INTRODUCTION
Due to a significant increase in cancer cases year to year, this disease has become a major global concern. Since 2010, cancer prevalence, mortality, and disease burden have increased by 26.3%, 20.9%, and 16%, respectively [1]. In 2019, cancer ranked among the top 10 leading causes of disease burden around the globe for people aged 50 and older [2]. However, the extensive plasticity and heterogeneity of cancer pose significant challenges for its treatment. Furthermore, cancer cells frequently develop resistance to anticancer medications [3]. Therefore, the exploration of new anticancer agents is urgently needed.
Breast cancer and prostate cancer ranked as the second and third most prevalent types of cancer in 2018. Among all cancers, breast cancer stands out as the leading cause of death in women [4]. Prostate cancer becomes more common with age, with the incidence rate reaching 60% among individuals aged 65 and older.
Several studies have reported the potential of Gnetum gnemon seeds as an anticancer agent. Gnetum gnemon is also known as “melinjo” in Indonesia. According to the Central Statistics Agency (BPS), Indonesia’s G. gnemon production reached 292,167 tons in 2021, marking a notable 14.13% increase from the previous year’s total of 255,985 tons. Lampung province consistently poses an annual increase in G. gnemon production, yielding 141,076, 141,848, and 172,238 quintals in 2019, 2020, and 2021, respectively. Stilbenoids are the major compound found in G. gnemon seeds, with six types of stilbene chemicals identified, including trans-resveratrol (3,5,4’-tryhydroxy-trans-stilbene), gnetin C, gnetin L, gnemonoside A, gnemonoside C, and gnemonoside D [5].
Gnetum gnemon seed extract has demonstrated its anticancer effects on various cancer cell types in vitro, including breast and prostate cancer cells [6]. In vitro experiments have also revealed the antioxidant and cytotoxic activity of ethyl acetate fraction [7] as well as ethanol fraction on colonic cancer (WiDr) cells when used as co-chemotherapy agents [8]. In vivo experiments have confirmed the anticancer effect of gnetin C in mice carrying prostate cancer cells. Notably, the anti-cancer effects of gnetin C and G. gnemon seed extract have been reported to be more potent than those of other potential anti-cancer agents, such as resveratrol and pterostilbene [9]. Furthermore, gnetin C is known to have bioavailability six times better than the tRV compound in red wine, making it a promising anticancer agent [10]. In addition to these findings, in silico studies also showed the potential activity of G. gnemon seeds in HeLa cervical cancer cells and as ACE inhibitors [7,11]. Gnetin C is known for its immunostimulating effects and anticancer efficacy. Supplementation with gnetin C significantly increased the number of circulating natural killer (NK) cells expressing the NKG2D and NKp46 activation receptors. NK cells in participants who received gnetin C for 2 weeks displayed more significant cytotoxicity against K562 target cells [12].
Several in vitro studies have demonstrated potential molecular targets of gnetin C, although a comprehensive understanding of this information still needs to be provided. According to reports, gnetin C inhibits prostate cancer by blocking the MTA1 pathway, which is crucial for the aggressiveness and spread of prostate malignancies. However, the anticancer action of gnetin C was also demonstrated in the same study, showing an anti-clonogenic impact in shMTA1 cells through a mechanism other than MTA1 [13].
Given the promising anticancer potential demonstrated by G. gnemon seeds, it is imperative to conduct a comprehensive study into the molecular targets of G. gnemon metabolites, that could contribute to the anticancer activity of these compounds. This study aims to explore the molecular target of G. gnemon metabolites in silico by bioinformatic study and molecular docking.
METHODS
Metabolomic study and pharmacokinetic profiling
Bioactive compounds of G. gnemon were obtained through a metabolomic approach using the KNapSAck family website (http://www.knapsackfamily.com/KNApSAcK_Family/) with the keyword “Gnetum gnemon.” Subsequently, each bioactive compound’s SMILES code was retrieved from the same website and subjected to pharmacokinetic profiling using http://www.swissadme.ch, following Lipinski’s Rule of Five criteria: the number of hydrogen donors (the number of hydrogen atoms attached to O and N, nOHNH ≤ 5), the number of hydrogen bond acceptors (nON ≤ 10), water–lipid partition coefficient (mLogp ≤ 5), and relative molecular mass (MW ≤ 500). These selected metabolites were then used as ligands for molecular docking studies [14].
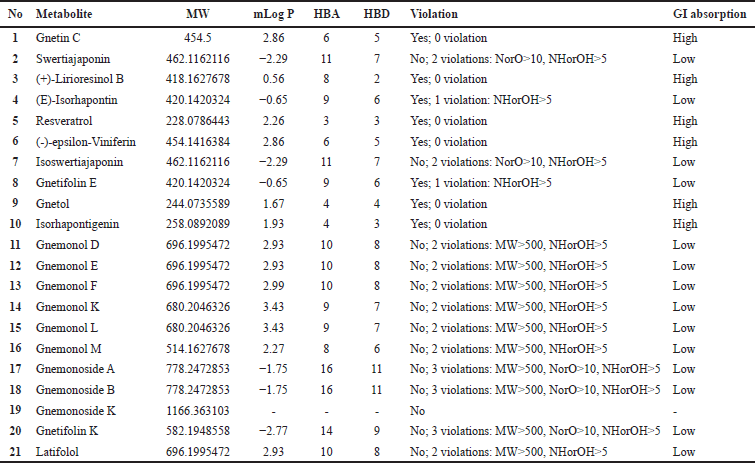 | Table 1. ADMET Prediction of bioactive compounds from G. gnemon. [Click here to view] |
Bioinformatic study
Bioinformatic analysis was used to identify potential protein targets involved in G. gnemon anticancer mechanism for breast and prostate cancer. This analysis used the keywords “resveratrol”, “breast cancer,” and “prostate cancer.” The “resveratrol” was chosen as a keyword because it represents the basic structure of stilbenes in G. gnemon and remains relevant.
