INTRODUCTION
The coronavirus disease 2019 (COVID-19) has caused an outbreak worldwide [1]. Even though WHO has declared the end of the COVID-19 pandemic, marking the transition into an endemic disease [2], new cases of COVID-19 are still emerging, accompanied by new mutations found in the viral genome of the causative virus SARS-CoV-2 [3]. SARS-CoV-2 infections generally cause mild to moderate symptoms, for instance, cough, sore throat, fever, and myalgia [4]. However, although different major clinical manifestations occur across SARS-CoV-2 variants, more persistent symptoms are observed for current Omicron variants such as Omicron BA.2, which may affect long-term routine activities [5,6]. On the other hand, the main focus of the global response to this disease has been to minimize the risk of SARS-CoV-2 transmission by vaccine development [7]. Nevertheless, as SARS-CoV-2 has evolved, developing therapeutic candidates that can prevent COVID-19 progression becomes more crucial.
Citrus (Citrus reticulata Blanco) peel is rich in hesperidin (HSD) [8,9]. Upon enzymatic hydrolysis, HSD is converted to its aglycon, hesperetin (HST) (Fig. 1A), which has a better membrane permeability and is more absorbable than HSD [10]. In-silico studies revealed that HSD and HST interact with human Angiotensin Converting Enzyme 2 (hACE2) and TMPRSS2 [11–13], two proteins involved in SARS-CoV-2 cellular entry [14,15], indicating their potential anti-SARS-CoV-2 effects. However, a report on laboratory-based in vitro assays showing HSD-HST antiviral effects is still lacking. On the other hand, since SARS-CoV-2 is considered a risk group 3 virus by the WHO [16], a biosafety level (BSL) 3 containment is needed to handle this virus. Therefore, the limited number of facilities providing BSL-3 containments becomes a challenge in studying SARS-CoV-2.
The pseudovirus model offers an alternative to performing a SARS-CoV-2 cellular entry assay. Pseudovirus is a virus whose main structure and envelope proteins are derived from two viruses [17]. The main structure of SARS-CoV-2 pseudovirus can be constructed from a lower-risk group virus, such as vesicular stomatitis virus (VSV), enabling it to be handled in a laboratory with a lower biosafety level [18]. As a primary structure, VSV can be engineered to incorporate spike proteins of SARS-CoV-2 as its outer proteins [19]. Thus, the SARS-CoV-2 pseudovirus model facilitates the study of HSD and HST effects on SARS-CoV-2 infection.
To observe the inhibitory effect of citrus peel extract, HSD, and HST on SARS-CoV-2 infection, we performed a SARS-CoV-2 pseudovirus internalization assay in 293T cells expressing hACE2/TMPRSS2 and in 293/hACE2 cells. A replication-incompetent pseudovirus spike*?G-GFP rVSV was used as a model of SARS-CoV-2 on the internalization assay.
METHOD
Preparation of orange peel extract, HSD, and HST
As previously described, orange peel extract was prepared by hydrodynamic cavitation of dried orange peel (HCV) [20]. The HCV extract was freeze-dried to obtain dry extract. HSD (cat no. PHR1794) and HST (cat no. SHBL8821) were acquired from Sigma (Sigma-Aldrich, St. Louis, USA). The HCV extract was dissolved in DMSO to prepare a 50 mg/mL stock solution for cell assay, while the flavonoid compounds were dissolved in DMSO to make a 50 mM stock solution. All solutions were stored at −20°C until used. Before cell treatment, the serial concentrations of samples were newly prepared in a culture medium with DMSO as cosolvent with a final concentration of less than 1%.
Cell culture
The 293T (ECACC 12022001) and 293 (ECACC 85120602) cell lines were obtained from the National Research and Innovation Agency (BRIN, Indonesia). The 293T cells were maintained in High-Glucose Dulbecco’s-modified Eagle’s medium (Gibco, Billings USA) with supplementation of 10% FBS (Sigma-Aldrich, St. Louis, USA and Biosera, Cholet, France) and antibiotics (100 µg/mL streptomycin and 100 U/mL penicillin) (Gibco, Billings USA). Recombinant 293 cells expressing hACE2 were generated by lentiviral vector and cultured in MEM medium (Sigma-Aldrich, St. Louis USA) with supplementation of 10% FBS, antibiotics (100 µg/mL streptomycin and 100 U/mL penicillin) (Invitrogen, Thermo), and 1% NEAA (Invitrogen, Thermo). The cell culture was stored in a humid incubator at 37°C with 5% CO2. The introduction of recombinant plasmid DNA into cells was carried out by using a polyethylenimine transfection reagent (PEI-Max, Polysciences).
HPLC analyses
Orange peel extract was dissolved in DMSO to a final concentration of 1,000 µg/mL, while HSD was dissolved in DMSO to a final concentration of 800 µM, and HST was dissolved in DMSO to a final concentration of 100 µM. The solution was filtered through a 0.22 µm nylon filter before HPLC analysis that was modified from [21]. Three microliters of the sample were injected into an HPLC instrument (UFLC Shimadzu, Shimadzu Corp., Kyoto, Japan) with a C18 reversed-phase column (Shim-pack GIST C18 5μm 4.6 × 25 mm, Shimadzu, Shimadzu Corp., Kyoto, Japan) using methanol: acetonitrile: 0.1% acetic acid (89.3:10:0.7) as mobile phase with a flow rate of 1 mL/minute at 35°C and detected using a photodiode array detector at 280 nm.
