INTRODUCTION
The epidemiology and etiology of liver injury vary among the population of different countries. The risk of drug-induced liver damage is low in the general population but higher in hospitalized patients and liver cirrhosis is often treated with pharmaceuticals, nutritional supplements, and botanical medicines. Recent evidence showed that oxidative stress has been linked to the major pathogenic mechanism of chemical-induced liver injury resulting in various disorders such as hepatotoxicity, cancer, diabetes, hypertension, various neurological disorders, etc. [1]. Hepatotoxicity is usually caused by a viral infection, excess alcohol consumption, non-alcoholic fatty liver disease (NAFLD), and drug toxicity [2]. Among the modulated genes in liver disease, Liver X receptors (LXRs) is a nuclear receptor that regulates lipid equilibrium and cytoplasmic cholesterol homeostasis. They have a key regulatory role in the immune system including de novo lipogenesis and inflammation. The binding of synthetic cholesterol derivatives, endogenous and certain oxysterols to LXR are widely recognized as the biologically active LXR ligands [3]. Two isoforms of LXR viz., NR1H3 and NR1H2, in which NR1H3 is highly expressed in liver tissue, with a lower level in the kidney, spleen, adipose tissue, and intestine [4,5]. The promoter region in LXRα conforms to the regulation of LXRα and LXRβ. Specific oxysterols or synthetic ligands modulate both LXR isoforms [6,7]. Expanding knowledge of LXR function in fatty acid and cholesterol homeostasis has been considered a major precursor from the pharmaceutical point of view.
Isoniazid (INH) is considered a first-line drug for the treatment of tuberculosis. Several cases of INH-induced hepatotoxicity have been detected in different patients. INH is metabolized in the liver by N-acetyl transferase 2 into acetyl-INH. A number of secondary metabolites, including monoacetyl hydrazine (MAH) and diacetyl hydrazine are produced during the metabolism of acetyl-INH. A covalent connection between acetyl hydrazine and liver macromolecules and the formation of free radicals from reactive metabolites of MAH have both been hypothesized as mechanisms by which INH metabolites may impact the rate of INH-induced hepatotoxicity.
To combat the drug-induced hepatotoxicity includes the utilization of antioxidant nutraceuticals to target liver in?ammation triggered by oxidative stress. From the plants, secondary metabolites have eminent pharmacological activity for the management of multiple ailments such as diabetes, hypertension, cancer, hepatotoxicity, allergy, etc., and these secondary metabolites protect the cells from free radicals, and thus they act as free radical scavenging activity. Recently researchers are more focussing on fruit materials. Which are considered a rich source of natural antioxidants, plant secondary metabolites such as phytosterols, terpenoids (carotenoids), polyphenols (phenolic acids, flavonoids). Cucurbita pepo seeds (C. pepo L.) are a rich source of unsaturated antioxidants, fatty acids, and fibers and known to have hepatoprotective and anti-atherogenic activities. Cucurbita pepo is widely used as functional food or medicine because of a rich source of carotenoids, polyunsaturated fatty acids. It has high contents of alkaloids, flavonoids, terpenoids, proteins, lipids, carbohydrates, minerals, and phytosterols. In general, hepatoprotective plants contains a variety of phytocompounds, such as phenols, glycosides, alkaloids, flavonoids, carotenoids coumarins, steroids, lignans, essential oils, monoterpenes, organic acids, carotenoids, xanthines, triterpenoids, etc. As such, on the basis of the traditional use, the present investigation was carried out to evaluate the molecular mechanism of C. pepo by network pharmacology approach supported by in vivo hepatoprotective studies.
MATERIALS AND METHODS
Computational approach
Retrieval of phytocompounds and proteins involved in liver disease
The reported phytocompounds from C. pepo were collected from phytochemical databases, and PubChem was employed to retrieve structural data on those compounds from several open-source libraries. The potential probable protein targets of phytocompounds for liver cirrhosis were then identified. Throughout the dataset’s construction, duplicate phytochemicals and proteins related to liver cirrhosis were removed. PubChem database was employed to retrieve each phytocompound’s canonical simplified molecular-input line-entry system (SMILES) [8]. SMILES were queried in SwissTargetPrediction [9] for the target prediction at a percentage similarity of >0.1%. With respect to the existing targets of liver disorders included in the therapeutic target database, the proteins implicated in liver cirrhosis were located [10]. Each protein identified as the cause of liver cirrhosis has its gene ID acquired from UniProt [11] and phytocompounds acting on liver targets were identified.
G-Profiler web server
The study of biological data frequently involves looking at the number of genes associated with different diseases. G: Profiler software server is frequently used to determine biological categories that are enriched in gene lists (https://biit.cs.ut.ee/profiler). GOSt uses g: Convert to map gene/protein IDs across different namespaces, g: Orth to map orthologous genes across species, and g: SNPense to map human single nucleotide polymorphisms identifiers to genes to perform the enrichment analysis of single or multiple gene lists [12].
