INTRODUCTION
Surface layer protein (SLP) is an array of crystalline proteins or glycoproteins found on the outermost cell exterior of unicellular microorganisms and a predominant protein secreted by bacteria or archaea [1,2]. It consists of monomeric subunits aligned in two-dimensional lattice symmetries [3]. The SLP was found in different locations depending on the host, for instance, on the peptidoglycan of Gram-positive bacteria, on the lipopolysaccharide of Gram-negative bacteria, or on the pseudomurein or other biopolymers—layers substituting peptidoglycan of the archaea [4]. The SLP directly interacts with the environment; thus, it offers protective, adhesive, antifouling, and adaptability towards extreme conditions, also plays a role in the virulence of pathogenic bacteria [4,5]. In addition, because of the presence of the pores (approximately 2–8 nm) which are formed by the array structures, the SLP is responsible in ion transport [2,4,6]. The percentage of the pores was 70% of the total SLP surface area [7].
The SLP was observed for the first time in Spirilium serpens, a Gram-negative bacterium [2]. It is visualized as hexagonal structure formed from monomeric subunit proteins. In the 1970s, it was reported that the protein could be effortlessly isolated using a high concentration (2 M) of guanidine hydrochloride because of its non-covalent binding with the cell wall [6,8]. However, the 2D hexagonal conformation is not retained by the detached SLP, yet it could be reconstituted in an appropriate substrate [8].
In correlation with the protection and adaptive function, SLP is a non-conserved protein that has undergone evolution, even being diverse at the same species level [7]. This diversity includes variety in the level of gene and protein; and phenotypic structure and function. Previously, these variances were not considered correlated with taxonomic identification; however, some evidences have been found in some species that might contribute to taxonomic classification [7]. Bharat and colleagues reported that SLP consists of 400–2,500 amino acid residues with low sequence similarity, which are predominantly hydrophobic and acidic [9]. The protein structure is generally the same in certain domains; for example, an anchoring domain is commonly built by trimers, pseudotrimers, or α-helix folds and a combination of beta-sandwich, beta-roll, beta-helix, and coiled-coil structures [9].
Interestingly, among those of either bacteria’s or archaea’s SLPs, SLPs from Lactobacillus sp show unique features, such as relatively higher pI value and smaller molecular size [2,10]. Heretofore, the smallest SLP is found in Lactobacillus sp. (approximately 23–71 kDa), whereas most of the SLP is 40–200 kDa [11,12]. The exploration of SLP from Lactobacillus sp. has been reported in the last three decades, exhibiting potential in medicine and nanotechnology [3]. The position of SLP among other outer proteins of Lactobacillus sp. is depicted in Figure 1. In this review, we focus on prospective applications of SLP from Lactobacillus sp., particularly as a drug nanocarrier.
METHODOLOGY
Publications that focused on SLP; Lactobacillus sp.; and nanomedicine or drug delivery were collected from search engines (Google Scholar, Scopus, and PubMed) until July 2024. The related publications were selected and reviewed.
RESULTS AND DISCUSSION
Features of SLP from Lactobacillus sp.
Lactobacillus is a genus of lactic acid bacteria (LAB), rod or coccobacilli-shaped, Gram-positive, low pH tolerant, facultatively anaerobic, non-sporing, and the largest genus among the other genera in the LAB [13–15]. The main characteristic of Lactobacillus sp. is the production of lactic acid (up to 85%) during carbohydrate fermentation [16]. These bacteria are commonly identified in healthy digestive and female genitourinary tracts [17]. Lactobacillus sp. is commonly found in plants and overripe foods [10]. Adhesion of the cell surface to its natural habitat is important for Lactobacillus sp. [3].
The characteristics of Lactobacillus SLP are the smallest size among SLP from other bacteria or archaea; high pI (approximately 9.4–10.4), positively charged; high content of hydrophobic residues and low content of sulfuric acid residues, mostly in oblique or hexagonal shape, and non-glycosylated [10,18]. Only L. kefir possesses glycosylated SLP, which was identified as O- and N-glycosylated [19,20]. Unlike other genera that display a correlation between the SLP genetic order and taxonomy classification, Lactobacillus has undetectable homogeneity [9]. However, SLP-encoding genes show similarity at the strain level [3]. Extracted SLP from Lactobacillus brevis KM3 dan L. brevis KM7 showed similar molecular mass, secondary structure (predominantly formed of β-sheet and less α-helix), and thermal analysis profile [21]. An alignment analysis of the amino acid residue sequences of Lactobacillus acidophilus, Lactobacillus casei, Lactobacillus helveticus, and Lactobacillus crispatus recognized a conserved domain at the C-terminus; meanwhile, the N-terminal was more variable. In particular, C-terminal SLPA_III shows conserved amino acid sequences in Lactobacillus, whereas the N-terminal domains SLPA_I and SLPA_II are responsible for the self-assembly process [22]. The β-sheet and α-helix structures were identified at the N-terminal domain of the SLP of L. acidophilus ATCC 4356, but only the α-helix structure was observed at the C-terminal domain [23]. Only the N-terminal domain is exposed to the nearby environment; however, the C-terminus plays a role in anchoring the bacterial cell wall [23]. This anchoring function is useful for surface antigen display for vaccine delivery [24].
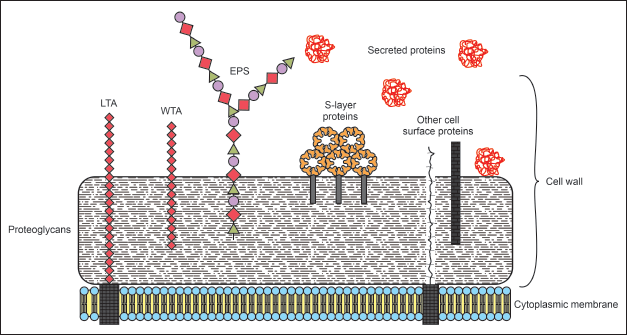 | Figure 1. Structure of Lactobacillus sp outer protein. LTA: lipoteichoic acid; WTA: wall teichoic acid; EPS: extracellular polymeric substances. S-layer proteins are attached on the outer part of the bacterial cell wall. [Click here to view] |
Genetic variation in the Lactobacillus sp led to secondary structural diversification among the strains. A comprehensive study of the SLP-encoding gene from numerous strains of Lactobacillus sp has been written by Hyn?nen et al. [3]. In general, SLPA-, SLPB-, SLPC-, and SLPD-encoding genes are commonly found in Lactobacillus sp. However, the expression of these genes varied depending on growth conditions. For instance, under an aerobic condition, SLP B and D were expressed in L. brevis ATCC 14869, whereas only SLP B was found under an anaerobic condition [18]. The secondary structure of SLP isolated from Lactobacillus kefiri HBA20 was dominant in the β-sheet conformation compared to the α-helix content [25]. SLPA is a major SLP found in Lactobacillus family. All of Lactobacillus species reported by Palomino et al. [24] own the SLPA gene, while only seven of 13 species harboring the SLPX gene, and only two of 13 species have the SLPB gene. L. acidophilus La14 was a species having all of these SLP-encoding genes [24]. The SLPX was more than 50-fold upregulated in L. acidophilus in gastrointestinal stimulated growth conditions. The overexpression of the SLPX corresponded to a survival rate of up to 93% [26].