The NCBI database (https://www.ncbi.nlm.nih.gov/gene) was utilized to identify regulatory genes associated with breast and prostate cancer. Direct target proteins of resveratrol were identified using the STITCH database (http://stitch.embl.de), while the indirect target proteins were identified through STRING-DB v11.0 (https://string-db.org). Subsequently, a Venn diagram was constructed using Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/fat/) to delineate overlapping genes between breast or prostate cancer regulators and protein targets of resveratrol. The genes appearing in the intersection of the Venn diagram represent the molecular targets of resveratrol, which play a role in regulating breast and prostate cancer [15].
To further understand the protein–protein interactions among the targets, STRING-DB v11.0 (https://string-db.org) was employed. The targets were ranked with Cytoscape software and the Cytohubba plugin, focusing on the degree score to identify the top 20 hub genes [16].
Molecular docking
The crystal structures of the target protein were initially searched for in the PDB database (Table 2), and their selection was based on several criteria: the presence of a structure bound to a native ligand or a small molecule inhibitor, as well as the absence of mutation at the catalytic or active site. Subsequently, the protein structures were downloaded and validated in Autodock, which generated RMSD value. Validation was considered optimum when the values were below 2 [17]. In parallel, ligand preparation was carried out by optimizing each G. gnemon bioactive compound using the density functional theory method to attain the most stable 3D configuration. The optimized ligands were then docked with the target protein using Autodock [17].
Analysis of interaction
We utilized the Biovia Discovery Studio 2021 Client version 21.1.0.0 to visualize the interactions between ligands and target proteins. The visualization provided a view of how each chemical functional group of the ligand interacts with every amino acid residue of the target protein.
RESULTS AND DISCUSSION
Metabolomic study and drug likeness analysis
The metabolomic website initially listed 21 bioactive compounds. To narrow down the selection to orally active drug-like compounds, these 21 compounds underwent a filtering process based on the pharmacokinetic parameters. Lipinski’s rules of five parameters were used to predict these parameters, including absorption, distribution, metabolism, and elimination of the compounds. The outcomes of this filtering process are presented in Table 1.
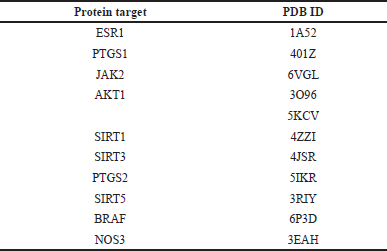 | Table 2. PDB ID of each selected target protein used in molecular docking. [Click here to view] |
Solubility and lipinski analysis on ligands
The solubility and permeability predictions for all drug candidates with optimized structures were based on their physicochemical properties. Notably, Lipinski’s rules of five, which encompass five parameters capable of predicting ligand solubility and permeability, were used in this assessment. These parameters include having no more than five hydrogen bond donors, no more than t10 hydrogen bond acceptors, a molecular weight not more than 500 Da, a log p value not more than 5, and polar surface area not exceeding 140 A, with less than 10 rotational bonds for the ligand [18]. In this study, we focused on four of Lippinski’s rules, specifically the molecular weight, hydrogen bond donors, hydrogen bond acceptor, and log P. Upon analysis, it was found that 15 compounds did not meet these criteria, while six compounds passed Lipinski`s rule of five according to the physicochemical analysis of the ligands. The molecular weights of the analyzed ligands ranged from 228 to 454 g/mol, log p values ranging from 0.56 to 2.86. Hydrogen bond donor values fell from 3 to 8, and hydrogen bond acceptor values ranged from 2 to 5 [19].
Figure 1 shows the six bioactive compounds that passed Lipinski’s rule. Notably, all six of these bioactive compounds are resveratrol derivatives. Consequently, these six compounds were selected as potential ligands for molecular docking.
Bioinformatic study
From our bioinformatics analysis of breast and prostate cancer, we identified sixteen target genes associated with gnetin C and resveratrol. These genes are JAK2, RAF1, CAMK2B, CAMK2G, AGTR1, AGTRAP, TP53, ESR1, PTGS1, PTGS2, AKT1, SIRT1, SIRT3, SIRT5, PPARγ, and NOS3 (Fig. 2). TP53 is excluded as this protein is a tumor suppressor; hence, it cannot be used as a target for inhibition. The crystal structure of RAF1, also known as CRAF, from Homo sapiens with a resolution of less than 3, cannot be found in PDB. Instead, BRAF, a member of the RAF family, activates or forms a dimer with CRAF and, therefore, is chosen for further steps. The crystal structures of AGTR1, AGTRAP, CaMK2B, and CaMK2G or relevant substitutes bound to small molecule inhibitors or ligands are unavailable in PDB and, therefore, have not proceeded to molecular docking. PPARγ also showed insufficient resolution on PDB data and was excluded from the target protein. Accordingly, 10 proteins were used for molecular docking, specifically JAK2, BRAF, ESR1, PTGS1, PTGS2, AKT1, SIRT1, SIRT3, SIRT5, and NOS3. The PDB ID of each of these 10 selected proteins is listed in Table 2.
