanapradit K, Sangkhathat S. Juvenile hyaline fibromatosis: report of a case with a novel ANTXR2 gene mutation. Am J Case Rep. 2022;23:1–6. CrossRef
47. McCombie WR, McPherson JD, Mardis ER. Next-generation sequencing technologies. Cold Spring Harb Perspect Med. 2019;9:a036798. CrossRef
48. Tripathi R, Sharma P, Chakraborty P, Varadwaj PK. Next-generation sequencing revolution through big data analytics. Front Life Sci. 2016;9:119–49. CrossRef
49. Bai J, Jhaney I, Wells J. Developing a reproducible microbiome data analysis pipeline using the amazon web services cloud for a cancer research group: proof-of-concept study. JMIR Med Inform. 2019;7:e14667. CrossRef
50. Gonzalo R, Sánchez A. Introduction to microarrays Technology and Data Analysis. 1st ed. Comprehen Anal Chem. 2018;82:37–69.
51. Miller S, Karaoz U, Brodie E, Dunbar S. Solid and suspension microarrays for microbial diagnostics. Methods Microbiol. 2015;42:395–431.
52. Ona S. Sanger sequencing [Internet]. BioRender. 2020. Available from: https://app.biorender.com/biorender-templates/figures/all/t-5ef132f6c7bcd500b388a9c3-sanger-sequencing
53. Ona S. Next Generation Sequencing (Illumina) [Internet]. BioRender. 2020. Available from: https://app.biorender.com/biorender-templates/figures/all/t-5ef134a11c72b100ad8d13ac-next-generation-sequencing-illumina
54. Agapito G. Computer Tools to Analyze Microarray Data. Methods Mol Biol. 2019;1986:267–82. CrossRef
55. Ziemann M, Reimann V, Liang Y, Shi Y, Ma H, Xie Y, et al. CvkR is a MerR-type transcriptional repressor of class 2 type V-K CRISPR-associated transposase systems. Nat Commun. 2023;14:1–16. CrossRef
56. Dugoff L, Norton ME, Kuller JA. The use of chromosomal microarray for prenatal diagnosis. Am J Obstet Gynecol. 2016;215:B2–B9. CrossRef
57. Pavši? J, Žel J, Milavec M. Assessment of the real-time PCR and different digital PCR platforms for DNA quantification. Anal Bioanal Chem. 2016;408:107–21. CrossRef
58. Auliah FN, Lawi A, Thamrin SA, Budiman E. Selection of informative genes to classify type 2 diabetes mellitus using support vector machine. 5th International Conference on Computing Engineering and Design, ICCED 2019. Singapore, Singapore: Institute of Electrical and Electronics Engineers Inc; 2019.
59. Kralik P, Ricchi M. A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front Microbiol. 2017;8:1–9. CrossRef
60. Park M, Won J, Choi BY, Lee CJ. Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR. Exp Mol Med. 2020;52:963–77. CrossRef
61. Patel JC, Lucchi NW, Srivastava P, Lin JT, Sug-Aram R, Aruncharus S, et al. Field evaluation of a real-time fluorescence loop-mediated isothermal amplification assay, realamp, for the diagnosis of Malaria in Thailand and India. J Infect Dis. 2014;210:1180–7. CrossRef
62. Ramírez JC, Cura CI, Da Cruz Moreira O, Lages-Silva E, Juiz N, Velázquez E, et al. Analytical validation of quantitative real-time PCR methods for quantification of trypanosoma cruzi DNA in blood samples from chagas disease patients. J Mol Diagn. 2015;17:605–15. CrossRef
63. Long S, Berkemeier B. Ultrasensitive detection and quantification of viral nucleic acids with Raindance droplet digital PCR (ddPCR). Methods. 2022;201:49–64. CrossRef
64. Whale AS, Jones GM, Pavši? J, Dreo T, Redshaw N, Akyürek S, et al. Assessment of digital PCR as a primary reference measurement procedure to support advances in precision medicine. Clin Chem. 2018;64:1296–307. CrossRef
65. Zhang H, Cao L, Brodsky J, Gablech I, Xu F, Li Z, et al. Quantitative or digital PCR? a comparative analysis for choosing the optimal one for biosensing applications. Trends Anal Chem. 2024;174:117676. CrossRef
66. Gutteridge A, Rathbone VM, Gibbons R, Bi M, Archard N, Davies KEJ, et al. Digital PCR analysis of circulating tumor DNA: a biomarker for chondrosarcoma diagnosis, prognostication, and residual disease detection. Cancer Med. 2017;6:2194–202. CrossRef
67. Chen B, Xie Y, Zhang N, Li W, Liu C, Li D, et al. Evaluation of droplet digital PCR assay for the diagnosis of candidemia in blood samples. Front Microbiol. 2021;12:700008. CrossRef
68. Kadri K. Polymerase chain reaction (PCR): principle and applications. In: Nagpal ML, Boldura OM, Balta C, Enany S, editors. Synthetic Biology - New Interdisciplinary Science. IntechOpen; 2020.