Recombinant plasmids
Plasmid pcDNA3.1-SARS2-Spike (Addgene #145032) [22], which expresses a spike of SARS-CoV-2 Wuhan/ wild-type variant, was used for pseudotyping to produce SARS-CoV-2 pseudovirus. We used pcDNA3.1-hACE (Addgene #145033) [22] and TMPRSS2 (Addgene #53887) [23] plasmids to engineer the target cells for the pseudovirus entry assay. Moreover, to generate recombinant 293/hACE2 cells, 293 cells were transduced with lentivirus produced utilizing pCMV-dR8.2-dvpr (Addgene #8455) [24]; pCAGGS-G-Kan (Kerafast EH1017) [25]; and (RRL.sin.cPPT.SFFV/Ace2.IRES-neo.WPRE(MT129) (Addgene#145840) [26] plasmids were co-transfected into 293T cells. The plasmids were propagated in E. coli and purified from the cell pellet using a plasmid maxi/midi-prep kit (Qiagen, Venlo, The Netherlands). The plasmid DNA concentration was determined using a micro-volume spectrophotometer (Thermo Fisher Scientific, Massachusetts, USA).
Immunofluorescence staining
About 3 × 104 293T cells per well of a 24-well plate were seeded on coverslips coated with 2% gelatin (Sigma-Aldrich, St. Louis, USA). The following day, cells were transfected with TMPRSS2 plasmids and incubated for about 18 hours. Cells were then washed with PBS and left overnight inside a CO2 incubator. For immunofluorescence staining, the cells were fixed with 4% paraformaldehyde (PFA) for about 15 minutes, permeabilized with 0.2% Triton-X for 10 minutes, then blocked with 1% BSA/PBS for 30 minutes-1 hour at room temperature (RT). Then, the cells were incubated in an anti-TMPRSS2 antibody (Novus Biologicals NBP2-93322, USA) diluted in blocking buffer at 1:250 overnight at 4°C. After washing with PBS, the cells were incubated in an antirabbit secondary antibody conjugated with Alexa Fluor™-488 (1:1,000) (Abcam ab150077, USA) for 1 hour at RT. Next, the excess antibody was washed with PBS, followed by nuclei staining and cell mounting using a DAPI-containing mountant (Abcam Ab104139, USA). The fluorescence signal was observed using an Olympus-IX83 microscope (Olympus, Tokyo, Japan).
Western blot
The pellet of cells was lysed with a cold RIPA buffer (Abcam ab288006, USA) containing a protease inhibitor cocktail (Nacalai Tesque, Japan). The total protein concentration of the lysates was determined by BCA assay (Thermo Scientific, USA) to prepare about 10–40 µg protein mixed in Laemmli buffer. The proteins were resolved by SDS-PAGE and transferred onto the PVDF membrane. The membrane was incubated in 5% skim milk/TBS/0.05% tween-20 or 5% skim milk/PBS/0.05% tween-20 for 1 hour to minimize nonspecific binding. Then, the membrane was incubated with primary antibody (anti-SARS-CoV-2 spike (Abcam, ab275759, USA) 1:1,000, anti-hACE2 (Sigma SAB 3500978, USA) 1:1,000, or anti-β-actin (Sigma A2228, USA) 1:2,000) overnight at 4°C. After washing, the membrane was incubated in HRP-conjugated anti-rabbit or anti-mouse secondary antibodies (Abcam ab205718 or ab6728) or ALP-conjugated anti-rabbit secondary antibodies (Abcam ab6722), 1:2,000 or 1:4,000, for about 2 hours at RT or overnight at 4°C. Chemiluminescence substrates (Abcam, USA) were added to visualize antibody reactions and observed using a chemiluminescence imaging system (Uvitec Cambridge, UK), while 1-Step™ NBT/BCIP Substrate Solution (Thermo Scientific 34042) was added to visualize antibody reaction based on colorimetric development.
SARS-CoV-2 pseudotyping
About 1.5 × 106 of 293T cells were seeded on a 100 mm dish and incubated overnight. The cells were then transfected with SARS-CoV-2 spike encoding DNA plasmid and incubated for about 18 hours. The next day, the cells were incubated with pseudotyped G*ΔG-GFP rVSV (Kerafast EH1024-PM) [25] at MOI ~3 for 1 hour inside a CO2 incubator [25]. Then, the medium containing pseudotyped rVSV was replaced with a culture medium added with anti-VSV-G antibody (1:2,000) (Invitrogen PA1-30138, Thermo Fisher Scientific, USA) to neutralize the excess of G*ΔG-GFP rVSV, and the cells were kept inside an incubator overnight. After pseudotyping, the cell-conditioned medium was collected and spun to remove cell debris. The supernatant containing SARS-CoV-2 spike*ΔG-GFP rVSV (SARS-CoV-2 pseudovirus) was stored at −80°C in aliquots.