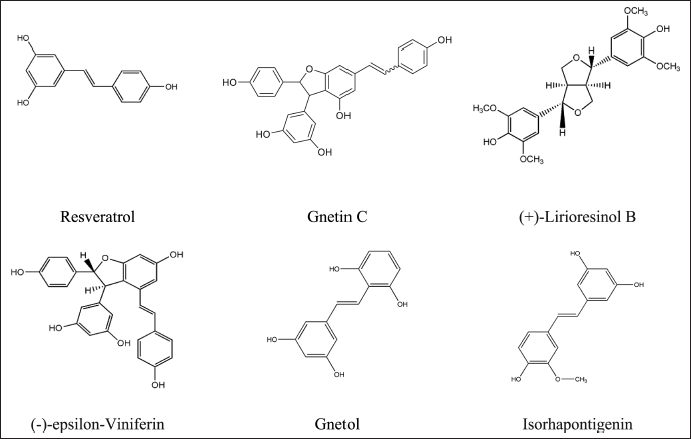 | Figure 1. Selected molecules from G. gnemon . [Click here to view] |
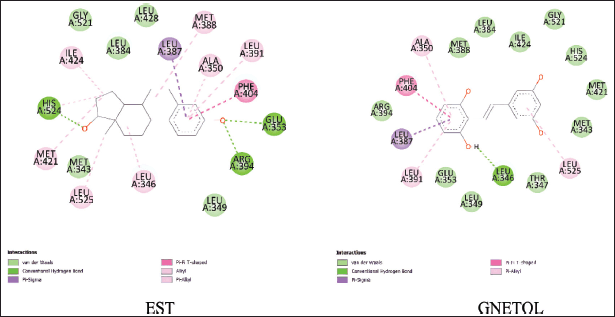 | Figure 2. Binding interaction between EST and gnetol with ESR1 protein. [Click here to view] |
Several hormones and cytokines can activate JAK2, which in turn phosphorylates multiple proteins, including the STAT transcription factor. The JAK2/STAT3 pathway promotes gene expression in cell proliferation. Overactivation of the JAK2/STAT3 pathway has been associated with promoting tumorigenesis. Constitutively activated STAT3 is expressed in numerous tumors, including breast and prostate cancers [20].
C-Raf (also known as Raf-1) and B-Raf are well-studied within the Raf families. They are pivotal effector in the ERK-induced cell growth pathway. Historically, C-Raf has been a primary target in cancer therapy due to its early discovery. However, with the identification of significantly higher occurrences of mutated B-Raf in various tumor types, the focus has shifted towards targeting B-Raf [21]. The most prevalent oncogenic mutation in B-Raf, known as V600E, leads to the overactivation of this protein [22].
ESR1 encodes for estrogen receptor alpha (ERα). Seventy percent of breast cancer patients express estrogen receptors [23]. In ER+ breast cancer cells, ERα plays a pivotal role in driving cancer progression through the PI3K/AKT/mTOR and PI3K/AKT/NF-κB signaling pathway [24]. Similarly, in both preclinical and epidemiological studies, estrogen has been implicated in initiating and advancing prostate cancer. Prostate tissue expresses ERα, and study indicates that the expression level of ERα, but not ERβ, is associated with the promotion of prostate carcinogenesis. In addition, it has been reported to correlate with the severity of prostate cancer [25].
PTGS1 and PTGS2 are genes responsible for encoding cyclooxygenase 1 (COX1) and 2 (COX2), respectively. COX1 is constitutively expressed, while the expression of COX2 is induced during inflammation. COX-1 expression is found to be higher in breast cancer than in normal tissue. In experiments with the MCF7 breast cancer cell line, treatment with a COX1 inhibitor resulted in apoptosis and cell growth arrest [26]. Another study on breast cancer patients revealed that COX-2 expression is elevated in metastasis camcer samples compared to nonmetastatic ones. Moreover, this high expression level of COX2 is associated with a higher mortality rate [27].
AKT1 is a kinase protein activated by growth factors, and it plays a crucial role in regulating a wide array of biological processes. These processes include the inhibition of apoptosis and the induction of cell proliferation. Notably, over-expression and over-activation of AKT1 have been documented in various cancer types, with approximately 40% of breast cancer cases and over 50% of prostate cancer [28].
Sirtuins are a group of proteins involved in numerous cellular processes, including cell differentiation and apoptosis. Upregulation of sirtuin 1-encoding gene SIRT1 is reported in breast cancer. Sirtuin 1 inhibits the activity of anti-tumorigenic proteins, such as FOXO3, and the tumor suppressor p21 and p53. Conversely, SIRT2 and SIRT3 are known for their dual roles, as they can act both as pro- and anti-tumorigenic factors [29,30].
NOS3 encodes for endothelial nitric oxide synthase (eNOS), an enzyme responsible for generating nitric oxide. Nitric oxide (NO) has been shown to have both tumorigenic and tumoricidal roles in cancer. However, most evidence supporting its tumoricidal effect comes from in vitro and has not been clearly observed in cancer patients. Furthermore, the tumoricidal effect is proposed to be associated with a high concentration of NO, resulting in cytotoxicity and apoptosis. This approach is particularly relevant in the early stages of cancer pathogenesis, as once cancer has fully developed, the NO concentration may not be high enough to exert an anti-tumorigenic effect. Clinically, the amount of NO and the activity of eNOS are reported to be higher in breast cancer tissue compared to normal breast tissue. The anti-apoptotic effect of NO is mediated by inhibiting caspases and cytochrome c release and inducing the expression of Bcl-2, Hsp70, and Hsp32. In addition, NO has been found to stimulate tumor blood flow and promote angiogenesis [31]. Based on the description above, the 10 target proteins identified have the potential as anti-breast or anti-prostate cancer. These proteins can be further analyzed for their interactions with G. gnemon metabolites.