Set of pathway and network analysis of C. pepo
STRING database was used to identify a set of proteins involved in liver disease [13] and enrichment analysis of gene was employed to infer the related pathways that are altered by the phytocompound. Detailed examination of the pathways implicated in liver pathogenesis was found using the KEGG database. The network linking phytocompounds, protein molecules, and identified pathways was built using Cytoscape 3.7.2 [14].
Docking studies
Protein retrieval and preparation
PubChem online database was utilized to retrieve the 3D structure of all phytocompounds and minimized using MMFF94 forcefield [15]. A literature search and Research Collaboratory for Structural Bioinformatics database [https://www.rcsb.org/] were used to identify the protein and Protein Data Bank ID for NR1H3 protein (1UHL) and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) protein (2R4F). Protein was prepared via protein preparation wizard panel of Schrodinger’s (2021–2), where water beyond 5 ? from Het groups was removed, zero bond order to metals was created, polar hydrogen was assigned Methionine residues were restored, and Restrain energy minimization was carried out using optimized potential for liquid simulation (OPLS3e) force field [16,17].
Ligand preparation
The ligprep panel of Schrodinger’s (2021-2) was used to generate possible states of ligands at the target pH using the Epik module. Chirality was retained for the selected ligand and at most one stereoisomer was generated for the given ligand, finally, energy was minimized using the OPLS3e force field [18].
Grid generation
The glide module of Schrodinger containing a receptor grid generation panel was utilized to create a grid at the binding site. The site of the co-crystal ligand was considered a binding site and a grid was generated.
Ligand docking
The glide module of Schrodinger’s was utilized for ligand docking and Maestro 12.3 was used to visualize the binding interactions. Extra precision mode for docking was carried out to understand the binding energy and orientation of ligands within the binding pocket. To visualize the interaction between the ligand and the protein following molecular docking, the posture scoring the lowest BE was chosen. The nuclear receptor superfamily’s NR1 subfamily includes the NR1H3 protein, which is encoded by the gene. NR1 family members are the primary modulators, controlling transcriptional regulation in the maintenance of lipid homeostasis and inflammation. As demonstrated, experimental research in mice lacking this gene suggests that it may be crucial for controlling cholesterol homeostasis [19].
Molecular dynamics
To evaluate the stability of complex formation, a 30 ns molecular dynamics (MD) simulation was performed on ligand-protein complexes using the Desmond software. Using the simple point charge water model, the system in a cubical box with 10 Å × 10 Å × 10 Å periodic boundary conditions was solvated. Na+/Cl- counter ions were also added, which further neutralized the solution. Bond angles and lengths of heavy atoms geometry were restrained by the SHAKE algorithm. Long-range interactions were calculated by “particle mesh Ewald” method. The cut-off value for “Lennard-Jones interactions” was set at 10.0 Å. Additionally, the system was loosened up or minimized using default settings. A Nosé-Hoover chain thermostat with a 1.0 ps relaxation time and the Martyana-Tobias-Klein barostat method with a 2.0 ps relaxation time were then used for the final 30 ns of the production run, with a Coulombic short range cutoff radius set to 9.0 Å and the temperature and pressure being set to 300 K and 1.01325 bar, respectively. To infer the structural stability, root-mean-square fluctuation (RMSF) of residues, root-mean-square deviation (RMSD) of complex, and protein radius of gyration were analyzed. In order to infer stable interactions within the ligand—protein throughout MD simulation, the ligand-protein percentage fraction interactions were analyzed during 30 ns simulation.
In vitro studies
Collection and authentication of C. pepo
The ripe fruits of C. pepo were purchased locally in June month of 2020 from the local area of Belagavi region, Karnataka then plant seeds were authenticated by Central Research Facility B.M.K. Ayurveda Mahavidyalaya, Belagavi. With voucher specimen number CRF/Auth./2020/1 and CRF/Auth./2020/2.
Preparation of plant extract and fraction
The C. pepo fruits were peeled, the seeds were carefully hand-removed from the pulp, cleaned, rinsed, and the damaged seeds were separated. The remaining seeds were then dried in an oven at 40°C for 24 hours. Dried seeds were then ground in an electrical grinder. 70% v/v ethanol was used as a solvent for the maceration of dried seeds and 5% chloroform (preservative) for 72 hours. The extract was filtered and the raw material that remained in the maceration was subjected to soxhlation (70% v/v ethanol). Filtrates of maceration and soxhlation were concentrated at 40°C under reduced pressure using a rotary evaporator which yield total extract. Finally, the extract was subjected to fractionation. Fractionation was performed as explained by Cos et al. [20]. The hydroalcoholic extract was dissolved in water and the dichloromethane mixture and dichloromethane fraction were further mixed with methanol (90%) to obtain the sterols fraction.
Yield of the extract (%) = (weight of extract/weight of dried seeds) × 100. The percentage yield of extract/ 100 g of powder was found to be 14% and the percentage yield of fraction using 20 g of hydroalcoholic extract of C. pepo was found to be 4%.