Several strains of L. brevis showed diverse expression levels of SLP. The L. brevis B144 from traditional fermented buffalo milk had the highest SLP concentration among the other strains. The amino acid sequence of this strain showed almost 50% peptide fragment coverage with L. brevis KB290 [27].
Lactobacillus sp. has been widely used as a probiotic and provides a number of health benefits, such as stimulation of the immune system and defence mechanisms against pathogens [28]. It competes with pathogens by occupying the binding sites on the gastrointestinal tissue via cell surface proteins, mucus-binding proteins, or moonlighting proteins [29]. SLP is one of the proteins that acts on mucus binding mechanisms and stimulates protection against viruses via a dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) interaction [29,30]. SLP of L. amylovorus is crucial for diarrhea recovery in post-weaning piglets [31]. In addition, SLPA and SLPB from several mutants of L. acidophilus were responsible for bacterial growth under different environmental stress conditions (salt, detergent, and alcohol) and enhanced human immunity by binding to uromodulin and DC-SIGN [32]. SLPA from L. helveticus showed anti-inflammatory activity by repressing inflammatory cytokines and biomarkers [33]. SLP of L. casei f05 protects intestinal tissue from Escherichia coli and Salmonella enterica by competing for mucus binding and inducing pathogen apoptosis [34]. A list of genes and molecules that are responsible for the protection and probiotic function of Lactobacillus sp was reviewed by Lebeer et al. [35].
Importantly, SLP maintains its bioactivity, even after being extracted from the host. Isolated SLP of L. plantarum L-91 decreased the adhesion percentage of pathogenic E. coli by more than 50% in an in vitro immobilized collagen assay [36]. The SLP which was isolated from L. helveticus R0052 successfully protected pathogenic E. coli–infected human cancer cells [37]. The extracted SLP of L. crispatus KT-11 combats rotavirus infection by inhibiting the release of viral-specific proteins in the Caco2 cell line [38]. In addition, SLP from L. acidophilus also acts in an autoimmune mechanism, for example, in inflammatory bowel disease (IBD), by maintaining gut homeostasis through the SLPA/SIGNR3 interaction [39]. Lactobacillus crispatus 2029 inhibited the interaction of Candida albicans on HeLa cells and protected the epithelial cells from pathogen infection [40]. Wang et al. [41] reported that SLP isolated from L. acidophilus NCFM® inhibited the growth of HTC116, a colon cancer cell line. The isolated SLP of L. acidophilus is recommended as an oral drug nanocarrier because of its impressive stability in stimulated gastric fluid with and without pepsin [42].
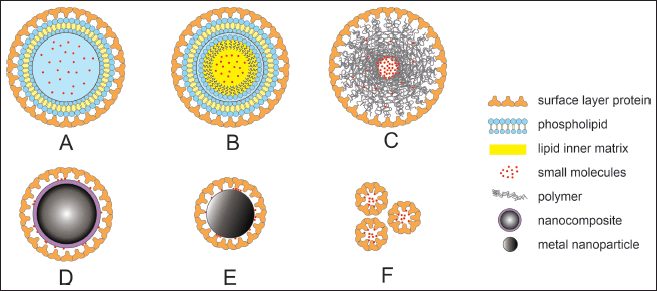 | Figure 2. Lactobacillus sp. SLP coatings on various types of drug carriers. (a) SLP-coated liposome. (b) SLP-coated emulsome. (c) SLP-coated polymeric nanoparticle. (d) SLP-coated nanocomposite. (e) SLP-coated metal nanoparticle. (f) small molecule loaded-SLP based carrier. [Click here to view] |
The development of recombinant Lactobacillus SLP was then considered to more efficiently exploit the advantages of using whole cells. The recombinant SLPA of L. brevis KCTC3102 expressed in E. coli BL21(DE3), was used to treat neonatal calves with diarrhea [43]. A SLP-encoding gene (including N- and C- fragments) was constructed with pHSAN, pHGFPSAC, and pHGFP expression plasmids, and then expressed in E. coli M15 [23]. A list of SLP promoters, secretion signal peptides, and other genetic analyses, which will be crucial for further recombinant SLP studies, has been reviewed [44,45].
The combination properties of maintaining activities after isolation (as native or a recombinant protein), self-assembly, regular and predictable structure, ability to form particles of nanosize, and high stability of SLP make it interesting to include this material in drug nanocarrier development [46]. Sagmeister et al. [22] observed that self-assembled Lactobacillus sp. SLP, forming lattice p2 symmetry with small pores. This structure is a potential characteristic of drug nanocarriers because it offers protection from harsh environments [22]. The interaction of each protein forming SLP remains unclear. However, although the diversity of the SLP amino acid sequences is high, the functional domains and their interactions, as well as their predicted domain folds, are similar. This even applies to SLP with some supplementary domains [22]. With regard to the properties required as a drug nanocarrier, SLP has certain regions with positive and negative charges, which in some cases are important for drug loading or entrapment. SLP (predominantly SLPA) maintains the mechanism of moonlighting protein once applied as a drug nanocarrier. It is beneficial for an active targeting delivery system, for instance, by specifically adhering to the intestinal or colon cells via target molecules (fibronectin, collagen I and IV; mucin; laminin; DC-SIGN) [24,47,48]. SLP can also precisely bind to the target cell via certain enzymes, such as triose phosphate isomerase, GPI, glucose-6 phosphate isomerase, or phosphoglycerate kinase [48,49]. Compilation of the features of Lactobacillus SLP is described in Table 1.
SLP as drug nanocarriers
The SLP matrix is composed of protein or glycoprotein molecules that form regular and typical arrangements [4]. They can assemble in liquid phases, solid phases, and at the air-liquid contact via an entropy-driven process [4,48,50]. The arrangement of the SLP matrix depends on the sequence of its amino acid residues. The SLP protein consists mostly of nonpolar amino acids, therefore, the early step of the assembly process mainly involves hydrophobic interactions [4,51]. The array is formed by the interaction of negatively charged carboxylic groups with positively charged amino groups at certain amino acid residues, without any covalent bonds [4]. Stabilization of the SLP matrix was enhanced by the bonding of acidic residues and cations (Ca2+ and Mg2+) [4,52,53]. In addition, after the SLP confirmation was completed, each monomer remained in its position because of the low free energy. Therefore, it could be used in nanoparticles (NPs) for therapeutic applications and is considered a building block of bionanomimetic NPs (Fig. 2) [54,55]. Table 2 shows a list of SLP applications for many types of drug nanocarriers.