MTT cell viability assay
About 7 × 103 of 293T cells/well were seeded onto a 96-well plate and incubated overnight inside a CO2 incubator. The following day, the culture medium was discarded. Cells were treated with HSD, HST, or HCV concentration series (1, 10, and 100 µM or 1, 10, and 100 µg/mL) for three replicates and then incubated for 24 hours. The cells were incubated with 100 μL of 0.5 µg/mL MTT reagent in a culture medium for 2 hours to measure cell viability based on enzymatic reaction. The MTT-containing medium was discarded, and then 100 μL DMSO was added to the well to dissolve the formazan crystal generated from MTT reduction. The absorbance of formazan was read with a microplate reader at a wavelength of 570 nm. The following formula was used to quantify cell viability:
Cell viability = (Abs treated cells—Abs blank) / (Abs nontreated cells—Abs blank) × 100%
Pseudovirus entry assay
About 4 × 104 of 293T cells/well were seeded onto a gelatin-coated 8-chamber slide (SPL Life Sciences, Pyeongtaek, South Korea) and incubated overnight. The next day, the cells were transfected with hACE2 and TMPRSS2 encoding plasmids and incubated for about 18 hours. The cells were pre-treated with HSD, HST, or orange peel extract for 30 minutes, then treated with a combination of tested sample and SARS-CoV-2 pseudovirus at 1:2 ratios in 300 μL total volume and incubated for another 16–18 hours. At the end of incubation, the cells were fixed using 4% PFA and mounted with a DAPI-containing mountant. Pseudovirus internalization, represented by observing GFP spots, was investigated using an Olympus IX83 motorized fluorescence microscope (Olympus, Tokyo, Japan). The GFP spots were counted from 8 images for each treatment by Fiji software (National Institute of Health). Pseudovirus entry assay was also performed utilizing 293/hACE2 stable cells generated using a lentiviral vector. The cells were seeded onto a gelatin-coated 8-chamber slide and incubated overnight. The following day, the cells were treated with tested samples described above. GFP spots were analyzed using cellSense dimension imaging software (Olympus, Tokyo, Japan).
Statistical analysis
Analyses were performed using GraphPad Prism software (GraphPad software, Inc., California, USA). Data were presented as mean ± SEM, as indicated in each figure. For pseudovirus entry assay in 293T/hACE2/TMPRSS2 cells, one-way ANOVA using Tukey’s test was used to determine the statistical differences with a p-value < 0.05 considered significant. While for pseudovirus entry assay in 293/hACE2 cells, statistical difference was analyzed using one-tail unpaired t-tests with a p-value < 0.05 considered significant.
RESULTS AND DISCUSSIONS
Determination of HSD and HST content in the orange peel extract
The orange peel contains bioflavonoids, especially HSD, and a smaller amount of its hydrolysis product, HST. We determined the HSD and HST contents using the HPLC method in tangerine peel extract prepared using hydrocavitation-based extraction (HCV). Based on Figure 1B, standard HSD appeared with a retention time of 5.06 minutes, while HSD in orange peel extract appeared with a retention time of 4.98 minutes. To ensure that the main peak in the extract was HSD, we spiked HSD in the sample extract and obtained a retention time of 4.99 minutes. By the appearance of the single main peak, it has been confirmed that the main peak in the HCV extract is HSD. The slight differences in the retention time can be due to the diverse extract content. Then, based on the area under curve data, we calculated the HSD content in the DMSO soluble fraction of the extract and obtained an HSD content of 4.34% w/w extract.
With the same HPLC condition, we observed the peak of standard HST appeared with a retention time of 2.51 minutes, while the main peak of the HCV extract existed with a retention time of 2.84 minutes. When we spiked the standard HST to be run with HCV extract, both prominent peaks appeared separately with consistent retention times (Fig. 1C).
Expression of recombinant SARS-CoV-2 spike, hACE2, and TMPRSS2
We performed Western blot and immunofluorescence staining to clarify the ectopic expression of the SARS-CoV-2 spike, hACE2, and TMPRSS2 in 293T cells. Using the anti-S1 sub-unit of spike WT antibody, we detected the expression of spike WT by Western blot (Fig. 2A). Moreover, we also observed the spike WT expression by immunofluorescence staining (Fig. 2B). In addition, hACE2 expression was also detected by Western blot (Fig. 2C), while TMPRSS2 expression was detected by immunostaining (Fig. 2D). Spike expression was expected for the pseudotyping of rVSV. Meanwhile, hACE2 and TMPRSS2 expressions were expected to modify the target cells for pseudovirus entry assay. SARS-CoV-2 spike pseudotyping analysis was already reported previously [27].