69. Findlay SD, Vincent KM, Berman JR, Postovit LM. A digital pcr-based method for efficient and highly specific screening of genome edited cells. PLoS One. 2016;11:1–17. CrossRef
70. Singh C, Roy-Chowdhuri S. Quantitative real-time PCR: recent advances. Methods Mol Biol. 2016;1392:161–76.
71. Ricchi M, Bertasio C, Boniotti MB, Vicari N, Russo S, Tilola M, et al. Comparison among the quantification of bacterial pathogens by qPCR, dPCR, and cultural methods. Front Microbiol. 2017;8:1174. CrossRef
72. Engstrom-Melnyk J, Rodriguez PL, Peraud O, Hein RC. Clinical applications of quantitative real-time PCR in virology. Methods Microbiol. 2015;42:161–97.
73. Levin RE, Ekezie F-GC, Sun D-W. DNA-based technique: polymerase chain reaction (PCR). In: Da-Wen Sun. Modern Techniques for Food Authentication. Amsterdam, The Netherlands: Elsevier; 2018. pp. 527–616.
74. Ellis JA, Ong B. The MassARRAY® system for targeted SNP genotyping. Methods Mol Biol. 2017;1492:77–94. CrossRef
75. Navarro E, Serrano-Heras G, Castaño MJ, Solera J. Real-time PCR detection chemistry. Clinica Chimica Acta. 2015;439:231–50. CrossRef
76. Chen R, Lu X, Li M, Chen G, Deng Y, Du F, et al. Polymerase chain reaction using “V” shape thermal cycling program. Theranostics. 2019;9:1572–9. CrossRef
77. VanInsberghe M, Zahn H, White AK, Petriv OI, Hansen CL. Highly multiplexed single-cell quantitative PCR. PLoS One. 2018;13:e0191601. CrossRef
78. Koessler T, Addeo A, Nouspikel T. Implementing circulating tumor DNA analysis in a clinical laboratory: a user manual. Adv Clin Chem. 2019;89:131–88.
79. Cao L, Cui X, Hu J, Li Z, Choi JR, Yang Q, et al. Advances in digital polymerase chain reaction (dPCR) and its emerging biomedical applications. Biosens Bioelectron. 2017;90:459–74. CrossRef
80. Sabatini LM, Mathews C, Ptak D, Doshi S, Tynan K, Hegde MR, et al. Genomic sequencing procedure microcosting analysis and health economic cost-impact analysis: a report of the association for molecular pathology. J Mol Diagn. 2016;18:319–28. CrossRef
81. Deharvengt SJ, Petersen LM, Jung HS, Tsongalis GJ. Nucleic acid analysis in the clinical laboratory. In: Clarke W, Marzinke M, editors. Contemporary practice in clinical chemistry. Amsterdam, The Netherlands: Elsevier Science; 2020. pp. 215–34.