Molecular docking
Molecular docking enables the rapid, high-throughput virtual screening of extensive compound libraries, significantly reducing the time and costs required to identify the first hit compounds. However, selecting potential hit compounds remains a reasonably random process, as there is yet to be a consensus on the binding energy and ligand efficiency. In practice, only 20%–30% of compounds identified through molecular docking are active in biological assays [32]. This study used 10 target proteins and evaluated six compounds from G. gnemon for their potential in breast and prostate cancer analysis.
Each of the potential target proteins was prepared and used them as target for molecular docking. The PDB for each protein is provided in Table 2, and we validated these proteins to obtain the coordinates for their binding site. To ensure the reliability of the docking results, we performed validation by redocking each unbound protein with its corresponding native ligand or reference drug, which had previously isolated from the PDB crystal structure [33]. Each ligand was prepared and optimized using Gaussian.
The primary objective of the molecular docking process is to predict the likely binding modes of small-molecule ligands to the target proteins. During the process, the docking program estimates the binding affinities, expressed as Gibb’s free binding energies or docking scores [34]. Predicted binding free energy is a critical parameter as it facilitates the initial selection of candidate molecules based on their likelihood to bind the target. Furthermore, molecular docking also involves estimating the free energy of binding between the reference drug and the target proteins. The results of the docking analysis are presented in Table 3. In conventional docking programs, the energy score function combines empirically derived terms to describe intramolecular conformational and nonbonded interaction energies between ligands and macromolecular targets [35]. The binding free energy is calculated as the final intermolecular energy, which includes van der Waals interactions, hydrogen bonds, desolvation energy, electrostatic energy, total internal energy, and torsional free energy, and is further adjusted by subtracting the unbound system’s energy [36].
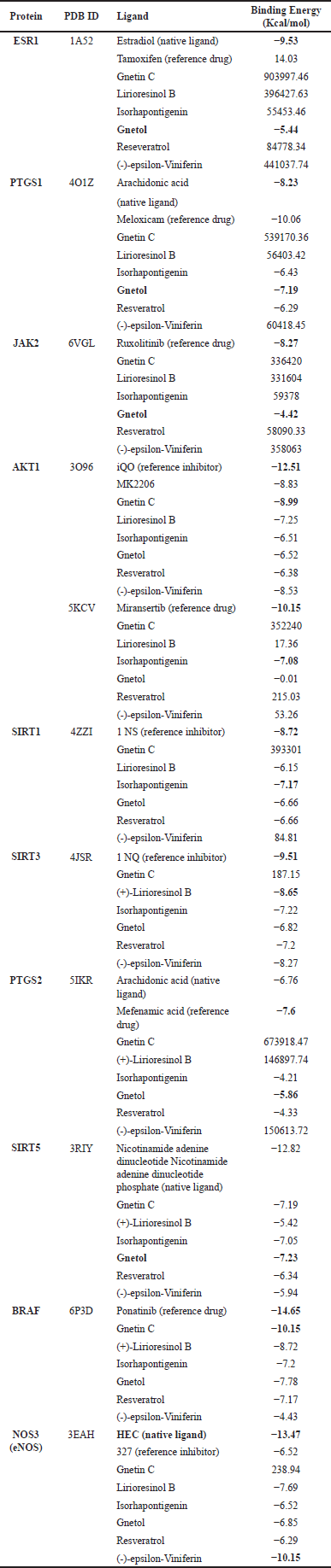 | Table 3. The binding energy of six candidate molecules from G. gnemon to selected breast and prostate cancer target proteins. [Click here to view] |
In the molecular docking analysis with ESR1, only gnetol demonstrated the ability to bind to the breast and prostate cancer regulatory protein. At the same time, the other compounds exhibited no interaction, characterized by notable binding energy values. This result suggests that among the compound from G. gnemon, gnetol is the sole potential candidate targeting ESR1. For PTGS1 (prostate cancer protein target) and PTGS2 (breast and prostate cancer protein target), three molecules—isorhapontigenin, gnetol, and resveratrol—showed favorable interactions, with gnetol exhibiting the lowest energy. This result also indicates gnetol as the most promising compound based on its interactions with PTGS1 and PTGS2. Similarly, for JAK2, a protein target for breast and prostate cancer, gnetol displayed significant interactions, while the other compounds did not. Gnetol has been previously tested in vitro on various cancer cell types, although its mechanism of action remains to be fully elucidated. The results of this docking and bioinformatic analysis provide valuable insights into the potential mechanism of action. Antiproliferative activities of gnetol were tested in multiple cell lines, including HCT-116 (colorectal carcinoma), Hep-G2 (hepatocellular carcinoma), MDA-MB-231 (triple negative breast adenocarcinoma), and PC-3 (prostate adenocarcinoma). Gnetol exhibited the most significant antiproliferative effects against colon cancer cells. While further exploration of gnetol’s mechanism of action on other cancer protein targets is possible, the tested target proteins may also provide insights into related mechanisms of action [37].
The target protein AKT1, relevant to both breast and prostate cancer, has been a focal point of previous research efforts. Blocking AKT, a pivotal component in the frequently disrupted PI3K/AKT signaling pathway, has long been regarded as an attractive therapeutic approach in cancer [38]. There are two categories of AKT inhibitors: ATP-competitive and allosteric. ATP-competitive inhibitors bind to the active site of AKT, preventing ATP from binding; whereas allosteric inhibitors bind to the PH domain to inhibit AKT phosphorylation and activation [39].