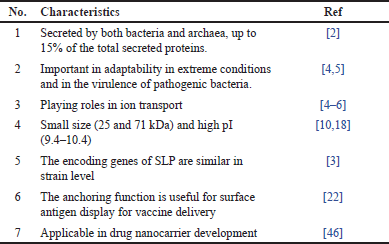 | Table 1. Characteristics of SLP from Lactobacillus sp. [Click here to view] |
SLP-coated lipid membrane liposomes and emulsome
Solid lipid NPs have been developed and used in various drug delivery systems. The delivery mode is particularly used to formulate poorly water-soluble drugs by incorporate the substance in the core of the nanocarrier that was formed in the multilamellar or unilamelar layer [56]. The lamellar system was formed through the ability of the lipids component that able to self-assemble into bilayers to form vesicle, this formation was affected by the charge and geometry of the phospholipids components. The spherical bilayer shape will encapsulate the drug substances and form vesicles with sizes ranging from 50 nm to 1 µm.
Liposomes
Liposomes are spherical vesicles consisting of a phospholipid bilayer shell with an aqueous core. It has been shown that liposomes are excellent drug vehicles because they can load and transport hydrophobic substances in their lipidic shells and hydrophilic compounds in their aqueous core [56]. Coating liposomes with SLP offers several advantages such as stabilizing lipid membrane, preventing aggregation, giving protection against the gastrointestinal tract environment, improving encapsulation efficiency, permeability, and bioavailability profile, and improving the release profile of the drug. SLP-coated liposomes are also comparable to the “artificial virus-like particles” or “artificial cell envelopes,” as they increase mucosal immunity of the vaccine preparation [57].
SLP lattices also stabilize the lipid membranes [58]. There are studies supporting the positive effects of protein-liposome interactions, including SLP protein from various Lactobacillus sp. [59–61]. The SLP coating effectively prevents the aggregation of positively charged liposomes, as it provides an immense barrier because of its bulky structure [3]. Additionally, SLP reduced the charge repulsion between stearyl amine molecules, leading to an increase in chain packaging and membrane rigidity [62].
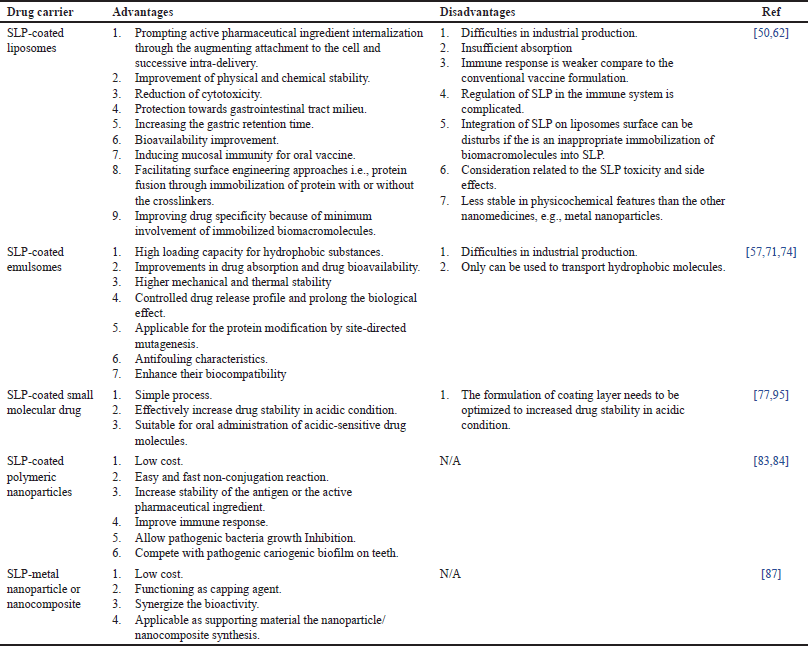 | Table 2. Application of S-layer protein (SLP) of Lactobacillus sp. in drug delivery system. [Click here to view] |
The presence of SLP neutralizes the charge at the membrane interface and increases the entrapment efficiency and permeability [63]. The membrane rigidity of liposomes was also improved when coated with SLP. Carboxyl groups in the interior of the SLP array electrostatically bind to the zwitterionic-lipidic parts of the liposome. Not all of the lipid molecules of the liposome bilayer had contact with the SLP, but only a maximum of three molecules had a protein-bilayer interaction. Therefore, it produces semi-fluidic membrane characteristics because unbound lipid molecules are allowed to freely diffuse in the liposomal bilayers [58].
The attachment of SLP did not only affect the hydrophobic lipid acyl chains, but also provided supporting scaffolding to the lipid membranes [64,65]. Glycosylated SLP has a better affinity to liposomes, but there was no difference in the stabilizing capacity of glycosylated and unglycosylated SLP in SLP-coated liposomes [60,62]. The interaction of SLP with the lipid monolayer of liposomes depends on the materials that determine the hydration state of the lipid interface. If the surface pressure was high, the membrane became more rigid, and surface polarity increased at high cholesterol ratios. Meanwhile, the membrane structure was more relaxed at low pressure because the monolayer was less packed and more hydrated. This indicated that protein insertion could modify the hydration state of the interface [60].
The SLP-coated liposome protects the drug substance within the micelles against the gastric environment condition, e.g., quick changes and variations in pH and temperature, gastric grinding process, and digestive enzymes. The cross-linking of the proteins and glutaraldehyde further enhances the effect [62]. SLP can adhere to the gastrointestinal mucosa, which increases drug retention time in the gastrointestinal tract and promotes drug internalization [50]. Their adhesive and immunomodulatory properties also account for their impressive potential as vaccine delivery platforms [47].
The interaction between SLP-coated calcein-loaded liposomes and human colon adenocarcinoma Caco-2 cells was evaluated under various conditions [60]. The SLPs used in this formulation were extracted from L. kefir JCM 5818. After incubation at 37°C for 30 minutes, the concentration of calcein internalized by Caco-2 cells from SLP-coated liposomes was significantly higher than that of the control liposomes. In the calcein dequenching analysis, a group of SLP-coated liposomes delivered up to 40% higher calcein than the group without the SLP coating [60].
SLP-coated liposomes can be developed as drug vehicles for oral administration, particularly for macromolecular substances [60]. Streptavidin and enhanced green fluorescent protein (EGFP) are macromolecular compounds that have been reported to be successfully entrapped by SLP-coated liposomes [66,67]. SLP-coated liposomes were also able to attach to ferritin via covalent bonds [64]. Biotinylation of SLP-coated liposomes has been reported to enhance the binding efficiency of streptavidin [67].
