INTRODUCTION
Although improvement in cancer treatment over decades has enhanced the survival rate in many countries, cancer is still one of the most common diseases responsible for millions of deaths globally. Chemotherapy is the most common method, which is used as first-line therapy to remove most of the cancer cells in the body. However, chemotherapy possesses many disadvantages. Since chemotherapy drugs are nonspecific, they directly affect both normal and abnormal cells. Therefore, they can cause damage and loss of function in normal cells. The observed serious side effects include hair loss, nausea, vomiting, and immune system destruction. Although chemotherapy has the potential to work systematically and kill even cancer stem cells, its toxicity needs special attention. Since cancer deaths continue to grow over the years, new discoveries about cancer treatments are still ongoing. Recently, the innovation of drug-targeted therapy has opened a new way of increasing specificity and reducing toxicity from chemotherapy.
Minicell is one of the potential drug carriers that has a size range between 100 and 400 nm. Minicells are nonliving cells that do not contain chromosomal DNA and do not have the ability to divide. Minicells have many advantages in drug delivery, such as having high packaging efficiency of molecular drugs (small interfering RNA) or chemotherapeutic drugs, being modified with bispecific antibodies to increase specificity [1], not being accumulated in organs since minicells are biodegradable, and having high safety concern with no drug leakage or serious side effects [2]. Minicells could be generated from both negative and positive bacteria, such as Bacillus subtilis [3], Salmonella typhimurium [4], Listeria monocytogenes [5], Leuconostoc mesenteroides [6], Escherichia coli [7], and Lactobacillus rhamnosus PN04 [8].
Recently, the use of probiotics has been rising in the pharmaceutical field because of its benefits to humans. Lactobacillus acidophilus is one of the essential probiotics that belongs to lactic acid bacteria (LAB). It shows many advantages in inhibiting some pathogens in the intestinal tract [9], increasing nutrient bioavailability, decreasing serum cholesterol [10], producing live vaccine vehicle [11], and so on. Currently, research about minicells of L. acidophilus has been carried out. Minicells of L. acidophilus VTCC-B871 were formed and packaged with paclitaxel and cephalosporin [12]. Studies about minC and minD in Lactobacillus species have explained the mechanism of minicell formation in this species.
CQ is well-known for treating malaria. Besides, CQ shows their ability to be anticancer [13]. The mechanism of action of CQ is mainly based on raising the surrounding pH when it passively diffuses into the cells and becomes protonated in some organelles, such as lysosomes. This can change the acid condition in lysosomes and cause dysfunction and apoptosis [14]. Moreover, chloroquine (CQ) was discovered to be able to stabilize wild-type p53, inducing p53-dependent pathway apoptosis [15] and inhibiting ATP-binding cassette [16]. However, as with other chemotherapeutic drugs, high doses and long use of CQ could cause toxicity to the myocardial, hypoglycemia in nondiabetic patients, vomiting, headache, nausea, and so on. Therefore, the report attempts to perform a study on CQ containing the ability of L. acidophilus ATCC 4356 minicells to solve the problems of toxicity and specificity and contribute information about bacterially derived LAB drug delivery to the pharmaceutical field.
MATERIALS AND METHODS
Bacteria strains and growth condition
Lactobacillus acidophilus was cultured in De Man, Rogosa and Sharpe (MRS) broth at 37°C for 48 hours in 5% CO2 condition. After that, the microorganism was transferred to an agar plate. Then, the plate was incubated at 37°C for 48 hours. One colony was inoculated into the broth and used as the stock for minicell formation.
Minicell formation and isolation
According to a study by Vinh et al. [12], 1% D-fructose gave the highest number of minicells formation. Therefore, L. acidophilus was cultured in the modified broth using 1% D-fructose at 37°C in 5% CO2. Lactobacillus acidophilus was inoculated to the D-fructose broth and grown at 37°C in 5% CO2. After 20–24 hours, it was centrifuged at 1,500, 2,000, and 2,500 rpm for 10 minutes to remove parent cells from minicells. Then, the supernatant was collected and checked for minicell purity using a light microscope with a magnification of 100×. Centrifugation of supernatant at 15,000 rpm for 20 minutes was used to collect minicells. After that, the pellet was washed three times with 1× PBS buffer (pH7) and resuspended in 1× Phosphate-buffered saline (PBS) buffer (pH 7) for further use. To avoid damage to minicells, centrifugation steps must be performed at 4°C. The stock of minicells was stored at a temperature of −80°C.