Some studies have indicated the potential binding of candidate molecules at the allosteric site [38]. Two PDB IDs corresponding to AKT1 bound to small molecule inhibitors in allosteric positions were identified, specifically 3O96 and 5KCV. The allosteric sites of protein 3O96 had previously been bound with the small molecule inhibitor IQO and the candidate drug in clinical study, MK2206. The allosteric AKT inhibitor MK-2206 primarily targets AKT1 and AKT2 and has demonstrated preclinical single-agent activity by inhibiting the phosphorylation of downstream AKT signaling in multiple cancer cell lines [40].
All tested compounds demonstrated interaction with this site, with gnetin C exhibiting a lower binding energy than MK2206. These compounds were all selected from G. gnemon, suggesting that the main structure of resveratrol plays a vital role in its interaction with AKT1. In the case of 5KCV, only isorhapontigenin displayed an interaction at this allosteric site. In addition, gnetin C’s role as an anticancer agent for prostate cancer has been studied through its involvement with metastasis-associated protein 1 (MTA1) in both in vitro and in vivo. Gnetin C exerts its anticancer activity by inhibiting the cancer-promoting co-operation between MTA1 and ETS2. This action leads to cytotoxicity, cell death, and a reduction in the metastatic potential of prostate cancer cells through MTA1-mediated mechanisms [9,13].
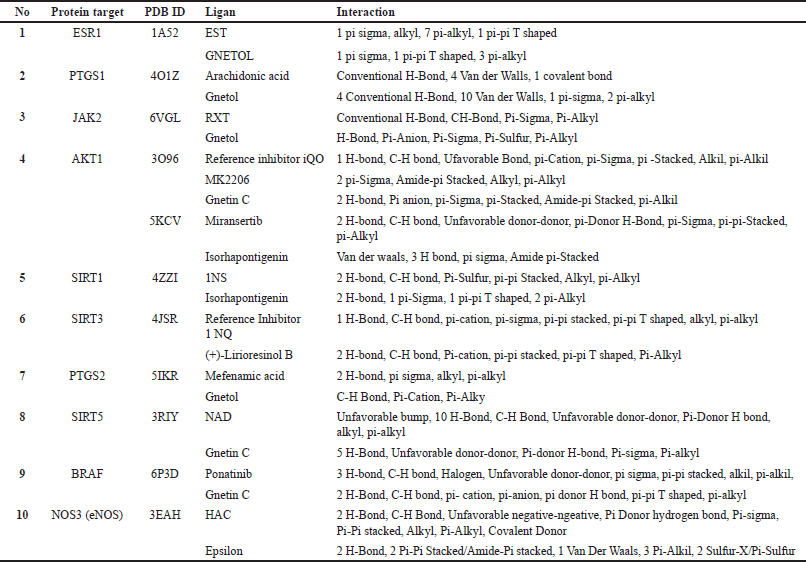 | Table 4. The interaction involved between ligand and protein target. [Click here to view] |
Sirtuins (SIRT1, SIRT3, and SIRT5) play a crucial role in the antitumor pathway, and inhibiting these enzymes opens up a new avenue in anticancer drug discovery. Notably, isorhapontigenin, lirioresinol B, and gnetol exhibit the most favorable interaction with SIRT1, SIRT3, and SIRT5, respectively. This finding suggests that, despite belonging to the same family, these three deacetylation enzymes interact selectively with specific resveratrol derivates.
In the case of BRAF, molecular docking in this study used a protein with the PDB ID 6P3D, targeting the ponatinib binding site. Ponatinib, an FDA-approved drug, is a potent inhibitor of BRAF monomers and dimers. It binds to the BRAF dimer and stabilizes different αC-helical conformations by interacting with a previously undiscovered allosteric site [41]. All six compounds interact in this allosteric site, with gnetin C demonstrating the most favorable interaction. In addition, the NOS3 inhibitor exhibits the most favorable interaction with (-)-epsilon-viniferin.
In summary, our computational docking analysis in this study has unveiled that among all the docking experiments involving six candidate molecules and 10 target proteins, gnetin C exhibits superior interactions with the target proteins associated with breast and prostate cancer compared to reference drugs. Gnetin C demonstrated a binding energy of −8.99 kcal/mol, whereas reference drug MK2206 displayed a binding energy of −8.83 kcal/mol, interacting through the allosteric site. In addition, gnetol also demonstrated the most favorable interactions with PTGS1, PTGS2, JAK2, and SIRT5.
Analysis of interaction
Drug development research relies on molecular docking and visualization. Molecular docking is instrumental in predicting how ligands bind to proteins, aiding in identifying and optimizing potential drugs. Meanwhile, visualization generates graphical representations of molecular structures and interactions, enhancing our understanding of the links between biomolecules and the outcomes of docking simulations [17].
To effectively comprehend and communicate the results of the docking simulations, it is imperative to employ visual representations that are both clear and informative [42]. With the help of the Discovery Studio software, the interaction patterns formed within the complex can be seen [43]. Interaction analysis provides information about the key amino acid residues responsible for binding to the ligand. Table 4 lists the interaction of six G. gnemon compounds with 10 protein targets and the type of chemical bond involved.
Hydrogen bonding plays a crucial role in determining the specificity of ligand binding. This significance is explicitly incorporated into a computational method designed to identify energetically favorable ligand binding sites on a selected target molecule with a known structure [44].
From Table 4, we can examine several compounds that share the same chemical bonds and key amino acids with the reference ligand. For instance, the interaction analysis of gnetol, the most promising compound bound to ESR1, reveals Van Der Walls and pi-alkyl interaction with critical residues of ESR1 (Fig. 2). Sixteen ESR1 residues are involved in interactions with either EST or gnetol. Among these residues are LEU A:384, GLY A:521, LEU A:349, MET A:343, which engage in Van Der Walls interaction, while LEU A:391, ALA A:350, LEU A:525 participate in pi-alkyl interactions.