Liquid chromatography-mass spectrometry (LC-MS) profile of the hydroalcoholic extract of C. pepo
When using the LC-MS 2010A (Shimadzu Japan) to run the sample, the following requirements were upheld. Methanol and water were employed in a 90:10 v/v ratio as the mobile phase, with a flow rate of 200μl/minute. The C18 column served as the stationary phase. The sample was injected (μl) after being dissolved in the mobile phase. The chemicals in the sample were identified using the electrospray ionization peaks [21].
Assessment of hepatoprotective activity of C. pepo by in vivo
The Institutional Animal Ethics Committee (Reg. No. 221/Po/Re/S/2000/CPCSEA at its meeting date of 21/01/2022) was evaluated the protocol of the study. Albino-Wistar rats weighing 160 to 180 g were housed in standard laboratory conditions of temperature (25°C ± 2°C), 12 hours in light and dark places, and with food and water ad libitum. INH (50 mg/kg p.o) was administered daily for 28 days in drug-induced liver toxicity model, to all animals except group 1. Silymarin (50 mg/kg, p.o) was used as standard. The animals were segregated into seven groups of six each as follows [22]. Group-I: received food and water. Group-II: (control) received INH (50 mg/kg) p.o. up to 28 days. Group III: received Silymarin 50 mg/kg orally and INH (50 mg/kg) p.o. up to 28 days. Group IV: received 500 mg/kg of C. pepo extract and INH (50 mg/kg). Group V: received 50 mg/kg of C. pepo steroid fraction and INH (50 mg/kg). Under ether anaesthesia, blood was collected from each rat via the retro-orbital plexus for biochemical analysis, including aspartate aminotransferase (AST), alanine transaminase (ALT), alkaline phosphatase (ALP), bilirubin estimate, high-density lipoprotein (HDL) low density lipoprotein (LDL), very low density lipoprotein (VLDL), triglyceride (TG) and total cholesterol (TC). The blood serum was separated by centrifugation at 2,500 rpm for 15 minutes after the blood had been allowed to coagulate at 37°C for 30 minutes. The animals were euthanized and the liver was promptly removed and processed for histological analysis.
Statistical analysis
Network analysis results were expressed in gene count and false discovery rate. Docking binding energy is expressed in kcal/mol. During MD analysis, RMSD and RMSF were considered for evaluation. In vivo experiments results were expressed as Mean ± SEM and the difference among the mean was evaluated using one-way ANOVA followed by Tukey’s test, using GraphPad Prism version 8.0.
RESULTS
In silico studies
Mining of phytoconstituents and their target prediction
135 phytoconstituents from C. pepo were mined from literature and public phytochemical databases. These 135 phytocompounds were predicted to be 408 protein molecules in the SwissTargetPrediction server.
Gene enrichment analysis of network
The gene set enrichment analysis of 408 targets revealed 26 targets that are potential to modulate 11 molecular pathways associated with liver cirrhosis. Out of 135 compounds, 18 compounds were found to target these 26 targets. These phytoconstituents majorly are of steroids. Among 11 pathways (Fig. 1) targeted by phytocompounds, Metabolic pathways, peroxisome proliferator-activated receptors (PPAR) signaling pathway, and insulin resistance are directly linked in the pathogenesis of liver disease against INH-induced hepatotoxicity (Table 1).
Druggability (absorption, distribution, metabolism and excretion, ADME) and side effects of compounds
Except (+)-Dehydrovomifoliol, Clerosterol, Codisterol, Isofucosterol, Cucurbitacin-D, Cucurbitacin-I, 22-Dihydrobrassicasterol the potential adverse consequences of the remaining phytosterols were projected (Table 2). The likelihood of phytosterol side effects is mostly nephrotoxicity with Pa 0.5, which indicates the least probability to induce the impact. The ADME profile of each phytosterol such as GI absorption, blood–brain barrier (BBB) permeation, P-gp substrate, CYP1A2 inhibitor, CYP2C19inhibitor, CYP2C9inhibitor, CYP2D6inhibitor, CYP3A4 inhibitor, LogKp (skin permeation) by Swiss ADME are mentioned in (Table 3)
Docking studies
The binding energy and mode of interaction of each phytocompounds against NR1H3 and HMGCR receptors are summarized in Tables 4 and 5. Among these Schottenol has the highest H-bond interaction with NR1H3 i.e. 24 viz Hie421, Gln424, Val425, Leu428, Leu439, Phe234, Phe257, Thr238, Leu260, Ala261, Ser264, Ile339, Phe326, Phe335, Leu331, Arg305, Thr302, Glh301, Met298, Ile295, Trp443, Thr314, Ile313, Tyr321 (Fig. 2) and Alpha-Spinasterol has the highest H-bond interaction with HMGCR i.e. 10 interactions viz., Lys691, Asp690, Met655, Met657, Asn658, Ser661, Lys662, Glu665, Arg590, Val683 in Figure 3.