82. Stubbs SCB, Blacklaws BA, Yohan B, Yudhaputri FA, Hayati RF, Schwem B, et al. Assessment of a multiplex PCR and Nanopore-based method for dengue virus sequencing in Indonesia. Virol J. 2020;17:1–13. CrossRef
83. Llamas B, Valverde G, Fehren-Schmitz L, Weyrich LS, Cooper A, Haak W. From the field to the laboratory: controlling DNA contamination in human ancient DNA research in the high-throughput sequencing era. Sci Technol Archaeol Res. 2017;3:1–14. CrossRef
84. Zaman GS. Introductory Chapter: History and Scope of Quality Control in Laboratories. Quality Control in Laboratory. 2018;1–8. CrossRef
85. Seo M, Takabatake R, Izumi S, Unno H, Kawashima Y, Ki U, et al. Novel bioprinting application for the production of reference material containing a defined copy number of target DNA. Anal Chem. 2019;91:12733–40. CrossRef
86. Pinheiro LB, O’Brien H, Druce J, Do H, Kay P, Daniels M, et al. Interlaboratory reproducibility of droplet digital polymerase chain reaction using a new DNA reference material format. Anal Chem. 2017;89:11243–51. CrossRef
87. Niu Q, Su X, Lian L, Huang J, Xue S, Zhou W, et al. Developing qualitative plasmid DNA reference materials to detect mechanisms of quinolone and fluoroquinolone resistance in foodborne pathogens. Foods. 2022;11:154. CrossRef
88. Mano J, Hatano S, Futo S, Yoshii J, Nakae H, Naito S, et al. Development of a reference material of a single DNA molecule for the quality control of PCR testing. Anal Chem. 2014;86:8621–7. CrossRef
89. Yao Z, Liu H, Xie F, Fischer S, Adkins RS, Aldridge AI, et al. A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex. Nature. 2021;598:103–10. CrossRef
90. Axtner J, Crampton-Platt A, Hörig LA, Mohamed A, Xu CCY, Yu DW, et al. An efficient and robust laboratory workflow and tetrapod database for larger scale environmental DNA studies. Gigascience. 2019;8:giz029. CrossRef
91. Debode F, Janssen E, Bragard C, Berben G. Detection by real-time PCR and pyrosequencing of the cry 1Ab and cry 1Ac genes introduced in genetically modified (GM) constructs. Food Additives Contaminants: Part A. 2017;34:1398–409. CrossRef
92. Rehder C, Bean LJH, Bick D, Chao E, Chung W, Das S, et al. Next-generation sequencing for constitutional variants in the clinical laboratory, 2021 revision: a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021;23:1399–415. CrossRef
93. Chen X, Ji Y, Li K, Wang X, Peng C, Xu X, et al. Development of a duck genomic reference material by digital PCR platforms for the detection of meat adulteration. Foods. 2021;10:1890. CrossRef
94. Ma Y, Duan X, Huang J. DNA Hydrogels as functional materials and their biomedical applications. Adv Funct Mater. 2024;34:1–20. CrossRef
95. Hardwick SA, Deveson IW, Mercer TR. Reference standards for next-generation sequencing. Nat Rev Genet. 2017;18:473–84. CrossRef
96. Zook JM, Catoe D, McDaniel J, Vang L, Spies N, Sidow A, et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data. 2016;3:160025. CrossRef
97. Kalman LV, Datta V, Williams M, Zook JM, Salit ML, Han JY. Development and characterization of reference materials for genetic testing: focus on public partnerships. Ann Lab Med. 2016;36:513–20. CrossRef
98. Li Z, Li X, Wang C, Song G, Pi L, Zheng L, et al. One novel multiple-target plasmid reference molecule targeting eight genetically modified canola events for genetically modified canola detection. J Agric Food Chem. 2017;65:8489–500. CrossRef
99. Li L, Chen J, Moore K, Jin W. Method validation. In: Shillito R, Shan G, editors. Application of sampling and detection methods in agricultural plant biotechnology. Amsterdam, The Netherlands: Elsevier; 2022. pp. 67–84.
100. Ye X, Lei B. The current status and trends of DNA extraction. BioEssays. 2023;45:e2200242. CrossRef
101. Qin Z, Wang C, Zhang J, Wang Z, Wei Y, Li Y, et al. DNA-based materials inspired by natural extracellular DNA. Adv Funct Mater. 2023;33. CrossRef
102. Ma W, Zhan Y, Zhang Y, Mao C, Xie X, Lin Y. The biological applications of DNA nanomaterials: current challenges and future directions. Signal Transduct Target Ther. 2021;6:1–28. CrossRef
103. Scott SA, Scott ER, Seki Y, Chen AJ, Wallsten R, Owusu Obeng A, et al. Development and analytical validation of a 29 gene clinical pharmacogenetic genotyping panel: Multi-Ethnic Allele and Copy Number Variant Detection. Clin Transl Sci. 2021;14:204–13. CrossRef
104. Gaedigk A, Turner A, Everts RE, Scott SA, Aggarwal P, Broeckel U, et al. Characterization of reference materials for genetic testing of CYP2D6 alleles. J Mol Diagn. 2019;21:1034–52. CrossRef