The SLP of L. acidophilus CICC6074 was applied to coat cholesterol-lowering peptide (LQPE)-loaded liposomes for increasing intestinal absorption. Even though SLP non-covalently binds to the lipid heads of liposomes, it could produce a firm and stable fusion. SLP facilitated liposome absorption across the intestinal mucosa by lowering the hydrophobicity and electrostatic interaction between them [68]. It contrasting with the study reported by Tan et al, SLPB-coated liposome reported that the uptake of the vesicles in the intestinal was not improved. While, it improved the stability of the preparation against harsh environments in the gastrointestinal tract, and facilitated specific uptake of vesicles into Peyer’s patches, and ultimately increasing the bioavailability of the drug by 427.65-fold [61]. Additionally, coating liposomes with SLP from L. buchneri reported to be able to increase the stability, maintain sustained release, and added antibacterial activity to carvacrol (Car)/β-cyclodextrin (β-CD)-loaded liposomes [69]. SLP from L. buchneri and L. kefir has been used to coat cationic liposomes and reduced the negativity of the liposome’s zeta potential. The SLP coating reduced the liposomes leakage percentage against high temperatures (50°C), light, and pH (3,7, and 9) [70].
Emulsomes
Emulsomes are liposome-like particle delivery systems with a solid lipid in the inner part that forms a colloidal solution as the final product [57,71,72]. The solid core of the emulsome can entrap a greater concentration of poorly water-soluble drug compounds and gives a prolonged release profile [71–73]. In the case of curcumin, a poorly water-soluble compound, the solubility increased 10,000-fold, from 0.07 to 0.11 mg/ml to ≈11 ng/ml [71]. Methods that have been utilized to prepare emulsomes involves a dehydration-rehydration process followed by temperature-controlled extrusion [71].
Ucisik et al. [71] studied the characteristics of SLP-coated emulsomes. It showed that the binding and crystallization of SLP on emulsomes also require intermolecular electrostatic interactions between the amine groups of the bilayers and the carboxyl groups inside the SLP matrix, similar to SLP-coated liposome production [71]. SLP coating decreased the cytotoxicity of positively charged emulsomes. Dipalmitoylphosphatidylcholine in emulsomes is toxic to the human liver carcinoma cell line (HepG2). Compared with the SLP-coated emulsomes at similar concentrations of dipalmitoylphosphatidylcholine, toxic effects were only observed in the group without SLP coating. The safety of SLP-coated emulsomes was still indicated even at 2.5 times higher concentration. They had solid electrostatic attraction with the negatively charged cell membrane (CM), which was assumed to be due to their high toxicity. The recrystallization of SLP on the surface of the emulsomes caused significant shifts in the positively charged vesicles to nearly zero or to a highly negative zeta potential [57,74]. According to visualization using a transmission electron microscope (TEM), it was observed that the SLP-coated emulsomes had already contact with the CM before being internalized by the cells. Therefore, SLP modifies the lipid surface of emulsomes and enhances their biocompatibility [57].
The S-layer fusion protein was shown to form a uniform monomolecular lattice on the surface of the emulsomes and the CurcuEmulsomes, altering the surface characteristics of the lipid-based nanocarrier and bestowing IgG binding functionality on the nanocarrier. Entrapped curcumin at a concentration of 30 μg·ml−1 did not influence the self-assembly characteristics of the S-layer protein. This study indicates that S-layer fusion technology is a highly effective approach for the immobilization of foreign proteins such as protein G domains on emulsomes. The distinct advantage of using S-layer proteins is that they can be recrystallized in an oriented fashion on a variety of supports including spherical surfaces covered by phospholipids [46]. Previous studies have also shown that mixtures of native S-layer proteins [47] and S-layer fusion proteins incorporating different functional domains [46] assemble into coherent monomolecular layers on different surfaces including liposomes [48].
Ucisik et al. [74] reported that SLP fusion protein (in this research SLP was fused with two protein G domain) able to form a uniform monomolecular lattice in the emulsome, but it alters the characteristics of the outward layer of the lipid nanoparticle while still able to entrapped the active compound. However, a high curcumin concentration (110 μg/ml) disturbed the self-assembly process of the SLP, in which SLP recrystallization was not detected [74]. It was assumed that the nonspecific absorption of curcumin on the outermost layer of the emulsome disturbed the recrystallization process of the SLPs or because the incorporation of a high concentration of curcumin influenced the rigidity or irregularity of the outermost phospholipid bilayer of the nanovesicle. SLP and SbpA are smooth cytophobic forms that exterminate the adsorption of human plasma proteins at basic pH [75] and cell adhesion (e.g., HepG2) [76]. Therefore, coating with SbpA might reduce the adhesion of opsonins and increase the half-life of the SLP-coated emulsome, resulting in antifouling characteristics. This study also showed that SLP is a valuable method for immobilizing foreign proteins, such as protein G domains, on emulsome.
SLP-coated small molecules drug
As previously mentioned, SLP can be attached directly to an active pharmaceutical ingredient without any other nanocarriers. Qamsari et al. encapsulated omeprazole, a proton pump inhibitor, with extracted SLP from L. acidophilus ATCC 4356 using a simple incubation method (25°C, 2 h, 100 rpm). SLP encapsulation reduced omeprazole instability and protected against gastric acid degradation [77].
SLP-coated polymeric NPs
Polymeric NPs are the most developed drug delivery systems. Various types of polymers have been applied to the system [76]. Natural or synthetically modified biodegradable polymers are preferred because of their low toxicity, side effects, high biocompatibility, and low immunogenicity [78]. Even so, a non-biodegradable polymer such as chemically synthesized dextran-methacrylate acid (dex-MA) has also been studied for a certain purpose of delivery with extreme barriers, such as the intracellular delivery of cytotoxic drugs [79]. Another advanced characteristic of polymeric NPs is that they are easily modified for a specific drug release-triggering factor, such as pH, temperature, oxidative stress level, or light [79–82]. Several polymeric NPs were coated with Lactobacili’s SLP to enhance their intrinsic bioactivity [83,84].
SLP isolated from Lactobacillus sp. was combined with doxorubicin (DOX)-loaded mouse melanoma B16F10 CM/polyethyleneimine-modified (2-hydroxylpropyl)-γ-cyclodextrin (HPAD) NPs to increase protection of the antigen and enhance the immune response. The addition of SLP to the outer part of the DOX/HPAD/CM NPs effectively and efficiently inhibited tumor growth and metastasis by improving the targeted microenvironment immune system [84]. SLP-coated polymeric NPs have also been used in dental care. Triclosan-loaded PLGA NPs were coated with SLP extracted from L. acidophilus. The presence of L. acidophilus SLP remarkably eliminated the Staphylococcus mutans biofilm and hindered the pathogenic formation of Sprague-Dawley rat teeth by sustaining the release of triclosan. In addition, taking advantage of the safety of L. acidophilus or generally recognized as safe (GRAS), the SLP-coated PLGA NPs are undoubtedly safe for dental treatment [84].