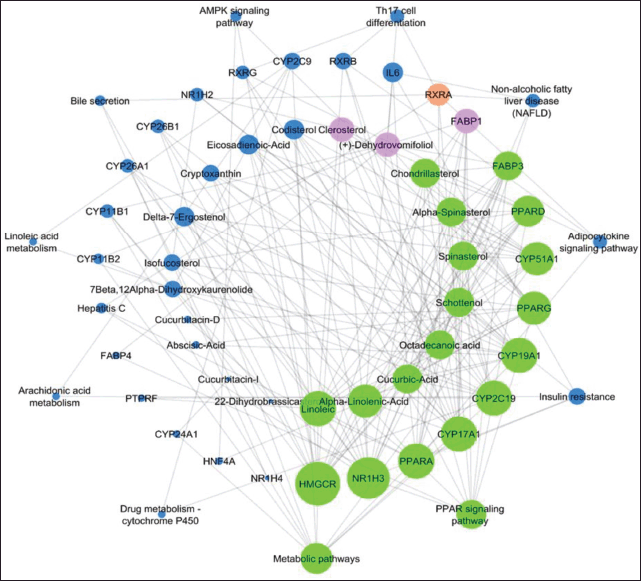 | Figure 1. Network analysis of the interaction between phytocompounds, modulated targets, and pathways involved in the pathogenesis of liver. Larger size nodes and higher edges represent highly interconnected compound, proteins, and pathways. While small seize nodes and edges represent, less modulated phytocompounds, proteins, and pathways. [Click here to view] |
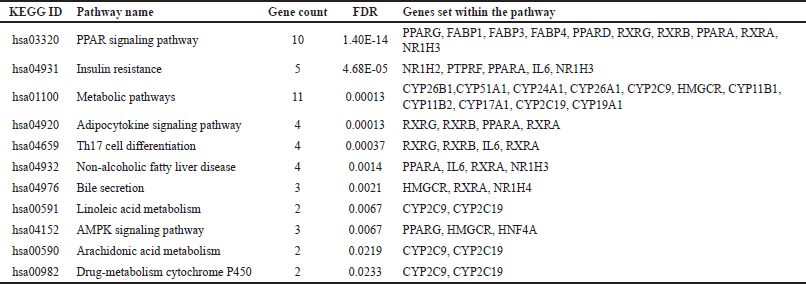 | Table 1. Pathways modulated by C. pepo compounds. [Click here to view] |
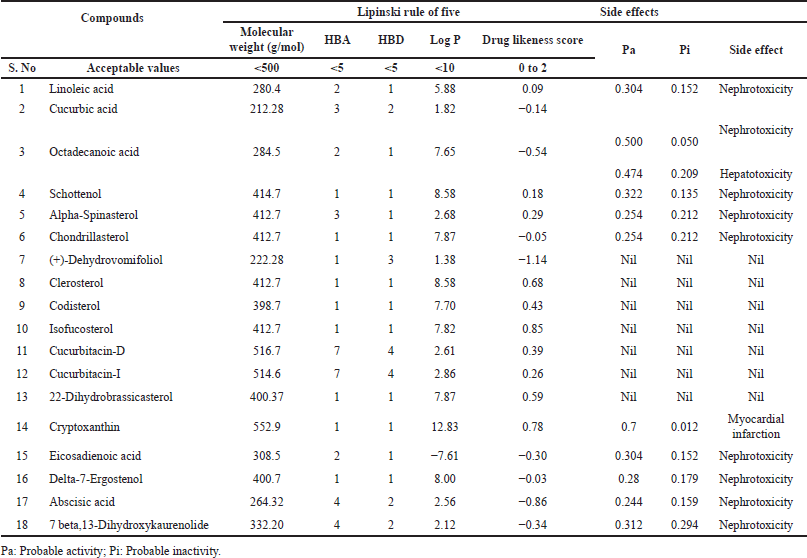 | Table 2. Druggability and rule of five drug-likeness score of phytocompounds. [Click here to view] |
Molecular dynamics
Stability of Schottenol with NR1H3
The RMSD of both backbone and complex was found to be stable throughout 30 ns MD simulation (Fig. 4). The average RMSD of the backbone was found to be ~2.0 Å and the complex was ~2.2 Å. The residues involving the ligand interaction, i.e., Phe329, Phe245, Leu274, Phe271, Ala275, Trp457, Met312 showed the least fluctuation compared to the N- and C- terminal residues. The compound “Schottenol” was found to interact with Phe329, Phe245, Leu274, Phe271, and Ala275, Trp457, and Met312 of NR1H3 throughout 30ns MD simulation.