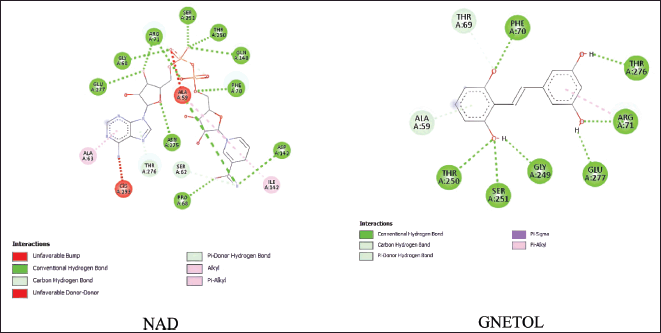 | Figure 3. Binding interaction between NAD and gnetol with SIRT5 protein. [Click here to view] |
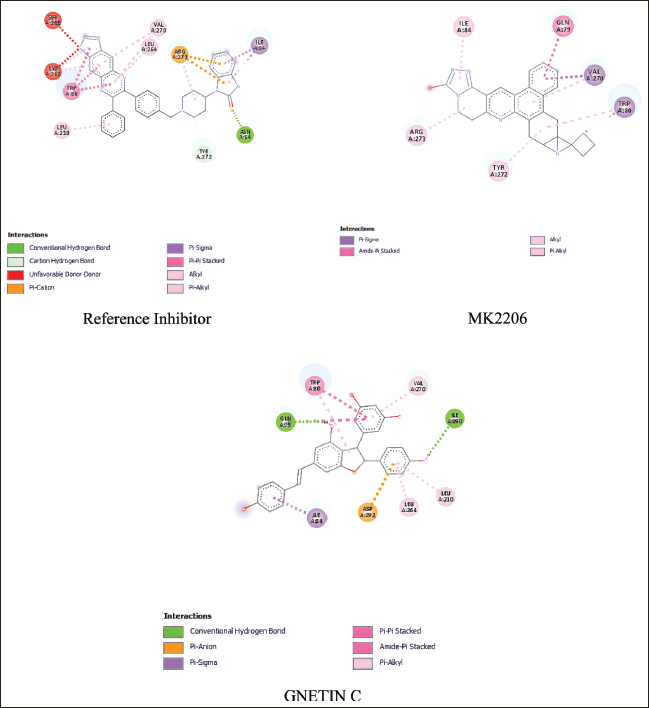 | Figure 4. Binding interaction between reference inhibitor, MK2206, and gnetin C with AKT1 protein [Click here to view] |
Furthermore, GLU A:353, ARG A:394, and HIS A:524 interact with EST through conventional hydrogen bond but engage in Van Der Walls interactions with gnetol. MET A:388, ILE A:424, and MET A:421 interact via pi-alkyl interactions with EST but exhibit Van Der Walls interactions with gnetol. In addition, residue LEU A:387 interacts in pi-sigma interaction, PHE A:404 in pi-pi T-shaped interaction, and LEU A:346 in pi-alkyl interaction with EST while forming conventional hydrogen bonds with gnetol. These findings will help further drug design attempts to develop anti-cancer drugs targeting ESR1. Another interaction analysis of ESR1 suggests that amino acids GLU A:353, LEU A:391, ARG A:394, and PHE A:404 may play essential roles in ligand-protein binding situations [45].
Our results also highlight gnetol as a potential compound targeting the SIRT5 protein. Based on Figure 3, this ligand formed a hydrogen bond with five key residues of SIRT5, which are THR A:250, SER A:251, GLU A:277, ARG A:71, and PHE A:70. Besides, THR A:276 exhibits a C-H bond interaction with NAD and hydrogen bond interaction with gnetol. Gnetol forms strong binding interaction, specifically with ESR1 and SIRT5 proteins, compared to all other docked compounds. These interactions were not observed with TYR A:102 and ARG A:105, which are essential residues [46]. However, gnetol does form an interaction with ARG A:71, suggesting its potential as a residue involved in the interaction with SIRT5 [47].
Another potential compound is gnetin C. Based on Figure 4, gnetin C exhibited a strong interaction with the AKT1 protein (PDB ID: 3O96), involving five identical residues in the same interaction. Furthermore, it displayed pi-alkyl interaction with residues in the active binding site, including LEU A:264, LEU A:210, VAL A:270, pi-sigma interaction with ILE A:84, and pi-pi stacked interaction with TRP A:80. The pi-pi stacked interaction with TRP A:80 plays an essential role in interaction with allosteric site AKT1 [48].
Based on the docking and interaction analysis, both gnetol and gnetin C show promise for development as anticancer agent, especially for breast cancer and prostate cancer. Gnetol exhibits potential in the ESR1 and SIRT5 protein target pathways, while gnetin C acts as an AKT1 allosteric inhibitor.
CONCLUSION
Based on this study, six metabolites from G. gnemon met Lipinski`s rule of five criteria. The molecular target proteins used were PTGS1, PTGS2, ESR1, SIRT1, SIRT3, SIRT5, AKT1, JAK2, BRAF, and NOS3. Free binding energy and binding interaction showed Gnetol and Gnetin C as the most potential metabolites to be developed for further studies as drug candidates for breast and prostate cancer.
ACKNOWLEDGEMENT
The project is a research collaboration between BRIN and ITERA. The authors are thankful to 2023 Drug Vaccine Program House, the Health Research Organization-BRIN for facilitating this research.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
There is no funding to report.