SLP-coated metal NPs/nanocomposites
Metal NPs (MNPs) of gold (Au), silver (Ag), titanium (Ti), palladium (Pd), copper (Pb), and zinc (Zn) are frequently studied as drug nanocarriers [85]. MNPs have a high loading capacity, easily conjugate with specific ligands through hydrogen bonds, covalent bonds, or electrostatic interactions, extend the half-life of the loaded drug in blood circulation, and avoid early renal elimination [86]. The application of SLP on MNPs could involve different functions, such as a capping agent, boosting bioactivity, or as a synthesis-supporting system [87–89].
Capping agents in NPs, specifically MNPs, are crucial for ensuring colloidal stability by preventing particle agglomeration [90]. MNPs stabilization is achieved by many interactions with the capping agent, for example, electrostatic, steric, van der Waals, or hydration forces [91]. Some capping agents also synergize with MNPs for several bioactivities such as anti-inflammatory, anticancer, antibacterial, antidiabetic, and wound healing activities [92]. A number of materials appear to be prospective MNPs capping agents, including polymeric materials (chitosan, polyethylene glycol, and polyvinyl alcohol), chelating agents (ethylenediaminetetraacetic acid or EDTA), plant extracts, and proteins (bovine serum albumin or BSA) [90].
SLP is considered for use in MNPs, nanocomposite coatings, or capping because appropriate interactions exist among these materials. SLP of L. buchneri was used to cap the AgNPs via electrostatic interactions and hydrogen bonding. The SLP-capped AgNPs resulted in satisfactory antibiofilm and antibacterial activity against S. enterica and Staphylococcus aureus because of increased permeability to the bacterial CM and lowered activity of respiratory chain dehydrogenase [87].
SLP of L. helveticus was applied to silver NPs (AgNPs) to enhance its antibacterial effect on pathogenic Pseudomonas aeruginosa PAO1. It was reacted with AgNO3 as a precursor, and the AgNPs were synthesized in an eco-friendly manner using an aqueous extract of Juglans regia green husk [88].
Isolated SLPs from L. kefiri CIDCA 8348 and 83111 were used as support systems for AgNPs. The SLPs were mixed with an AgNO3 solution before chemical reduction. As a supporting system, the SLP is low-cost, simple, and “greener” to use, and has an incredible bio-dimensional matrix, yet notably, no additional stabilizer is needed because it prevents the AgNPs aggregation and improves the particle catalytic activity [92]. In other studies, similar SLPs were used as a supporting system in combination with acrylic particles to synthesize platinum NPs (PtNPs), resulting in multiple reused catalytic MNPs [93,94].
CONCLUSION
As a food-grade and potential probiotic organism, Lactobacillus genus is an excellent source of SLP that will be developed as one of the raw materials in the drug formulation. SLP is a natural material with a typical structure and stability. SLP can be used directly as drug nanocarriers, antigens, drug substances, or other medically important molecules. It can also be integrated with other well-developed drug nanocarriers such as liposomes, emulsomes, polymeric NPs, and metal NPs/nanocomposites. The integration of SLP into the surface layer of a drug nanocarrier can improve its physical, chemical, and biological properties.
ACKNOWLEDGEMENT
The authors would like to thank to Directorate of Repositories, Multimedia, and Scientific Publishing, National Research and Innovation Agency for supporting us in plagiarsm and grammar checking.
AUTHOR CONTRIBUTIONS
RDP and MWF contributed in idea conception and design; RDP, MWF, YY, and TA wrote the manuscript draft; all authors equally contributed in improving and revising the manuscript; agreed to submit to the current journal; approved the final version.
FINANCIAL SUPPORT
Funding Programs from Organization of Health (5/III.9/HK/2024) and Organization of Nanotechnology and Materials (20/III.10/HK/2024), National Research and Innovation Agency.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
ABBREVIATIONS
2D: two dimensions; Ag: argentum; AgNO3: argentum nitrat; AgNPs: argentum nanoparticles; Au: aurum; BSA: bovine serum albumin; C-terminal: carboxyl terminal; DCSIGN: dendritic cell-specific ICAM-3-grabbing nonintegrin; dex-MA: dextran-methacrylate acid; DOX: doxorubicin; EDTA: ethylenediamine tetraacetic acid; EGFP: enhanced green fluorescent protein; GPI: glucose phosphate isomerase; GRAS: generally recognized as safe; HPAD: (2-hydroxylpropyl)-γ-cyclodextrin; IBD: inflammatory bowel disease; kDa: kilo Dalton; LAB: lactic acid bacteria; MNPs: metal nanoparticles; NPs: nanoparticles; N-terminal: amino terminal; Pd: palladium; pH: power of hydrogen; pI: isoelectric point; PLGA: polylactic-co-glycolic acid; PtNPs: platinum nanoparticles; SLP: surface layer protein; TEM: transmission electron microscopy; Ti: titanium.
REFERENCES
1. Fagan RP, Fairweather NF. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol. 2014;12:211–22. CrossRef
2. Sa´ra M, Sa´ra S, Sleytr UB. S-Layer Proteins. J Bacteriol. 2000;182:859–68.
3. Hynönen U, Palva A. Lactobacillus surface layer proteins: structure, function and applications. Appl Microbiol Biotechnol 2013;97:5225–43. CrossRef
4. Sleytr UB, Schuster B, Egelseer EM, Pum D. S-layers: principles and applications. FEMS Microbiol Rev 2014;38:823–64. CrossRef
5. Sleytr UB, Beveridge T. Bacterial S-layers. Trends Microbiol. 1999;7:253–60.
6. Desvaux M, Dumas E, Chafsey I, Hébraud M. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol Lett. 2006;256:1–15. CrossRef
7. Zhu C, Guo G, Ma Q, Zhang F, Ma F, Liu J, et al. Diversity in S-layers. Prog Biophys Mol Biol. 2017;123:1–15. CrossRef
8. Glaeser RM, Chiu W, Grano D. Structure of the surface layer protein of the outer membrane of Spirillum serpens. J Ultrastruct Res. 1979;66:235–42.