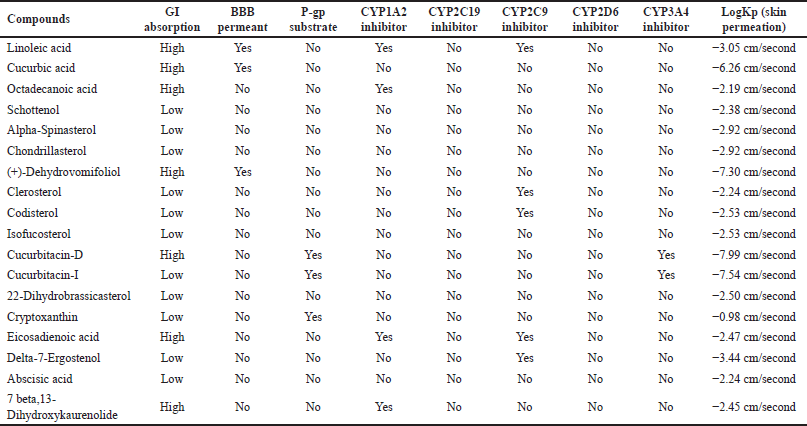 | Table 3. The ADME profile of each phytocompounds of C. pepo by Swiss ADME. [Click here to view] |
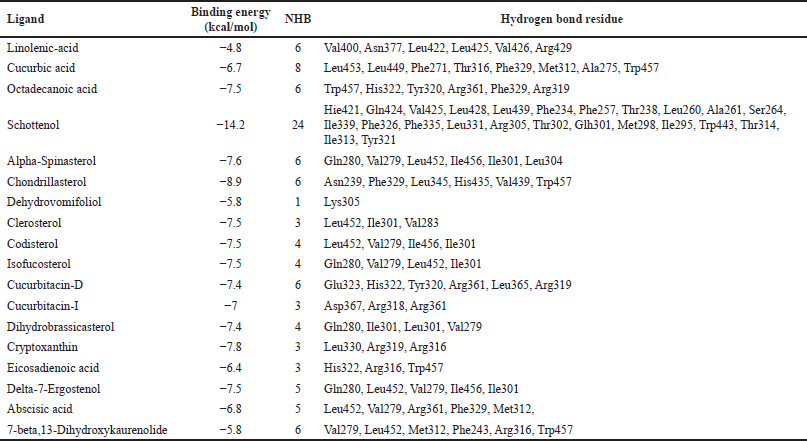 | Table 4. Binding energy and mode of interaction of each phytocompounds against NR1H3. [Click here to view] |
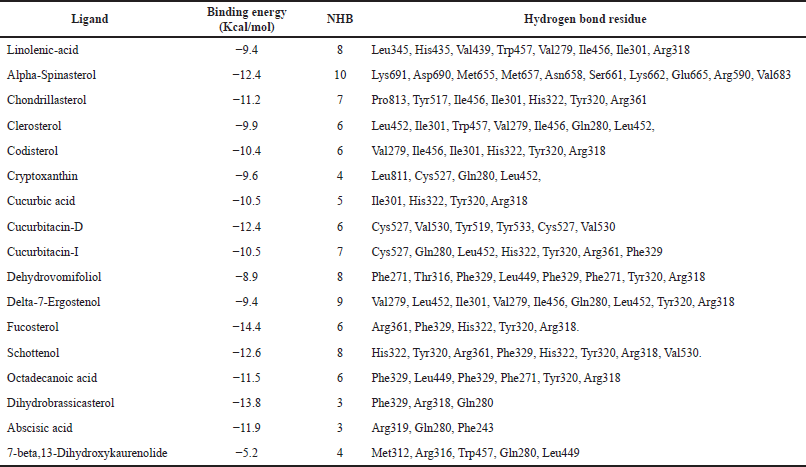 | Table 5. Binding energy and mode of interaction of each phytocompounds against HMGCR. [Click here to view] |
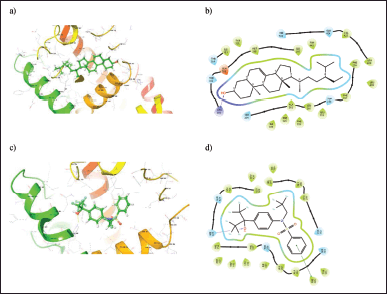 | Figure 2. (a) 3D and (b) 2D binding orientation of Schottenol ligand in NR1H3 binding pocket. (c) 3D and (d) 2D binding orientation of cocrystal ligand in NR1H3 binding pocket. [Click here to view] |
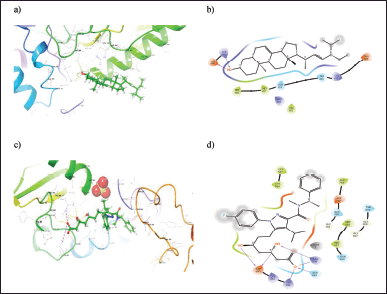 | Figure 3. (a) 3D and (b) 2D binding orientation of Alpha-Spinasterol ligand in HMGCR binding pocket. (c) 3D and (d) 2D binding orientation of cocrystal ligand in HMGCR binding pocket. [Click here to view] |
Experimental wet lab studies
Cucurbita pepo phytocompounds profile
Physico-chemical investigations of C. pepo were represented in Table S1. The phytochemical screening of C. pepo showed the presence of alkaloids, glycosides, saponins, steroids, tannins, and flavonoids (Table S2). LC-MS profile showed alpha-linolenic-acid, alpha-spinasterol, chondrillasterol clerosterol, codisterol, isofucosterol, dihydrobrassicasterol, and cryptoxanthin (Table S3 and Fig. S1).