CONFLICTS OF INTEREST
This study has no conflicts of interest to declare.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL, Harvey JD, et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019 a systematic analysis for the Global Burden of Disease Study 2019. JAMA Oncol. 2022;8(3):420–44. CrossRef
2. Abbafati C, Abbas KM, Abbasi-Kangevari M, Abd-Allah F, Abdelalim A, Abdollahi M, et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396(10258):1204–22. CrossRef
3. Nussinov R, Tsai CJ, Jang H. Anticancer drug resistance: an update and perspective. Drug Resist Updat. 2021;59:100796. CrossRef
4. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. CrossRef
5. Kato E, Tokunaga Y, Sakan F. Stilbenoids isolated from the seeds of melinjo (Gnetum gnemon L.) and their biological activity. J Agric Food Chem. 2009;57(6):2544–9. CrossRef
6. Narayanan NK, Kunimasa K, Yamori Y, Mori M, Mori H, Nakamura K, et al. Antitumor activity of melinjo (Gnetum gnemon L.) seed extract in human and murine tumor models in vitro and in acolon-26 tumor-bearing mouse model in vivo. Cancer Med. 2015;4(11):1767–80. CrossRef
7. Savitri RI, Arifin NH, Febriansah R. Antioxidant, cytotoxic activity and protein target inhibition of ethyl acetate fraction Melinjo seed (Gnetum gnemon L.) by in vitro and in silico studies on HeLa cervical cancer cells. HAYATI J Biosci. 2023;30(5):864–73. CrossRef
8. Arifin NH, Febriansah R, Octavia MA, Kenyori IK. Activity of ethanol fraction Melinjo (Gnetum Gnemon L.) seed on colonic cancer (Widr) cells as co-chemotherapy agent. Indones J Cancer Chemoprevention. 2023;14(1):49. CrossRef
9. Gadkari K, Kolhatkar U, Hemani R, Campanelli G, Cai Q, Kumar A, et al. Therapeutic potential of gnetin c in prostate cancer: a pre-clinical study. Nutrients. 2020;12(12):1–15. CrossRef
10. Ota H, Akishita M, Tani H, Tatefuji T, Ogawa S, Iijima K, et al. Trans -resveratrol in Gnetum gnemon protects against oxidative- stress-induced endothelial senescence. J Nat Prod [Internet]. 2013;76(7):1242–7. CrossRef
11. Triputra MA, Yanuar A. Analysis of compounds isolated from gnetum gnemon L. seeds as potential ace inhibitors through molecular docking and molecular dynamics simulations. J Young Pharm. 2018;10(2):s32–9. CrossRef
12. Nakagami Y, Suzuki S, Espinoza JL, Quang LV. Immunomodulatory and metabolic changes after gnetin-C supplementation in humans. Nutrients [Internet]. 2019;11(6):1–16. Available from: 10.3390/nu11061403 CrossRef
13. Kumar A, Dholakia K, Sikorska G, Martinez LA, Levenson AS. Mta1-dependent anticancer activity of gnetin c in prostate cancer. Nutrients. 2019;11(9):90–5. CrossRef
14. Veber DF, Stephen R J, Hung-Yuan C, Brian R S, Keith W W, Kenneth D K. Molecular properties that influence the oral bioavailability of drug candidates. Chemtracts. 2003;16(7):439–42.
15. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–8. CrossRef
16. Fauziyya R, Auli WN, Suprahman NY, Sarmoko S, Ashari A, Alsadila K, et al. Bioinformatic and molecular docking study of zerumbone and its derivates against colorectal cancer. Indones J Cancer Chemoprevention. 2023;14(1):39. CrossRef
17. Meng X-Y, Zhang H-X, Mezei M, Cui2 M. Overview on molecular docking: a powerful approach for structure based drug discovery. Int J Pharm Sci Rev Res. 2022;77(2):146–57. CrossRef
18. Brito MA de. Pharmacokinetic study with computational tools in the medicinal chemistry course. Brazilian J Pharm Sci. 2011;47(4):797–805. CrossRef
19. Syahputra G, Gustini N, Bustanussalam B, Hapsari Y, Sari M, Ardiansyah A, et al. Molecular docking of secondary metabolites from Indonesian marine and terrestrial organisms targeting SARS-CoV-2 ACE-2, Mpro, and PLpro receptors. Pharmacia. 2021;68(3):533–60. CrossRef
20. Ayele TM, Muche ZT, Teklemariam AB, Kassie AB, Abebe EC. Role of JAK2/STAT3 signaling pathway in the tumorigenesis, chemotherapy resistance, and treatment of solid tumors: a systemic review. J Inflamm Res. 2022;15(February):1349–64. CrossRef
21. Zhao J, Luo Z. Discovery of Raf Family is a milestone in deciphering the Ras-mediated intracellular signaling pathway. Int J Mol Sci. 2022;23(9):5158. CrossRef
22. Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, et al. Raf family kinases: old dogs have learned new tricks. Genes Cancer. 2011;2(3):232–60. CrossRef
23. Arnesen S, Blanchard Z, Williams MM, Berrett KC, Li Z, Oesterreich S, et al. Estrogen receptor alpha mutations in breast cancer cells cause gene expression changes through constant activity and secondary effects. Cancer Res [Internet]. 2021;81(3):539–51. Available from: 10.1158/0008-5472.CAN-20-1171 CrossRef
24. Liu Y, Yao J. ERα, A key target for cancer therapy.pdf. 2020;13:2183–91. CrossRef