9. Bharat TAM, von Kügelgen A, Alva V. Molecular logic of prokaryotic surface layer structures. Trends Microbiol. 2021;29:405–15. CrossRef
10. Åvall-Jääskeläinen S, Palva A. Lactobacillus surface layers and their applications. FEMS Microbiol Rev. 2005;29:511–29. CrossRef
11. Johnson B, Selle K, O’Flaherty S, Goh YJ, Klaenhammer T. Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology (United Kingdom) 2013;159:2269–82. CrossRef
12. Suhr M, Lederer FL, Günther TJ, Raff J, Pollmann K. Characterization of three different unusual s-layer proteins from Viridibacillus arvi JG-B58 that exhibits two super-imposed S-layer proteins. PLoS One 2016;11:e0156785. CrossRef
13. Salvetti E, Torriani S, Felis GE. The genus Lactobacillus: a taxonomic update. Probiotics Antimicrob Proteins 2012;4:217–26. CrossRef
14. Goldstein EJC, Tyrrell KL, Citron DM. Lactobacillus species: taxonomic complexity and controversial susceptibilities. Clin Infect Dis 2015;60:S98–107. CrossRef
15. Claesson MJ, Van Sinderen D, O’Toole PW. The genus Lactobacillus—a genomic basis for understanding its diversity. FEMS Microbiol Lett 2007;269:22–8. CrossRef
16. Tannock GW. A special fondness for lactobacilli. Appl Environ Microbiol 2004;70:3189–94. CrossRef
17. Martinez RM, Hulten KG, Bui U, Clarridge JE. Molecular analysis and clinical significance of Lactobacillus spp. Recovered from clinical specimens presumptively associated with disease. J Clin Microbiol 2014;52:30–6. CrossRef
18. Jakava-Viljanen M, Avall-Jaaskelainen S, Messner P, Sleytr UB, Palva A. Isolation of three new surface layer protein genes (SLP) from Lactobacillus brevis ATCC 14869 and characterization of the change in their expression under aerated and anaerobic conditions. J Bacteriol 2002;184:6786–95. CrossRef
19. Cavallero G, Malamud M, Casabuono A, Serradell M, Couto A. A glycoproteomic approach reveals that the S-layer glycoprotein of Lactobacillus kefiri CIDCA 83111 is O- and N-glycosylated. J Proteom. 2017;162:20. CrossRef
20. Malamud M, Cavallero GJ, Casabuono AC, Lepenies B, de los Ángeles Serradell M, Couto AS. Immunostimulation by Lactobacillus kefiri S-layer proteins with distinct glycosylation patterns requires different lectin partners. J Biol Chem. 2020;295:14430–44. CrossRef
21. Mobarak Qamsari E, Kasra Kermanshahi R, Erfan M, Ghadam P, Sardari S, Eslami N. Characteristics of surface layer proteins from two new and native strains of Lactobacillus brevis. Int J Biol Macromol. 2017;95:1004–10. CrossRef
22. Sagmeister T, Gubensäk N, Buhlheller C, Grininger C, Eder M, Ðordi? A, et al. The molecular architecture of Lactobacillus S-Layer: Assembly and attachment to teichoic acids n.d. CrossRef
23. Smit E, Oling F, Demel R, Martinez B, Pouwels PH. The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterisation of domains responsible for S-protein assembly and cell wall binding. J Mol Biol. 2001;305:245–57. CrossRef
24. Palomino MM, Allievi MC, Gordillo TB, Bockor SS, Fina Martin J, Ruzal SM. Surface layer proteins in species of the family Lactobacillaceae. Microb Biotechnol. 2023;16:1232–49. CrossRef
25. Fu M, Mao K, Gao J, Wang X, Sadiq FA, Li J, et al. Characteristics of surface layer protein from Lactobacillus kefiri HBA20 and the role in mediating interactions with Saccharomyces cerevisiae Y8. Int J Biol Macromol. 2022;201:254–61. CrossRef
26. Shi Z, Li X, Fan X, Zeng X, Zhang T, Wu Z, et al. The SLPX protein plays a crucial role in the intestinal juice tolerance of Lactobacillus acidophilus CICC6074. Food Biosci. 2024;59:103865. CrossRef
27. Pratiwi RD, Sembiring ER, Zanjabilla S. Isolation and characterization of Lactobacillus brevis’ surface layer protein (SLP) from Indonesian Culture Collection. IOP Conf Ser Earth Environ Sci, 439: 012045, Institute of Physics Publishing; 2020. CrossRef
28. Jeong JJ, Park HJ, Cha MG, Park E, Won SM, Ganesan R, et al. The Lactobacillus as a probiotic: focusing on liver diseases. Microorganisms 2022;10:1–20. CrossRef
29. Muscariello L, De Siena B, Marasco R. Lactobacillus cell surface proteins involved in interaction with mucus and extracellular matrix components. Curr Microbiol. 2020;77:3831–41. CrossRef
30. Acosta MP, Geoghegan EM, Lepenies B, Ruzal S, Kielian M, Martinez MG. Surface (S) layer proteins of Lactobacillus acidophilus block virus infection via DC-SIGN interaction. Front Microbiol 2019;10:810. CrossRef
31. Hynönen U, Kant R, Lähteinen T, Pietilä TE, Beganovic J, Smidt H, et al. Functional characterization of probiotic surface layer protein-carrying Lactobacillus amylovorus strains. BMC Microbiol 2014;14:1–16. CrossRef
32. Wakai T, Kano C, Karsens H, Kok J, Yamamoto N. Functional role of surface layer proteins of Lactobacillus acidophilus L-92 in stress tolerance and binding to host cell proteins. Biosci Microbiota Food Health 2021;40:33–42. CrossRef
33. Taverniti V, Stuknyte M, Minuzzo M, Arioli S, De Noni I, Scabiosi C, et al. S-Layer protein mediates the stimulatory effect of Lactobacillus helveticus MIMLH5 on innate immunity. Appl Environ Microbiol 2013;79:1221–31. CrossRef
34. Meng J, Wang YY, Hao YP. Protective function of surface layer protein from Lactobacillus casei fb05 against intestinal pathogens in vitro. Biochem Biophys Res Commun 2021;546:15–20. CrossRef
35. Lebeer S, Vanderleyden J, De Keersmaecker SCJ. Genes and molecules of Lactobacilli supporting probiotic action. Microbiol Mole Biol Rev 2008;72:728–64. CrossRef
36. Yadav AK, Tyagi A, Kaushik JK, Saklani AC, Grover S, Batish VK. Role of surface layer collagen binding protein from indigenous Lactobacillus plantarum 91 in adhesion and its anti-adhesion potential against gut pathogen. Microbiol Res 2013;168:639–45. CrossRef
37. Johnson-henry KC, Hagen KE, Gordonpour M, Tompkins TA, Sherman PM. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell Microbiol 2007;9:356–67. CrossRef
38. Kawahara T, Shimizu I, Tanaka Y, Tobita K, Tomokiyo M, Watanabe I. Lactobacillus crispatus strain KT-11 S-layer protein inhibits rotavirus infection. Front Microbiol 2022;13:783879. CrossRef
39. Lightfoot YL, Selle K, Yang T, Goh YJ, Sahay B, Zadeh M, et al. SIGNR 3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J 2015;34:881–95. CrossRef
40. Abramov VM, Kosarev IV, Machulin AV, Priputnevich TV, Deryusheva EI, Panin AN, et al. Protective properties of S-layer protein 2 from Lactobacillus crispatus 2029 against Candida albicans infections. Biomolecules 2023;13:1740. CrossRef
41. Wang H, Cheng X, Zhang L, Xu S, Zhang Q, Lu R. A surface-layer protein from: Lactobacillus acidophilus NCFM induces autophagic death in HCT116 cells requiring ROS-mediated modulation of mTOR and JNK signaling pathways. Food Funct 2019;10:4102–12. CrossRef
42. Eslami N, Kasra Kermanshahi R, Erfan M. Studying the stability of S-layer protein of Lactobacillus acidophilus ATCC 4356 in simulated gastrointestinal fluids using SDS-PAGE and circular dichroism. Shaheed Beheshti University of Medical Sciences and Health Services Iranian J Pharm Res 2013;12:47–56.