CQ containing and releasing test
The purified minicells were incubated with CQ (10 mg/ml) at different times (1, 2, 3, and 4 hours) at 37°C. After incubation, the solution was centrifuged at 15,000 rpm for 20 minutes at 4°C to collect the minicells. The pellet and supernatant after centrifugation were separated. The remaining amount of CQ after incubation was measured based on the supernatant. Therefore, the concentration of CQ that was packaged into minicells was calculated by subtracting the initial concentration and the residue concentration of CQ. Minicells packaged CQ was then resuspended in PBS with the gastric intestinal tract pH (pH 3) and plasma pH (pH 7) at 0.5, 1, 2, 3, 4, 5, and 6 hours. Then, the solution was centrifuged at 15,000 rpm for 20 minutes at 4°C. After that, the supernatant was taken out and the amount of CQ released in different conditions was measured by spectrophotometry. The experiment was duplicated.
CQ determination using high-performance liquid chromatography (HPLC) analysis
CQ in the supernatant after being incubated with minicells at 1 hour and after being released from minicells containing CQ at 2 hours was quantified and qualified by HPLC-UV/Vis Spectroscopy using the method of Miranda et al. [17].
Functional group determination using Fourier transform infrared spectroscopy (FTIR) analysis
Three samples, including L. acidophilus minicells, CQ, and L. acidophilus minicells containing CQ, were analyzed by FTIR to check the alteration in the functional group on minicell cell wall before and after incubation minicells with CQ.
In silico experiment for CQ containing the ability of L. acidophilus ATCC 4356 minicells
Molecular docking for interaction prediction between CQ and minicell cell wall components was performed based on the tools used in the study of Abdelsattar [18]. The 3D structures of all the substrates were obtained from the Protein Data Bank RCSB and the PubChem website. All the docking was performed by AutoDock Tools 4.2 and visualized by Discovery Studio Visualizer (2D,3D). CQ was chosen as the ligand. There were eight proteins used to predict the binding of CQ. Two Rib (resistance to protease, immunity, group B), RibS and RibL, were chosen from the domain architecture of Q5FIM8 protein from L. acidophilus [19]. Other proteins, such as mucus-binding proteins (Mubs), penicillin-binding proteins (PBP2b, PBP2x, and PBP1a), MraY, and MurG, were chosen from other bacteria. For those structures that do not come from L. acidophilus, protein alignment was performed to check for identities and positive scores before docking.
Statistical analysis
Data were presented as mean ± SD from three replications. One-way ANOVA was used to compare the average value with 95% confidence levels and p-value < 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
Minicell formation and isolation
After 24 hours in 1% D-fructose-containing broth, L. acidophilus was observed under the microscope to check for minicell formation (Fig. 1). As can be seen in Figure 2, the minicells had a round shape and could still be stained as normal cells. The concentration of minicells was counted using the Neubauer hemocytometer and gave the number of 107 minicells/ml.
Changing the carbon sources from D-glucose to D-fructose created stress on sugar, which led to the formation of minicells. The results showed that L. acidophilus could grow in this medium and could form minicells within a 4-compound broth compared to the original Lactobacilli MRS broth [20] that had 10 compounds. By minimizing the ingredients used in the culture medium, preparation steps will be more convenient, and it will also help to lower the cost of purchasing chemicals. Therefore, this report proposed a more convenient way of preparing the medium for minicell formation.
In the study of Vinh et al. [12], L. acidophilus VTCC-B-871 strain was incubated in a modified broth for 30 hours. However, in this study, from about 20 to 24 hours, minicells were formed. An explanation for the difference in formation time could be based on the strain, the culture medium, and the technique. Therefore, the study recommended that formation time needed to be focused on to have a better understanding of minicell formation and how to isolate them at the right time.
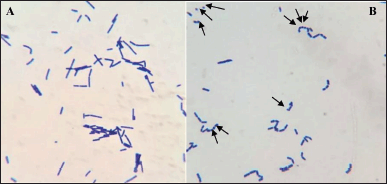 | Figure 1. Gram staining of L. acidophilus under light microscope 100× magnification. (A) L. acidophilus in D-glucose broth; (B) minicells formation of L. acidophilus in D-fructose broth. [Click here to view] |
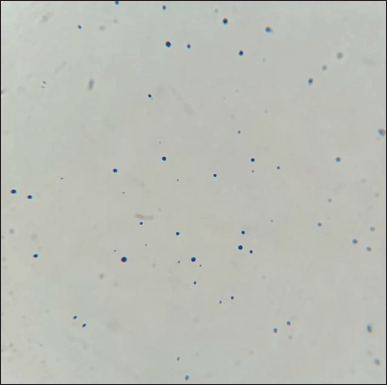 | Figure 2. Gram staining of isolated L. acidophilus minicells under light microscope 100× magnification. [Click here to view] |
CQ containing ability of L. acidophilus minicells
To test the encapsulation efficiency, L. acidophilus minicells in 1× PBS (pH 7) were incubated with CQ for 1, 2, 3, and 4 hours. The highest drug-containing efficiency was observed at 1 hour with 12.8785% ± 0.3442% (Table 1).Polar and nonpolar drugs are hypothesized to cross the membrane through a fast one-dimensional diffusion process [21] and outer membrane channels or nonspecific porin, such as the FadL family [22,23]. The mechanism of how CQ can diffuse and is trapped in the cells was explained by the previous study [24]. Unprotonated CQ can diffuse freely and quickly across cell and organelle membranes. It becomes protonated and becomes trapped in acidic organelles like the lysosome, where it cannot diffuse out. Thus, diffusion down a drug concentration in minicells proposed a simpler method than other methods in encapsulation drugs, such as liposomes.