In vivo experimental validation
The hepatoprotective effect of hydroalcoholic extract and steroid fraction of C. pepo studied on INH (50 mg/kg, p.o) induced hepatotoxicity in Wistar rats. Initially, body weight and liver weight of the animals were examined. As a result, INH showed no significant effect on the rat body weight compared to normal and treated groups. However, a significant increase in the liver weight was observed in INH treated group and a slight decrease in the liver weight after treatment with Silymarin and C. pepo (Table S4). Hepatoprotective activity was monitored by estimating the serum transaminases such as ALP, serum bilirubin, and lipid profile in the livers of experimental rats. Pre-treatment of rats with different concentrations of test drugs and standard leads to normalization of INH-induced changes in the levels of all the biochemical parameters (Fig. 5 and Table S5). Further, the effect of C. pepo on cholesterol also revealed normalization of their level, in which enriched fraction was found to reduce cholesterol levels significantly compared to INH group (Fig. 6 and Table S6). According to the current investigation, fraction was shown to be more efficient than extract at preventing liver damage brought on by INH-induced oxidative stress. The results of this study give evidence in favor of the therapeutic usefulness of medications that can be used to treat liver damage caused by INH. In comparison to the extract, the bioactive directed fraction exhibits superior impact.
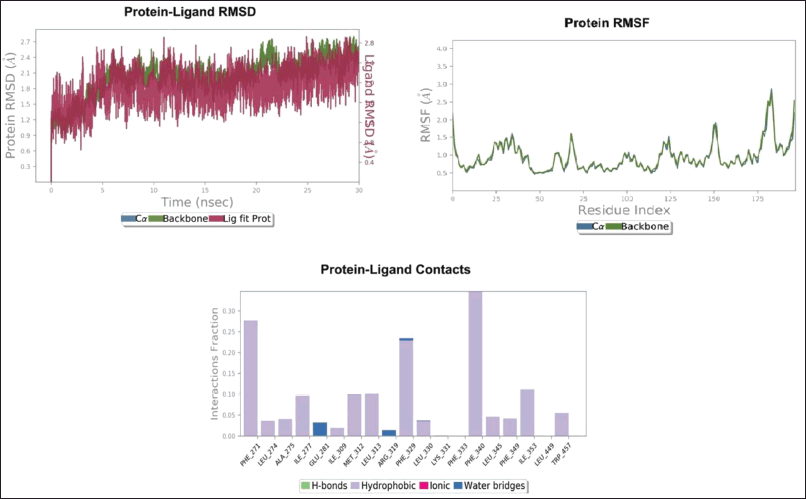 | Figure 4. Stability of Schottenol with NR1H3. [Click here to view] |
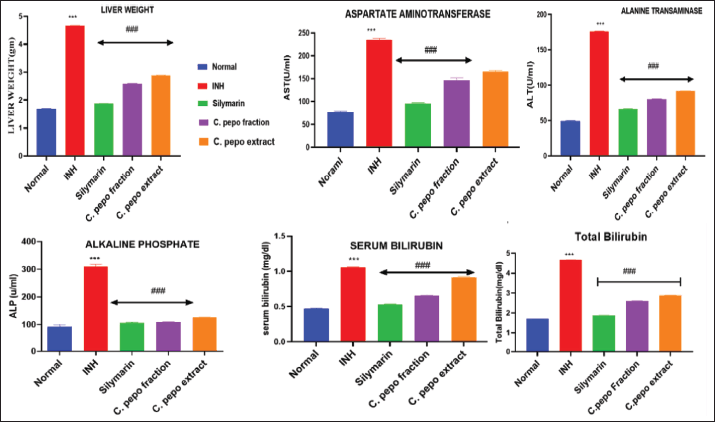 | Figure 5. Effect of C. pepo on liver weight, AST, ALT, ALP, serum bilirubin, and total bilirubin. Data are presented as Mean ± SEM (n = 6), one way ANOVA followed by Tukey’s test was applied. *p < 0.05, **p < 0.01, ***p < 0.001 compared with normal group, #p < 0.05, ##p < 0.01, ###p < 0.001 compared with INH treated group. [Click here to view] |
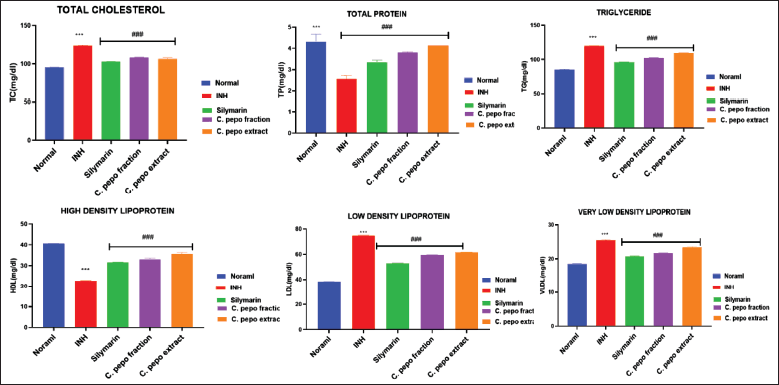 | Figure 6. Effect of C. pepo on total cholesterol, total protein, TG, HDL, LDL, and VLDL. Data are presented as Mean ± SEM (n = 6), one way ANOVA followed by Tukey’s test was applied. *p < 0.05, **p < 0.01, ***p < 0.001 compared with normal group, #p < 0.05, ##p < 0.01, ###p < 0.001 compared with INH treated group. [Click here to view] |
DISCUSSION
The present work described phytosterols from C. pepo can be preferred as liver protective agents in liver cirrhosis by preventing oxidative damage and inflammation caused by AntiTB drugs. Claims from the traditional system and the available literature, seeds of the C. pepo plant are used to treat liver disease. However, there is a lack of data on the molecular mechanism of C. pepo for the treatment of liver illness. Hence, the present study was designed to identify the potential bioactives and their molecular mechanisms for hepatoprotective activity. This study will extend from In silico research to wet-lab evaluations to determine the hepatoprotective activity of the secondary metabolites from C. pepo. LXR is a major nuclear regulatory transcription gene of cholesterol sensing in liver. Once it gets activated, the other genes are highly regulated by binding to different cholesterol molecules and play a significant role in inflammation, immune regulation, lipid, and glucose metabolism. Some selective LXR activators such as desmosterol [23], the intestine-specific ligand GW6340, and the LXR-623, show adverse effects. Oxysterols and intermediates from the cholesterol derivatives [24] activate NR1H3 signaling. The other enzyme, HMG-CoA reductase is the rate-controlling enzyme of the mevalonate pathway in cholesterol synthesis and its regulation is via a negative feedback mechanism by sterols and non-sterol isoprenoids derived from mevalonate, and the reaction is catalyzed by reductase. Cholesterol derivatives are major suppressors of this enzyme in mammalian cells and are responsible for the degradation of LDL via the LDL receptor.
Phytocompounds–target network was constructed through network pharmacology and analyzed by molecular docking. The underlying mechanism of network-predicted hepatoprotective effect of C. pepo is a novel approach for the management of liver disease against INH-induced hepatotoxicity. This research showed that the biological network method was more effective in illuminating the connections between illnesses and phytocompounds.