43. Khang YH, Park HY, Jeong YS, Kim JA, Kim YH. Recombinant S-layer proteins of Lactobacillus brevis mediating antibody adhesion to calf intestine alleviated neonatal diarrhea syndrome. J Microbiol Biotechnol 2009;19:511–9.
44. Klotz C, Barrangou R. Engineering components of the Lactobacillus S-layer for biotherapeutic applications. Front Microbiol 2018;9:1–12. CrossRef
45. Klotz C, Goh YJ, O’Flaherty S, Barrangou R. S-layer associated proteins contribute to the adhesive and immunomodulatory properties of Lactobacillus acidophilus NCFM. BMC Microbiol 2020;20:1–13. CrossRef
46. Schuster B, Pum D, Sára M, Sleytr UB. S-layer proteins as key components of a versatile molecular construction kit for biomedical nanotechnology. Rev Med Chem 2006;6:909–20.
47. Gaur N, Sharma A, Singhal B. Bacterial Surface layer proteins: from moonlighting to biomimetics: a new horizonto lead. Adv Biosci Biotech 2018;09:352–72. CrossRef
48. Pum D, Toca-Herrera JL, Sleytr UB. S-layer protein self-assembly. Int J Mol Sci 2013;14:2484–501. CrossRef
49. Nishiyama K, Sugiyama M, Mukai T. Adhesion properties of lactic acid bacteria on intestinal mucin. Microorganisms 2016;4:34. CrossRef
50. Luo G, Yang Q, Yao B, Tian Y, Hou R, Shao A, et al. SLP-coated liposomes for drug delivery and biomedical applications: potential and challenges. Int J Nanomedicine 2019;14:1359–83. CrossRef
51. Györvary E, Schroedter A, Talapin DV, Weller H, Pum D, Sleytr UB. Formation of nanoparticle arrays on S-layer protein lattices. J Nanosci Nanotechnol 2004;4:115–20. CrossRef
52. Baranova E, Fronzes R, Garcia-Pino A, Gerven N Van, Papapostolou D, Péhau-Arnaudet G, et al. SbsB structure and lattice reconstruction unveil Ca2+ triggered S-layer assembly. Nature 2012;487:119–22. CrossRef
53. Norville JE, Kelly DF, Knight TF, Belcher AM, Walz T. 7 Å projection map of the S-layer protein sbpA obtained with trehalose-embedded monolayer crystals. J Struct Biol 2007;160:313–23.
54. Chung S, Shin S-H, Bertozzi CR, De Yoreo JJ. Self-catalyzed growth of S layers via an amorphous-to-crystalline transition limited by folding kinetics. PNAS 2010;107:16536–1651. CrossRef
55. Comolli LR, Siegerist CE, Shin SH, Bertozzi C, Regan W, Zettl A, et al. Conformational transitions at an S-layer growing boundary resolved by cryo-TEM. Angewandte Chemie-International Edit. 2013;52:4829–32. CrossRef
56. Monteiro N, Martins A, Reis RL, Neves NM. Liposomes in tissue engineering and regenerative medicine. J R Soc Interface 2014;11:1–24. CrossRef
57. Ucisik MH, Küpcü S, Debreczeny M, Schuster B, Sleytr UB. S-layer coated emulsomes as potential nanocarriers. Small 2013;9:2895–904. CrossRef
58. Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM. S-layers as a tool kit for nanobiotechnological applications. FEMS Microbiol Lett 2007;267:131–44. CrossRef
59. Hollmann A, Delfederico L, De Antoni G, Semorile L, Disalvo EA. Interaction of bacterial surface layer proteins with lipid membranes: Synergysm between surface charge density and chain packing. Colloids Surf B Biointerf. 2010;79:191–7. CrossRef
60. Hollmann A, Delfederico L, Santos NC, Disalvo EA, Semorile L. Interaction of S-layer proteins of Lactobacillus kefir with model membranes and cells. J Liposome Res. 2018;28:117–25. CrossRef
61. Tan LZ, Yamamoto N. In vivo stability and biodistribution of liposome coated with SLPB from Levilactobacillus brevis. BioRvix 2023. CrossRef
62. Hollmann A, Delfederico L, Glikmann G, De Antoni G, Semorile L, Disalvo EA. Characterization of liposomes coated with S-layer proteins from Lactobacilli. Biochim Biophys Acta Biomembr. 2007;1768:393–400. CrossRef
63. Ma Y, Poole K, Goyette J, Gaus K. Introducing membrane charge and membrane potential to T cell signaling. Front Immunol. 2017;8:1–11. CrossRef
64. Schuster B, Sleytr UB. The effect of hydrostatic pressure on S-layer-supported lipid membranes. Biochim Biophys Acta. 2002;1563:29–34.
65. Schuster B, Sleytr UB. S-layer-supported lipid membranes. Rev Mole Biotechnol. 2000;74:233–54.
66. Mader HS, Link M, Achatz DE, Uhlmann K, Li X, Wolfbeis OS. Surface-modified upconverting microparticles and nanoparticles for use in click chemistries. Chem Eur J. 2010;16:5416–24. CrossRef
67. Guo X, Wu Z, Guo Z. New method for site-specific modification of liposomes with proteins using sortase a-mediated transpeptidation. Bioconjug Chem 2012;23:650–5. CrossRef
68. Jiang X, Pan D, Tao M, Zhang T, Zeng X, Wu Z, et al. New nanocarrier system for liposomes coated with Lactobacillus acidophilus S-layer protein to improve Leu-Gln-Pro-Glu absorption through the intestinal Epithelium. J Agric Food Chem. 2021;69:7593–602. CrossRef
69. Rao SQ, Hu X, Hu Y, Zhao MH, Dai CF, Gu RX, et al. Lactobacillus buchneri S-layer protein-coated liposomes loaded with β-cyclodextrin–carvacrol inclusion complexes for the enhancement of antibacterial effect. Food Res Int. 2022;160:111623. CrossRef
70. Meng J, Wang YY, Hao YP. Application of two glycosylated Lactobacillus surface layer proteins in coating cationic liposomes. World J Microbiol Biotechnol. 2023;39:108. CrossRef
71. Ucisik MH, Küpcü S, Schuster B, Sleytr UB. Characterization of CurcuEmulsomes: nanoformulation for enhanced solubility and delivery of curcumin. J Nanobiotechnol. 2013;11:1–13.