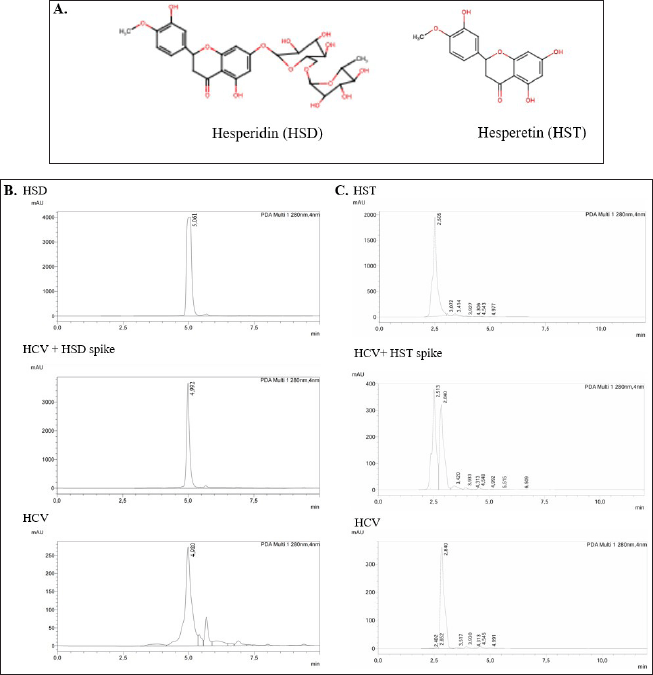 | Figure 1. Hesperidin (HSD) and hesperetin (HST) contents in the orange peel (HCV) extract. A. Structure of HSD and HST. B. Representative HPLC chromatograms of standard HSD, HCV extract, and HSD spiked-HCV extract. C. Representative chromatograms of standard HST, HCV extract, and HST spiked-HCV extract. [Click here to view] |
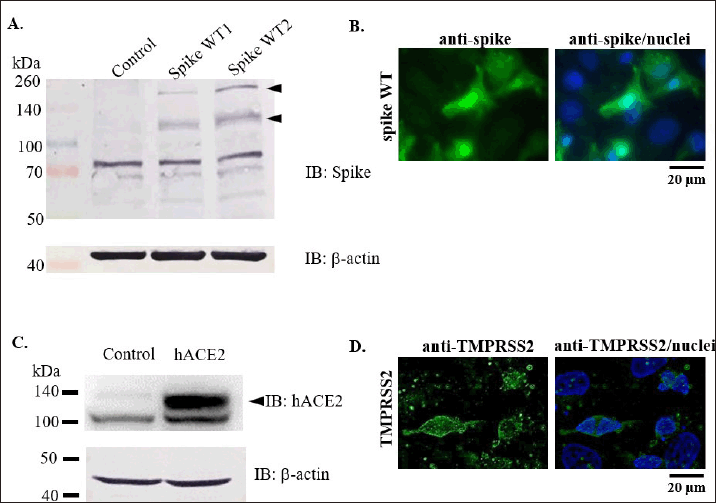 | Figure 2. Confirmation of spike, hACE, and TMPRSS2 over-expression in 293T cells. A-B. Expression check of SARS-CoV-2 spike by Western blot and immunofluorescence staining. C. Detection of hACE2 expression by Western blot. D. TMPRSS2 detection by immunofluorescence staining. [Click here to view] |
MTT cell viability assay
We conducted a cell viability assay to determine the noncytotoxic concentration of HSD, HST, and HCV for further assays in 293T cells since we used a 293T cell model to perform a pseudovirus entry assay. We used the noncytotoxic concentration of the tested samples, which, based on the MTT assay, did not reduce the cell viability to less than 70%. As a result, the cell viability after treatment with HSD and HST 1, 10, 100 µM was 86.83%, 84.62%, 72.35%, 96.39%, 84.81%, and 75.71%, respectively. In addition, HCV treatment 1, 10, and 100 µg/mL resulted in cell viability of 93.59%, 97.59%, and 72.34% (Fig. 3).
Pseudovirus entry assay
The pseudovirus model was chosen to target the entry point of SARS-CoV-2. The pseudovirus also possesses spike peptide in its outer layer, which mediates binding to hACE2 as the target receptor, thus enabling the pseudovirus cellular entry. Moreover, the presence of TMPRSS2 will help pseudovirus internalization by priming the S1 subunit of the spike peptide [14]. Then, upon internalization and release of the viral RNA genome, the GFP reporter will be expressed, generating a GFP spot. On the other hand, nonvirulent pseudoviruses will not generate competent new pseudoviruses (Fig. 4A). We used 10 and 100 μM of HSD and HSD for 4 pseudovirus entry assay with an incubation time of about 16–18 hours, shorter than the incubation time for MTT assay to reduce cytotoxicity. As shown in Figure 4B, the target cells were pretreated with a test solution before co-treatment with pseudovirus. Next, we investigated the GFP spots in the cellular area and analyzed the number of infected cells. As a result, the number of infected cells for control cells, HSD 10 and 100 µM, and HST 10 and 100 µM were 43.07%, 14, 21%, 18.25%, and 15.83%, respectively. Here, we showed that compared to control cells, HSD and HST treatment 10 and 100 μM can inhibit pseudovirus internalization at about 22%–29% (p < 0.001) (Fig. 4C).