In the current study, we relied on a plethora of publicly available data sources to mine the phytocompounds present in the C. pepo and the PASS server was used to pick the compounds with Pa >0.5, or 50%. Based on the constructed network, we selected the most highly modulated compounds from C. pepo and observed their interactions with highly modulated proteins. In docking, the lead hit molecule is determined by the three key points one) BE, which reflects binding affinity, 2) the number of H-bond interactions, and 3) the H-bond residues.
Oxidative stress, mitochondrial dysfunction. LDH leakage and apoptosis are critical factors in drug-induced hepatotoxicity and occurs due to the activation of the reactive oxygen species system and metal ions transition, which modify homeostatic proteins is one prime basis for the pathogenesis of INH-induced hepatotoxicity [25]. Phytosterols from C. pepo provide nutrition to the hepatocytes and contain a high amount of unsaturated fatty acids, a hydrogen-donating property that can stabilize and delocalize unpaired electrons by creating a hydrogen bond with free radicals and stopping the Fenton reaction. Finally, we constructed the network interaction between phytosterols of C. pepo their targets and probable pathways. The result reflects that phytocompounds mainly phytosteroids to regulate the fatty acids interactions with the multiple protein molecules involved in the pathogenesis of liver within the network.
CONCLUSION
The present research proposes that, C. pepo steroidal fraction may be utilized as a hepatoprotective agent. The presence of secondary metabolites in this fraction may operate synergistically to have a potential therapeutic impact by targeting multiple proteins and modulating different pathways implicated in liver pathogenesis by reducing oxidative stress and cytotoxicity. This research provides strong in silico support for in vivo experiments investigating INH-induced hepatotoxicity. More specifically, it provides direction for promoting clinical studies involving human participants.
ACKNOWLEDGEMENTS
The authors would like to thank the Principal, KLE College of Pharmacy, Belagavi, KLE Academy of Higher Education and Research (KAHER), Belagavi, for providing support to carry out the research.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
The Institutional Animal Ethics Committee (Reg. No. 221/Po/Re/S/2000/CPCSEA at its meeting date of 21/01/2022) was approved the protocol of the study.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Souza CF, Baldissera MD, Descovi SN, Zeppenfeld CC, Garzon LR, da Silva AS, et al. Serum and hepatic oxidative damage induced by a diet contaminated with fungal mycotoxin in freshwater silver catfish Rhamdia quelen: involvement on disease pathogenesis. Microb Pathog. 2018;124:82–8. doi: https://doi.org/10.1016/j.micpath.2018.08.041
2. Nirmala M, Girija K, Lakshman K, Divya T. Hepatoprotective activity of Musa paradisiaca on experimental animal models. Asian Pac J Trop Biomed. 2012;2:11–5. doi: https://doi.org/10.1016/S2221-1691(11)60181-0
3. Fu X, Menke JG, Chen Y, Zhou G, MacNaul KL, Wright SD, et al. 27-hydroxycholesterol is an endogenous ligand for liver X receptor in cholesterol-loaded cells. J Biol Chem. 2001;276:38378–87. doi: https://doi.org/10.1074/jbc.M105805200
4. Apfel R, Benbrook D, Lernhardt E, Ortiz MA, Salbert G, Pfahl M. A novel orphan receptor specific for a subset of thyroid hormone-responsive elements and its interaction with the retinoid/thyroid hormone receptor subfamily. Mol Cell Biol. 1994;14:7025–35. doi: https://doi.org/10.1128/mcb.14.10.7025-7035.1994
5. Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–45. doi: https://doi.org/10.1101/gad.9.9.1033
6. Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXRα. Nature. 1996;383:728–31. doi: https://doi.org/10.1038/383728a0
7. Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, et al. Structural requirements of ligands for the oxysterol liver X receptors LXRα and LXRβ. Proc Natl Acad Sci. 1999;96:266–71. doi: https://doi.org/10.1073/pnas.96.1.266
8. Kim S, Thiessen PA, Bolton BB, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;4:D1202–13. doi: https://doi.org/10.1093/nar/gkv951
9. Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. Swiss target prediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;1:W32–8.