25. Di Zazzo E, Galasso G, Giovannelli P, Di Donato M, Castoria G. Estrogens and their receptors in prostate cancer: therapeutic implications. Front Oncol. 2018;8(JAN):1–7. CrossRef
26. Pannunzio A, Coluccia M. Cyclooxygenase-1 (COX-1) and COX-1 inhibitors in cancer: a review of oncology and medicinal chemistry literature. Pharmaceuticals. 2018;11(4):1–20. CrossRef
27. Richardsen E, Uglehus RD, Due J, Busch C, Busund LT. COX-2 is overexpressed in primary prostate cancer with metastatic potential and may predict survival. A comparison study between COX-2, TGF-β, IL-10 and Ki67. Cancer Epidemiol [Internet]. 2010;34(3):316–22. CrossRef
28. Song M, Bode AM, Dong Z, Lee MH. AKt as a therapeutic target for cancer. Cancer Res. 2019;79(6):1019–31. CrossRef
29. Onyiba CI, Scarlett CJ, Weidenhofer J. The mechanistic roles of sirtuins in breast and prostate cancer. Cancers (Basel). 2022;14(20):5118. CrossRef
30. Hernandez-Quiles M, Broekema MF, Kalkhoven E. PPARgamma in metabolism, immunity, and cancer: unified and diverse mechanisms of action. Front Endocrinol (Lausanne). 2021;12(February):1–17. CrossRef
31. Korde Choudhari S, Chaudhary M, Bagde S, Gadbail AR, Joshi V. Nitric oxide and cancer: a review. World J Surg Oncol [Internet]. 2013;11(1):1. Available from: CrossRef
32. Ivanova L, Karelson M. The impact of software used and the type of target protein on molecular docking accuracy. Molecules. 2022;27(24):9041. CrossRef
33. Tallei TE, Tumilaar SG, Niode NJ, Fatimawali, Kepel BJ, Idroes R, et al. Potential of plant bioactive compounds as SARS-CoV-2 main protease (Mpro) and spike (S) glycoprotein inhibitors: a molecular docking study. Scientifica (Cairo). 2020;2020:6307457. CrossRef
34. Bender BJ, Gahbauer S, Luttens A, Lyu J, Webb CM, Stein RM, et al. A practical guide to large-scale docking. Nat Protoc. 2021;16(10):4799–832. CrossRef
35. Malmstrom RD, Watowich SJ. Accuracy of virtual screening predictions. J Chem Inf Model [Internet]. 2011;51(7):1648–55. Available from: 10.1021/ci200126v CrossRef
36. Azad I. We are IntechOpen , the world ’ s leading publisher of open access books built by scientists, for scientists TOP 1 %. Intech [Internet]. 2010;34(8):57–67. Available from: https://doi.org/10.1007/s12559-021-09926-6%0Ahttps://www.intechopen.com/books/advanced-biometric-technologies/liveness-detection-in-biometrics%0Ahttp://dx.doi.org/10.1016/j.compmedimag.2010.07.003
37. Remsberg CM, Martinez SE, Akinwumi BC, Anderson HD, Takemoto JK, Sayre CL, et al. Preclinical pharmacokinetics and pharmacodynamics and content analysis of gnetol in foodstuffs. Phytother Res. 2015;29(8):1168–79. CrossRef
38. Shariati M, Meric-Bernstam F. Targeting AKT for cancer therapy. Expert Opin Investig Drugs. 2019;28(11):977–88. CrossRef
39. Bhutani J, Sheikh A, Niazi AK. Akt inhibitors: mechanism of action and implications for anticancer therapeutics. Infect Agent Cancer. 2013;8(1):12–5. CrossRef
40. Sangai T, Akcakanat A, Chen H, Tarco E, Wu Y, Do KA, et al. Biomarkers of response to Akt inhibitor MK-2206 in breast cancer. Clin Cancer Res. 2012;18(20):5816–28. CrossRef
41. Cotto-Rios XM, Agianian B, Gitego N, Zacharioudakis E, Giricz O, Wu Y, et al. Inhibitors of BRAF dimers using an allosteric site. Nat Commun [Internet]. 2020;11(1):4370. CrossRef
42. Baroroh U, Muscifa ZS, Destiarani W, Rohmatullah FG, Yusuf M. Molecular interaction analysis and visualization of protein-ligand docking using biovia discovery studio visualizer. Indones J Comput Biol. 2023;2(1):22. CrossRef
43. Gholam GM, Firdausy IA, Artika IM, Abdillah RM, Firmansyah RP. Molecular docking: bioactive compounds of Mimosa pudica as an inhibitor of Candida albicans Sap 3. bioRxiv. 2022;2022:09. CrossRef
44. Wade RC, Goodford PJ. The role of hydrogen-bonds in drug binding. Prog Clin Biol Res. 1989;289:433–44.
45. Hung TC, Lee WY, Chen KB, Chan YC, Chen CYC. Investigation of estrogen receptor (ESR1) for breast cancer from traditional Chinese medicine. Biomed Res Int. 2014;2014:321486. CrossRef
46. Liu S, Ji S, Yu ZJ, Wang HL, Cheng X, Li WJ, et al. Structure-based discovery of new selective small-molecule sirtuin 5 inhibitors. Chem Biol Drug Des. 2018;91(1):257–68. CrossRef
47. Abdul NS, Nagiah S, Anand K, Chuturgoon AA. Molecular docking and mechanisms of fusaric acid induced mitochondrial sirtuin aberrations in glycolytically and oxidatively poised human hepatocellular carcinoma (HepG2) cells. Toxicon [Internet]. 2020;173(September 2019):48–56. CrossRef
48. Yilmaz OG, Olmez EO, Ulgen KO. Targeting the Akt1 allosteric site to identify novel scaffolds through virtual screening. Comput Biol Chem [Internet]. 2014;48:1–13. CrossRef