Additionally, we performed a pseudovirus entry assay using 293/hACE2 stable cells. As in the previous experiment, cells were pretreated with the test material for 30 minutes, then treated with a mixture of pseudovirus and test material for 16–18 hours (Fig. 5A). In this assay, we used lower concentrations of HSD and HST to minimize cytotoxic effects. Based on this assay, HSD demonstrated inhibition of pseudovirus entry at both 1 and 10 μM concentrations with the value of 34.28% and 26.44% (Fig. 5B). Meanwhile, HST showed the potential to inhibit pseudovirus entry at a concentration of 10 μM with a value of 30.69% (p < 0.05). When comparing the inhibitory effects of HSD and HST, HSD 1 μM, and HST 1 μM show significantly different inhibitory effects on pseudovirus entry with 34.28% and 46.30% (p < 0.05). HSD shows a more significant inhibitory potential than HST. Interestingly, HCV also showed inhibition, as did these two flavonoids. However, the inhibitory effect of 1 µg/mL extract was higher than that of 10 µg/mL extract concentration, with values of 28.25% and 48.18%, respectively (Fig. 5B). The representative images of cells after pseudovirus entry assay were shown in Figure 5C.
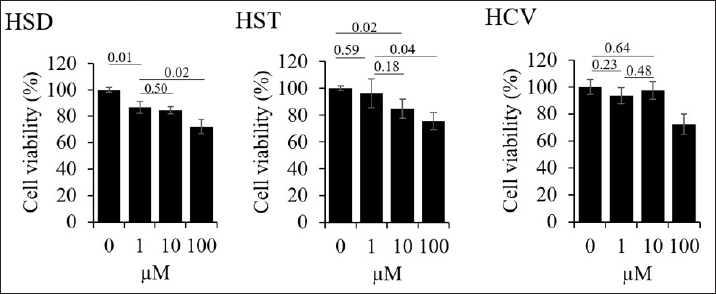 | Figure 3. Cell viability assay of hesperidin (HSD), hesperetin (HST), and orange peel extract (HCV) toward 293T cells. Cells were seeded onto a 96-well plate and incubated overnight. The next day, cells were treated with HSD, HST, or HCV serial concentrations, as shown in the graphs and incubated for 24 hours. Cell viability was analyzed based on MTT assay. N = 3 replication. Data shown as cell viability + SD. [Click here to view] |
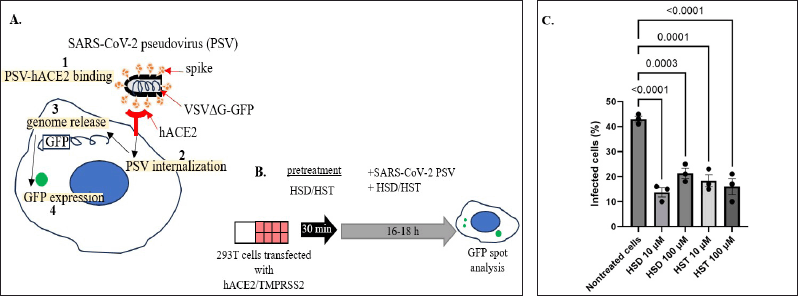 | Figure 4. Effect of hesperidin, hesperetin, and orange peel extract on SARS-CoV-2 pseudovirus entry. A. Mechanism of SARS-CoV-2 pseudovirus cell entry and GFP expression. B. Schematic representation of pseudovirus entry assay. C. Quantification of SARS-CoV-2 (WT) pseudovirus entry assay in 293T cells transfected with hACE2/TMPRSS2. HSD = hesperidin, HST = hesperetin, PSV = pseudovirus. Data presented as mean + SEM (n = 3 microscope fields). [Click here to view] |
HSD and HST show the binding potential to the hACE2 receptor through 3 binding sites with docking energies of −4.21 kcal/mol and −6.09 kcal/mol, respectively, based on in silico molecular docking studies [28]. In addition, other molecular docking studies reported the binding potential of HSD and HST to spike glycoprotein-RBD (PDB: 6LXT) and the RBD-hACE2/PD-ACE2 complex (PDB: 6VW1). When compared to previous docking studies, this in silico study shows that the binding potential of HSD is higher than HST, both in the RBD spike wt or the RBD-hACE2 complex with HSD docking scores of −9.61 and −9.50 kcal/mol, while the docking score of HST is −9.08 and −6.71 kcal/mol. Compared with other citrus flavonoids, the binding potential of HSD and HST at these two receptors is more remarkable than tangeretin, nobiletin, and naringenin [11].
Referring to the pseudovirus entry assay results on HSD and HST, these two compounds exhibit pseudovirus entry inhibitory activity in hACE2/TMPRSS2 expressing 293T cells with comparable effects without showing a statistically significant difference (p > 0.05) (Fig. 3). However, the inhibitory effect of pseudovirus entry on 293/hACE2 stable cells from HSD and HST appeared significantly different at a low concentration of 1 μM (Fig. 4). In addition, HCV containing HSD and HST showed potential for pseudovirus inhibition at a 1 µg/mL concentration. Even though HSD and HST inhibit PSV entry dose-dependently, it is not for HCV. It might be that HCV, besides containing HSD, also includes other components that might interfere with the biological effects of HCV. However, we need to study this phenomenon further.