72. Schuster B, Sleytr UB. Biomimetic interfaces based on S-layer proteins, lipid membranes and functional biomolecules. J R Soc Interface 2014;11:20140232. CrossRef
73. Vyas SP, Subhedar R, Jain S. Development and characterization of emulsomes for sustained and targeted delivery of an antiviral agent to liver. J Pharm Pharmacol 2010;58:321–6. CrossRef
74. Ucisik MH, Küpcü S, Breitwieser A, Gelbmann N, Schuster B, Sleytr UB. S-layer fusion protein as a tool functionalizing emulsomes and CurcuEmulsomes for antibody binding and targeting. Colloids Surf B Biointerfaces 2015;128:132–9. CrossRef
75. Picher MM, Küpcü S, Huang C-J, Dostalek J, Pum D, Sleytr UB, et al. Nanobiotechnology advanced antifouling surfaces for the continuous electrochemical monitoring of glucose in whole blood using a lab-on-a-chip. Lab Chip 2013;13:1780–9. CrossRef
76. Rothbauer M, Küpcü S, Sticker D, Sleytr UB, Ertl P. Exploitation of S-layer anisotropy: PH-dependent nanolayer orientation for cellular micropatterning. ACS Nano 2013;7:8020–30. CrossRef
77. Qamsari EM, Kermanshahi RK, Erfan M, Ghadam P. Microencapsulation of omeprazole by Lactobacillus acidophilus atcc 4356 surface layer protein and evaluation of its stability in acidic condition. Iranian J Pharm Res 2020;19:240–54. CrossRef
78. Su S, Kang PM. Systemic review of biodegradable nanomaterials in nanomedicine. Nanomaterials 2020;10:656. CrossRef
79. Kordalivand N, Li D, Beztsinna N, Sastre Torano J, Mastrobattista E, van Nostrum CF, et al. Polyethyleneimine coated nanogels for the intracellular delivery of RNase A for cancer therapy. Chem Eng J 2018;340:32–41. CrossRef
80. Muttaqien S El, Nomoto T, Dou X, Takemoto H, Matsui M, Nishiyama N. Photodynamic therapy using LCST polymers exerting pH-responsive isothermal phase transition. J Control Release 2020;328:608–16. CrossRef
81. Palanikumar L, Al-Hosani S, Kalmouni M, Nguyen VP, Ali L, Pasricha R, et al. pH-responsive high stability polymeric nanoparticles for targeted delivery of anticancer therapeutics. Commun Biol 2020;3:95. CrossRef
82. Salkho NM, Awad NS, Pitt WG, Husseini GA. Photo-induced drug release from polymeric micelles and liposomes: phototriggering mechanisms in drug-delivery systems. Polym (Basel) 2022;14:1286. CrossRef
83. Wu M, Liu X, Bai H, Lai L, Chen Q, Huang G, et al. Surface-layer protein-enhanced immunotherapy based on cell membrane-coated nanoparticles for the effective inhibition of tumor growth and metastasis. ACS Appl Mater Interf. 2019;11:9850–9. CrossRef
84. Weng L, Wu L, Guo R, Ye J, Liang W, Wu W, et al. Lactobacillus cell envelope-coated nanoparticles for antibiotic delivery against cariogenic biofilm and dental caries. J Nanobiotechnol. 2022;20:356. CrossRef
85. Chandrakala V, Aruna V, Angajala G. Review on metal nanoparticles as nanocarriers: current challenges and perspectives in drug delivery systems. Emergent Mater. 2022;5:1593. CrossRef
86. Alalaiwe A. The clinical pharmacokinetics impact of medical nanometals on drug delivery system. Nanomedicine 2019;17:47–61. CrossRef
87. Rao SQ, Zhang RY, Chen R, Gao YJ, Gao L, Yang ZQ. Nanoarchitectonics for enhanced antibacterial activity with Lactobacillus buchneri S-layer proteins-coated silver nanoparticles. J Hazard Mater 2022;426:128029. CrossRef
88. Rahimzadeh F, Ghadam P, Kasra-Kermanshahi R, Zarrabi M. In-situ production of silver nanobiocomposite using surface layer protein of Lactobacillus helveticus and aqueous extract of dried Juglans regia green husk and investigation of antibacterial activity. Polymer Bulletin 2022;79:8353–67. CrossRef
89. Huggias S, Bolla PA, Azcarate JC, Serradell MA, Casella ML, Peruzzo PJ. Noble metal nanoparticles-based heterogeneous bionano-catalysts supported on S-layer protein/polyurethane system. Catal Today 2021;372:98–106. CrossRef
90. Javed R, Zia M, Naz S, Aisida SO, Ain NU, Ao Q. Role of capping agents in the application of nanoparticles in biomedicine and environmental remediation: recent trends and future prospects. J Nanobiotechnol. 2020;18:1–15. CrossRef
91. Ajitha B, Kumar Reddy YA, Reddy PS, Jeon HJ, Ahn CW. Role of capping agents in controlling silver nanoparticles size, antibacterial activity and potential application as optical hydrogen peroxide sensor. RSC Adv 2016;6:36171–9. CrossRef
92. Sidhu AK, Verma N, Kaushal P. Role of biogenic capping agents in the synthesis of metallic nanoparticles and evaluation of their therapeutic potential. Front Nanotechnol 2022;3:801620. CrossRef
93. Huggias S, Bolla PA, Serradell MA, Casella M, Peruzzo PJ. Platinum nanoparticles obtained at mild conditions on S-layer protein/polymer particle supports. Langmuir 2020;36:1201–11. CrossRef
94. Bolla PA, Sanz A, Huggias S, Ruggera JF, Serradell MA, Casella ML. Regular arrangement of Pt nanoparticles on S-layer proteins isolated from Lactobacillus kefiri: synthesis and catalytic application. Mole Catal 2020;481:110262. CrossRef
95. Schuster B. S-layer protein-based biosensors. Biosensors (Basel) 2018;8:2–25. CrossRef