10. Li YH, Yu CY, Li XX, Zhang P, Tang J, Yang Q, et al. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018;4:D1121–7. doi: https://doi.org/10.1093/nar/gkx1076
11. Apweiler R, Bairoch A, Wu CT. The universal protein resource (UniProt). Nucleic Acids Res. 2007;36:D190–5. doi: https://doi.org/10.1093/nar/gki070
12. Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, et al. g: profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019;47:W191–8. doi: https://doi.org/10.1093/nar/gkz369
13. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016:gkw937. doi: https://doi.org/10.1093/nar/gkw937
14. Tubachi SS, Rasal VP, Ugare SR, Khatib NA, Ojha PS, Patil VS. Evaluation of Ylang Ylang essential oil on alcohol induced hepatotoxicity in rats. Adv Trad Med. 2022;1:2381. doi: https://doi.org/10.1007/s13596-022-00630-w
15. Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17:490–519. doi: https://doi.org/10.1002/(sici)1096-987x(199604)17:5/6<490::aid-jcc1>3.0.co;2-p
16. Svensson S, Östberg T, Jacobsson M, Norström C, Stefansson K, Hallén D, et al. Crystal structure of the heterodimeric complex of LXRα and RXRβ ligand-binding domains in a fully agonistic conformation. EMBO J. 2003;22:4625–33. doi: https://doi.org/10.1093/emboj/cdg456
17. Pfefferkorn JA, Choi C, Larsen SD, Auerbach B, Hutchings R, Park W, et al. Substituted pyrazoles as hepatoselective HMG-CoA reductase inhibitors: discovery of (3 R, 5 R)-7-2-(4-Fluoro-phenyl)-4-isopropyl-5-(4-methyl-benzylcarbamoyl)-2 H-pyrazol-3-yl]-3, 5-dihydroxyheptanoic acid (PF-3052334) as a candidate for the treatment of hypercholesterolemia. J Med Chem. 2008;51:31–45. doi: https://doi.org/10.1021/jm070849r
18. David TI, Adelakun NS, Omotuyi OI, Metibemu DS, Ekun OE, Inyang OK, et al. Molecular docking analysis of phyto-constituents from Cannabis sativa with pfDHFR. Bioinformation. 2018;14:574. doi: https://doi.org/10.6026/97320630014574
19. Zhang Z, Burch PE, Cooney AJ, Lanz RB, Pereira FA, Wu J, et al. Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome. Genome Res. 2004;14:580–90. doi: https://doi.org/10.1101/gr.2160004
20. Cos P, Vlietinck AJ, Berghe DV, Maes L. Anti-infective potential of natural products: how to develop a stronger in vitro ‘proof-of-concept’. J Ethnopharmacol. 2006;106:290–302. doi: https://doi.org/10.1016/j.jep.2006.04.003
21. Parekh JM, Sutariya DK, Vaghela RN, Sanyal M, Yadav M, Shrivastav PS. Sensitive, selective and rapid determination of bupropion and its major active metabolite, hydroxybupropion, in human plasma by LC-MS/MS: application to a bioequivalence study in healthy Indian subjects. Biomed Chromatogr. 2012;26:314–26. doi: https://doi.org/10.1002/bmc.1660
22. Patel J, Reddy V, Kumar GS, Prasad J, Krishna S. Pharmacognostic study of some important hepatoprotective plants. J Pharmacogn Phytochem. 2016;5:75–9.
23. Fessler MB. The challenges and promise of targeting the liver X receptors for treatment of inflammatory disease. Pharmacol Ther. 2018;181:1–2. doi: https://doi.org/10.1016/j.pharmthera.2017.07.010
24. Olkkonen VM, Béaslas O, Nissilä E. Oxysterols and their cellular effectors. Biomolecules. 2012;2:76–103. doi: https://doi.org/10.3390/biom2010076
25. Uttara B, Singh AV, Zamboni P, Mahajan R. Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Curr Neuropharmacol. 2009;7:65–74. doi: https://doi.org/10.2174/157015909787602823
SUPPLEMENTARY MATERIAL
Supplementary data can be downloaded from the journal’s website