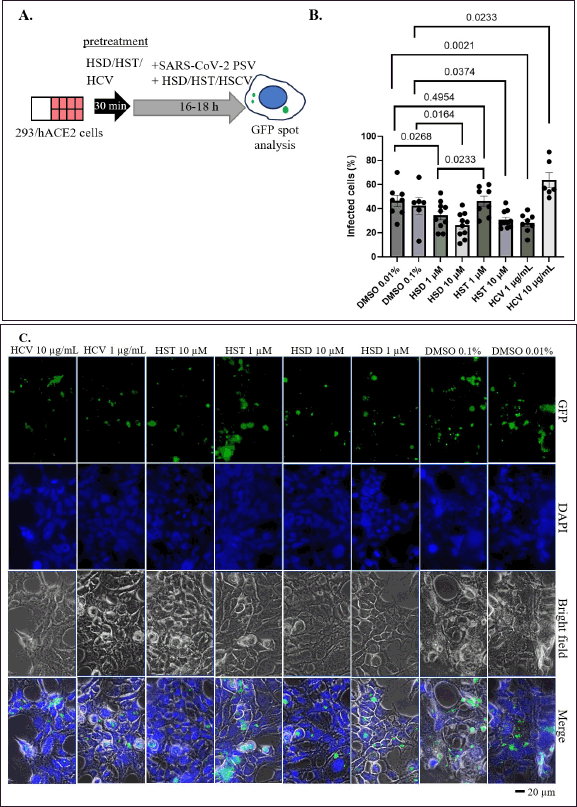 | Figure 5. Effect of HSD, HST, and HCV on SARS-CoV-2 pseudovirus entry. A. Schematic representation of pseudovirus entry assay. B. Quantification of SARS-CoV-2 (WT) pseudovirus entry assay 293 cells stably expressing hACE2. C. Representative images showed GFP spots. HSD = hesperidin; HST = hesperetin; HCV = orange peel extract, PSV = pseudovirus. Data presented as mean + SEM (n = 6–10 microscope fields). [Click here to view] |
A noncompetent pseudovirus is a nonvirulent virus model that is very advantageous for studying the antiviral effect of targeting the viral entry point. The pseudovirus entry assay will represent the interaction between SARS-CoV-2 spike proteins incorporated in the outer part of the VSV backbone with hACE2 of the target cell [19]. This specific mechanism restricts the use of pseudovirus assay for studying viral replication. However, based on molecular docking studies, HSD and HST are predicted to interact with the SARS-CoV-2 protease (PDB ID: 6LU7), which is expected to inhibit virus replication [28,29]. A lab-based assay is still needed to prove the inhibitory effect of those compounds on SARS-CoV-2 replication. Apart from HSD and HST, standardized orange peel extract will be beneficial to reduce the risk of SARS-CoV-2 viral infection. The docking studies are mainly conducted against the SARS-CoV-2 wt variant, and we performed a pseudovirus entry assay against the SARS-CoV-2 wt pseudovirus. Thus, the effect of HSD, HST, and HCV must also be tested against other SARS-CoV-2 variants.
In the previous report, we evaluated the acute toxicity of a single oral administration of HCV extract on Sprague Dawley rats. We applied several doses in 5 rat groups, with the highest dose of 5,000 mg/kg body weight (bw). The results showed no abnormalities in the fur, skin, eyes, feces consistency, somatomotor activity, and behavior patterns, while mortality did not occur during the 15-day test. Higher HCV doses of 2,000 and 5,000 mg/kg bw cause renal abnormalities such as inflammation and congestion in rats, but not in lower doses. However, the acute toxicity report proves that the orange peel HCV is practically not toxic since its lethal dose 50 (LD50) is more than 5,000 mg/kg bw [20]. Moreover, another study showed a sub-chronic toxicity study of orange peel extract was performed with daily doses of up to 540 mg/kg bw. We did not mention the extract as an HCV extract since the extract was not prepared using the HCV extraction method. Based on the report, during the 90-day observation, the orange peel extract administration did not significantly affect body weight, and no mortality event was found. A 540 mg/kg bw daily dose of orange peel extract caused nephropathy symptoms only in male rats but not in female rat groups. Therefore, 540 mg/kg bw can be administered to female rats, while a lower dose should be applied to male rats [30]. As a precaution, renal impairment should be considered a possible side effect of orange peel HCV dry extract.
Being the 4th world’s main producer of medicinal plants, in 2020, Indonesia exported herbal medicinal products, mostly from ginger and turmeric, which reached a value of USD 9.64 million [31]. According to the European Medicines Agency (2018), those medicinal products are commonly marketed as dry extracts and tinctures [32]. Considering the advantage of the HCV technique, since the extract would have no residual ethanol, the dry extract is preferred over tincture to be marketed as a bulk extract product. In addition, another common formulation of orange peel extract supplement currently available in the market is manufactured as capsules.
For the local regulation, the registration of the herbal product should comply with the Indonesia National Agency of Drug and Food Control (NA-DFC) regulation No. 25 year 2023 to ensure safety, efficacy, and quality. According to this guideline, pre-clinical and clinical studies should be performed to claim it as a preventive and remedy for COVID-19 [33]. Lastly, traditional medicine GMP certification is also required to obtain market authorization from NA-DFC [34].