CQ releasing of L. acidophilus minicells containing CQ
The pellet of minicells containing CQ at 1 hour was used to perform drug release at two different pHs 3 and 7, for 0.5, 1, 2, 3, 4, 5, and 6 hours. Both pH 3 and pH 7 conditions illustrated the highest releasing rate after 2 hours, at 3.1112 ± 0.0348 and 3.5847 ± 0.0179, respectively (Table 2). Testing drug release at pH 3 and pH 7 was performed to examine the development of oral drugs or intravenous drugs since pH 3 and pH 7 are near the pH in the human intestinal tract and blood plasma, respectively.
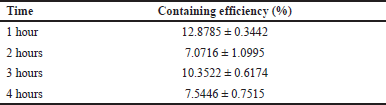 | Table 1. The CQ-containing efficiency of L. acidophilus ATCC 4356 minicells in 1, 2, 3, and 4 hours. [Click here to view] |
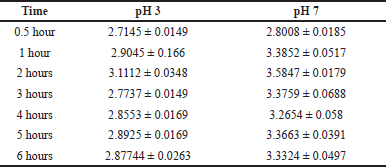 | Table 2. The CQ-containing efficiency of L. acidophilus ATCC 4356 minicells containing CQ at 1 hour in 0.5, 1, 2, 3, 4, 5, and 6 hours. [Click here to view] |
According to Flemming [2], the process of drug uptake in minicells through diffusion was proved to be unidirectional, meaning that no drug could be leaked. Moreover, unprotonated CQ, after entering the cells and becoming protonated, was proven not to leak out. However, a low percentage of CQ was detected in the release test at both pH 3 and pH 7, so that CQ release could prolong. Besides, it meant that minicells were not affected by pH levels. Normally, other drug delivery systems (DDSs) swell to let chemicals release when pH changes in a wide range. In the minicell case, although the cell did not swell, CQ was released from minicells in both pH levels, which did not affect minicells. CQ binding might be onto the minicells and then CQ was released reversibly. The protonated form of CQ might not diffuse freely across the membrane as the unprotonated form. Since minicells cell walls could comprise most of the components of parent cells, such as peptidoglycan, (lipo)teichoic acids, and protein [25], these characteristics provide different functional groups on the outside surface that can have different reactions or interactions with other groups [26] in the protonated CQ. However, the interaction or binding between CQ and the cell wall was not strong enough to keep CQ permanently bound to the cell surface. Thus, these bonds could be broken and release CQ into the different pH. The wild-type cannot pass the gastrointestinal membrane due to its big size (>1 μm) [27]. The study aimed to find out more evidence of minicell delivery. Therefore, the study did not mention wild-type ability. The study did not focus on pH 1 and pH 2 because the study aimed to develop minicells in a formulation that can deliver a drug in the stomach and cross the stomach wall at pH 3 and around gastric pH. For pH 7, the study was to know whether minicells could deliver drugs stably in plasma and human cells around pH 7. From that, the study pointed out the capacity of the CQ package and release. The reasons for the leak and release shortly might be the membrane structure [27] or vesicles in the membrane [28]. This study focused on minicell delivery and did not compare it to the in vivo dosage because the study aimed to know the effective ways. Moreover, DDS is important to reduce side effects and increase bioavailability. The in vivo dose will be a combination of DDS-CQ after studying well. Therefore, the study did not compare to the in vivo dosage.