CONCLUSION
Citrus reticulata peel contains HSD as a major component [9]. We can also confirm the significant HSD content in the orange peel HCV extract we tested in this study. Since we could not confirm the HST content in the extract, the HSD content can be used to standardize the HCV extract for further research. In addition, besides being absorbed as HST, HSD can also be absorbed via the intestine without undergoing hydrolysis. This evidence was shown by detecting conjugated or nonconjugated HSD in the urine of rats fed with HSD [34].
Another flavonoid in orange peel, naringin, has been studied for its potential to prevent cytokine storms by decreasing the expression of LPS-induced proinflammatory cytokines COX-2, iNOS, IL-1β, and IL-6 [9]. Naringin also inhibits SARS-CoV-2 pseudovirus entry at 100 µM in hACE2-overexpressing 293T cells [27]. Apart from that, in the tests carried out in the pseudovirus entry, these orange peel HCV extracts inhibited SARS-CoV-2 infection. Therefore, it strengthens the potential for using orange peel extract to prevent SARS-CoV-2 infection or to reduce the severity of disease prognosis in COVID-19 patients.
ACKNOWLEDGMENTS
We thank the iLab facility of the National Research and Innovation Agency (BRIN) for performing HPLC analyses. The article was uploaded to a preprint server prior to publication in this journal. The link and DOI of the article displayed at bioRxiv is given below; https://www.biorxiv.org/content/10.1101/2024.05.03.592493v1, Doi: https://doi.org/10.1101/2024.05.03.592493
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FUNDING SOURCES
We are grateful for the financial support from LPDP/BRIN (grant no. 102/FI/P-KCOVID-19.2B3/IX/2020 and RIIM No. KEP-5/LPDP/LPDP.4/2022) and Research Organization for Life Sciences and Environment, BRIN (Research Program (DIPA Rumah Program) 2022).
CONFLICTS OF INTEREST
The authors declared no potential conflict of interest.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Dhama K, Khan S, Tiwari R, Sircar S, Bhat S, Malik YS, et al. Coronavirus disease 2019-COVID-19. Clin Microbiol Rev. 2020;33(4):e00028–20. CrossRef
2. Sarker R, Roknuzzaman ASM, Nazmunnahar, Shahriar M, Hossain MJ, Islam MR. The WHO has declared the end of pandemic phase of COVID-19: way to come back in the normal life. Health Sci Rep. 2023;6(9):e1544. CrossRef
3. Islam MA, Marzan AA, Arman MS, Shahi S, Sakif TI, Hossain M, et al. Some common deleterious mutations are shared in SARS-CoV-2 genomes from deceased COVID-19 patients across continents. Sci Rep. 2023;13(1):18644. CrossRef
4. Hammad WAB, Al Beloushi M, Ahmed B, Konje JC. Severe acute respiratory syndrome (SARS) coronavirus-2 infection (COVID-19) in pregnancy—an overview. Eur J Obstet Gynecol Reprod Biol. 2021;263:106–16. CrossRef
5. Whitaker M, Elliott J, Bodinier B, Barclay W, Ward H, Cooke G, et al. Variant-specific symptoms of COVID-19 in a study of 1,542,510 adults in England. Nat Commun. 2022;13:6856.
6. Torabi SH, Riahi SM, Ebrahimzadeh A, Salmani F. Changes in symptoms and characteristics of COVID-19 patients across different variants: two years study using neural network analysis. BMC Infect Dis. 2023 Nov 28;23(1):838. CrossRef
7. Dong Y, Dai T, Wang B, Zhang L, Zeng LH, Huang J, et al. The way of SARS-CoV-2 vaccine development: success and challenges. Signal Transduct Target Ther. 2021;6(1):387. CrossRef
8. Kumar D, Ladaniya MS, Gurjar M, Kumar S, Mendke S. Quantification of flavonoids, phenols and antioxidant potential from Dropped Citrus reticulata Blanco Fruits Influenced by Drying Techniques. Molecules. 2021;26(14):4159. CrossRef
9. Liu W, Zheng W, Cheng L, Li M, Huang J, Bao S, et al. Citrus fruits are rich in flavonoids for immunoregulation and potential targeting ACE2. Nat Prod Bioprospect. 2022;12(1):4. CrossRef
10. Wdowiak K, Walkowiak J, Pietrzak R, Bazan-Wo?niak A, Cielecka-Piontek J. Bioavailability of hesperidin and its aglycone hesperetin-compounds found in Citrus fruits as a parameter conditioning the pro-health potential (Neuroprotective and Antidiabetic Activity)-mini-review. Nutrients. 2022;14(13):2647. CrossRef
11. Utomo RY, Ikawati M, Meiyanto E. Revealing the potency of Citrus and Galangal constituents to halt SARS-CoV-2 infection. Preprints 2020, 2020030214. CrossRef
12. Utomo RY, Ikawati M, Putri DDP, Salsabila IA, Meiyanto E. The chemopreventive potential of diosminand hesperidin for COVID-19 and its comorbid diseases. Indonesian J Cancer Chemoprevention. 2020;11(3):154–67. CrossRef