CQ determination using HPLC
To determine whether minicells could package CQ or not, HPLC was used to qualify and quantify CQ in the supernatant after incubation. Similarly, CQ released to the supernatant in the drug release test was also examined by HPLC (Table 3). The standard CQ used for incubation with minicells was also analyzed to ensure precise concentration.
 | Table 3. The HPLC result of the supernatant obtained from 1-hour incubation and 2 hours of release at the pH 7 test. [Click here to view] |
Compared with supernatant after 1-hour incubation, there was a 1.34 mg/ml decrease. This indicated Minicell could package CQ. After 2 hours of incubating minicells-CQ in 1 × PBS (pH 7), a sample of supernatant was determined to have a CQ concentration of 0.517 mg/ml. The result showed that CQ from minicells that have already packaged CQ could be leaked. Moreover, in this test, a small amount of drug was released compared to the amount of drug that was packaged.
Functional group determination
FTIR analysis was applied to three samples to determine whether or not CQ could bind to the minicell, minicell incubated with CQ and pure CQ (Fig. 3). There were changes in peaks of minicells before and after CQ incubation in the IR spectrum. This proved that CQ was outside of minicell, which meant that CQ could be bound to the minicell cell wall. There were two noticeable changes in the IR spectrum between the three samples. First, the appearance of amine groups in sample minicells-CQ at the peak of around 3,438 cm−1 (%T = 53.42). Meanwhile, minicell alone exhibited a spectrum at 3,647.33 cm−1 (%T = 81.91) compared to the peak at around 3,712.45 cm−1 (%T = 59.85) of CQ
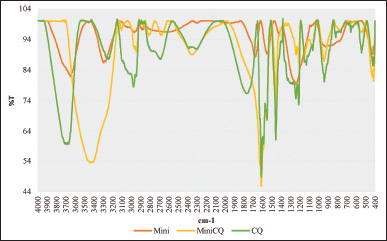 | Figure 3. FTIR spectrum of minicell, minicell containing CQ, and CQ [Click here to view] |
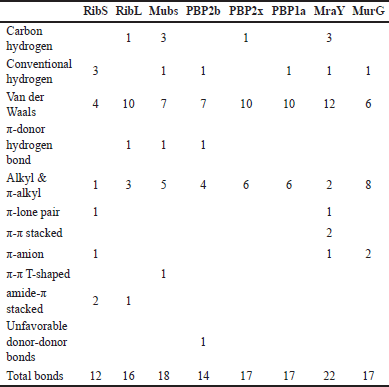 | Table 4. Number of interactions in the prediction between CQ and cell surface protein. [Click here to view] |
Moreover, after incubation, the aromatic ring (peak at around 1,600 cm−1) appeared in CQ and minicells containing CQ, while minicells did not exhibit it.
In silico experiment for CQ containing ability of L. acidophilus ATCC 4356 minicells
About 11 types of noncovalent bonds were displayed among eight proteins. Although noncovalent bonds were supposed to have weaker binding than covalent bonds, more than 10 amino acids involved in the interaction with more than 10 bonds could reinforce the point that CQ can bind to the minicell surface (Table 4).
Along with the FTIR analysis, molecular docking could be used as a reference to explain which interaction and bond might occur during the change in the functional group before and after drug incubation.
CONCLUSION
In this report, minicells from L. acidophilus ATCC 4356 have been successfully generated and contained CQ. Based on our in silico screening, minicells containing CQ can be modified to be used as a new targeted drug for cancer treatment. This research contributes to innovation in pharmaceutical science. More studies should be carried out to better understand drug delivery by L. acidophilus minicells.
AUTHOR CONTRIBUTIONS
Yen T.H. Tran and Tu H.K. Nguyen contributed to the study of conception and design. All authors contributed to performing research and collecting and analyzing data. All authors prepared the first draft of the manuscript. Critical revision of the manuscript was edited by Yen T.H. Tran and Tu H.K. Nguyen. Technical and material support was done by Tu H.K. Nguyen. All authors read and approved the final manuscript.