13. Zalpoor H, Bakhtiyari M, Shapourian H, Rostampour P, Tavakol C, Nabi-Afjadi M. Hesperetin as an anti-SARS-CoV-2 agent can inhibit COVID-19-associated cancer progression by suppressing intracellular signaling pathways. Inflammopharmacology. 2022;30(5):1533–9. CrossRef
14. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80.e8. CrossRef
15. Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022;23(1):3–20. CrossRef
16. Kaufer AM, Theis T, Lau KA, Gray JL, Rawlinson WD. Laboratory biosafety measures involving SARS-CoV-2 and the classification as a Risk Group 3 biological agent. Pathology. 2020;52(7):790–5. CrossRef
17. Sanders DA. No false start for novel pseudotyped vectors. Curr Opin Biotechnol. 2002;13(5):437–42. CrossRef
18. Salazar-García M, Acosta-Contreras S, Rodríguez-Martínez G, Cruz-Rangel A, Flores-Alanis A, Patiño-López G, et al. Pseudotyped vesicular stomatitis virus-severe acute respiratory syndrome-coronavirus-2 spike for the study of variants, vaccines, and therapeutics against coronavirus disease 2019. Front Microbiol. 2022;12:817200. CrossRef
19. Septisetyani EP, Prasetyaningrum PW, Anam K, Santoso A. SARS-CoV-2 Antibody neutralization assay platforms based on epitopes sources: live virus, pseudovirus, and recombinant S Glycoprotein RBD. Immune Netw. 2021;21(6):e39. CrossRef
20. Putri DDP, Maran GG, Kusumastuti Y, Susidarti RA, Meiyanto E, Ikawati M. Acute toxicity evaluation and immunomodulatory potential of hydrodynamic cavitation extract of citrus peels. J Appl Pharm Sci. 2022;12(04):136–45. CrossRef
21. Cahyono B, Afiyah N, Suzery M, Bima DN, Amalina ND. Optimization condition of HPLC method to determine hesperidin contents from various orange peel extracts. AIP Conf Proc. 2023;2738(1):050004. CrossRef
22. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–4. CrossRef
23. Edie S, Zaghloul NA, Leitch CC, Klinedinst DK, Lebron J, Thole JF, et al. Survey of human chromosome 21 gene expression effects on early development in Danio rerio. G3 (Bethesda). 2018;8:2215–23. CrossRef
24. Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9(4):493–501. CrossRef
25. Whitt MA. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods. 2010;169:365–74. CrossRef
26. Rebendenne A, Valadão ALC, Tauziet M, Maarifi G, Bonaventure B, McKellar J, et al. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J Virol. 2021;95(8):e02415–20. CrossRef
27. Septisetyani EP, Prasetyaningrum PW, Paramitasari KA, Suyoko A, Himawan ALI, Azzahra S, et al. Naringin Effect on SARS-CoV-2 Pseudovirus entry and spike mediated syncytia formation in hACE2-overexpressing Cells. HAYATI J Biosci. 2023;31(2):336–47. CrossRef
28. Kashyap P, Thakur M, Singh N, Shikha D, Kumar S, Baniwal P, et al. In Silico evaluation of natural flavonoids as a potential inhibitor of coronavirus disease. Molecules. 2022;27(19):6374. CrossRef
29. Cheng FJ, Huynh TK, Yang CS, Hu DW, Shen YC, Tu CY, et al. Hesperidin is a potential pnhibitor against SARS-CoV-2 infection. Nutrients. 2021;13(8):2800. CrossRef
30. Nakajima A, Nemoto K, Ohizumi Y. An evaluation of the genotoxicity and subchronic toxicity of the peel extract of ponkan cultivar ‘Ohta ponkan’ (Citrus reticulata Blanco) that is rich in Nobiletin and Tangeretin with anti-dementia activity. Regul Toxicol Pharmacol RTP. 2020;114:104670. CrossRef
31. Embassy of the Republic of Indonesia (RI) in Brussels. Indonesia’s Potential Herbal Products Gain in the European Market [Internet]. 4th ed. 2021. Available from: https://kemlu.go.id/download/L1NoYXJlZCUyMERvY3VtZW50cy9icnVzc2VsL3Jlc2Vhcm NoJTIwc2VyaWVzL1Jlc2VhcmNoJTIwU2VyaWVzJTIwSW5kb25lc2lhJTIwUG90ZW50aWFsJTIwSGVyYmFsJTIw UHJvZHVjdHMucGRm.
32. European Medicines Agency (EMA). Assessment report on Curcuma longa L., rhizoma [Internet]. Amsterdam, The Netherlands: EMA; 2018. Report No.: EMA/HMPC/749518/2016. Available from: https://www.ema.europa.eu/en/documents/herbal-report/final-assessment-report-curcuma-longa-l-rhizoma-revision-1_en.pdf.
33. The National Agency of Drug and Food Control (NA-DFC) of Indonesia. Regulation of The National Agency of Drug and Food Control No. 25 year 2023 about Criteria and Procedures for Registration of Natural Ingredients Medicines. Jakarta, Indonesia 2023. (in Indonesian).
34. Yamada M, Tanabe F, Arai N, Mitsuzumi H, Miwa Y, Kubota M, et al. Bioavailability of glucosyl hesperidin in rats. Biosci Biotechnol Biochem. 2006;70(6):1386–94. CrossRef