FUNDING
The study does not receive any financial support. The authors want to acknowledge the Lansium-Vietnam for their encouragement for conducting this research.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHERS NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. MacDiarmid JA, Mugridge NB, Weiss JC, Phillips L, Burn AL, Paulin RP, et al. Bacterially derived 400 nm particles for encapsulation and cancer cell targeting of chemotherapeutics, Cancer Cell. 2007;11:431–5. CrossRef
2. Flemming A. Minicells deliver lethal load to tumours. Nat Rev Drug Discov. 2007;6:519. CrossRef
3. Reeve JN, Mendelson NH, Coyne SI, Hallock LL, Cole RM. Minicells of Bacillus subtilis. J Bacteriol. 1973;114(2):860–73. CrossRef
4. Sheehy RJ, Allison DP, Curtiss III R. Cryptic plasmids in a minicell-producing strain of Salmonella typhimurium. J Bacteriol. 1973;114:439–2. CrossRef
5. Setlow JK, Boling ME, Allison DP, Beattie KL. Relationship between prophage induction and transformation in Haemophilus influenzae. J Bacteriol. 1973;115:153–61. CrossRef
6. Luan VK, Dung NA, Santa Romero J, Tu NHK. Tinidazole delivery improved by nanosized minicells originated from Leuconostoc mesenteroides, J Nanomaterials. 2019;2019:7684795, 9. CrossRef
7. Jaffe A, Dari R, Hiraga S. Minicell-forming mutants of Escherichia coli: production of minicells and anucleate rods. J Bacteriol. 1988;170:3094–101. CrossRef
8. Nguyen HKT, Doan TTV. Effects of carbon sources on cell differentiation of Lactobacillus rhamnosus PN04 and applications. Biomed Pharmacol J. 2015;6(2):197–203. CrossRef
9. Gilliland SE, Speck ML. Antagonistic action of Lactobacillus acidophilus toward intestinal and foodborne pathogens in associative cultures (1). J Food Prot. 1977;40:820–3. CrossRef
10. Galdeano LE, Rossi LA, Santos CB, Dantas RA. Nursing diagnosis in the perioperative period of cardiac surgery. Rev Esc Enferm USP. 2006;40:26–33. CrossRef
11. Moeini H, Rahim RA, Omar AR, Shafee N, Yusoff K. Lactobacillus acidophilus as a live vehicle for oral immunization against chicken anemia virus. Appl Microbiol Biotechnol. 2011;90:77–88. CrossRef
12. Vinh DT, Khue NT. Study on minicell generation of Lactobacillus acidophilus VTCC-B-871 for drug delivery. J Appl Pharm Sci. 2019;3:33–6.
13. PubChem Compound Summary for CID 2719. Chloroquine. 2021 [cited 2021 Jan 9]. Available from: https://pubchem.ncbi.nlm.nih.gov/compound/Chloroquine
14. Homewood CA, Warhurst DC, Peters W, Baggaley VC. Lysosomes, pH and the anti-malarial action of chloroquine. Nature. 1972;235:50–2. CrossRef
15. Kim EL, Wüstenberg R, Rübsam A, Schmitz-Salue C, Warnecke G, Bücker EM, et al. Chloroquine activates the p53 pathway and induces apoptosis in human glioma cells, Neuro Oncol. 2010;12:389–400. CrossRef
16. Vezmar M, Georges E. Reversal of MRP-mediated doxorubicin resistance with quinoline-based drugs. Biochem Pharmacol. 2000;59:1245–52. CrossRef
17. Miranda TA, Silva PH, Pianetti GA, César IC. Simultaneous quantitation of chloroquine and primaquine by UPLC-DAD and comparison with a HPLC-DAD method. Malar J. 2015;14:29. CrossRef
18. Abdelsattar AS, Dawoud A, Helal MA. Interaction of nanoparticles with biological macromolecules: a review of molecular docking studies. Nanotoxicology. 2021;15(1):66–95. CrossRef
19. Whelan F, Lafita A, Griffiths SC, Cooper REM, Whittingham JL, Turkenburg JP, et al. Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection. Proc Natl Acad Sci U S A. 2019;116(52):26540–8. CrossRef
20. de Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of Lactobacilli. J Appl Bacterial. 1960;23:130–5. CrossRef
21. Schulz GE. Bacterial porins: structure and function. Curr Opin Cell Biol. 1993;5:701–7. CrossRef
22. Graeme-Cook KA, May G, Bremer E, Higgins CF. Osmotic regulation of porin expression: a role for DNA supercoiling. Mol Microbiol. 1989;3:1287–94. CrossRef
23. van den Berg, B. The FadL family: unusual transporters for unusual substrates. Curr Opin Struct Biol. 2005;15:401–7. CrossRef
24. Al-Bari MA. Chloroquine analogues in drug discovery: new directions of uses, mechanisms of actions and toxic manifestations from malaria to multifarious diseases. J Antimicrob Chemother. 2015;70:1608–21. CrossRef
25. Delcour J, Ferain T, Deghorain M, Palumbo E, Hols P. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie Van Leeuwenhoek. 1999;76:159–84. CrossRef
27. Schär-Zammaretti P, Ubbink J. The cell wall of lactic acid bacteria: surface constituents and macromolecular conformations. Biophys J. 2003;85(6):4076–92. CrossRef
28. Dean SN, Leary DH, Sullivan CJ, Oh E, Walper SA. Isolation and characterization of Lactobacillus-derived membrane vesicles. Sci Rep. 2019;9:877. CrossRef