INTRODUCTION
Among cancer, the third most common cancer worldwide is cancer of the colon with an incidence rate and mortality rate of 6.1% and 9.2% when combined for both sexes, respectively (Bray et al., 2018). Although there are several chemotherapies and surgeries for colon cancer patients, the rate of recurrence after surgery is high (Hellinger and Santiago, 2006). Thus, exploration of an effective clinical treatment for the treatment of colon cancer is essential. It is seen that herbal phytoconstituents have been broadly used for the treatment of various tumors such as colon carcinoma (Benarba and Pandiella, 2018). Chrysin is a herbal phytoconstituent that is found to have anti-inflammatory activity (Nunes et al., 2020) by effectively impairing cisplatin-influenced expression for iNOS and COX-2 (Rehman et al., 2014), neuroprotective activity by triggering nuclear factor-kB and inducing the expression of nitric oxide synthase (Zhang et al., 2015), and antidiabetic activity by significantly inhibiting advanced glycation end products-receptor for advanced glycation end products conciliated oxidative stress, activation of PPAR-g resulting into inflammation (Rani et al., 2016), and antidepressant activity by culmination in the uplift of nerve growth factor and BDNF levels (Filho et al., 2015). Reviews of previously published scientific literature have disclosed that the results of phytoconstituent Chrysin showed improved cytotoxic effects against colorectal cancer on various in vitro and in vivo studies (Bahadori et al., 2016). One study showed that in Wistar rats, Chrysin guarded against colon cancer which was induced by cisplatin through amelioration of oxidative stress and apoptosis which caused a significant decrease in cis-diamminedichloroplatinum-induced deterioration of the goblet cells in the crypts of the colon (Khan et al., 2012). However, Chrysin is found to be efficacious in the treatment of colon carcinoma (Lin et al., 2018). To date, there is no data available on Chrysin action on multiple protein targets and its pharmacological mechanisms on colon cancer. Thus, the drug targets and pharmacological mechanism can be predicted using network pharmacology analysis which provides insight for improving the discovery of various drugs for various diseases (Shi et al., 2020).
In the current study, the inherent mechanism of phytoconstituent Chrysin for the treatment of colon cancer was scrutinized with the help of a network-based systematic study. Briefly, the drug-target network was constructed considering the target sets of Chrysin and colon cancer. The genes which overlapped between Chrysin and colon cancer were inspected by comparative analysis with the aid of KEGG pathway enrichment analysis. Hence, the current study was designated to access and document the Chrysin probable protein targets and their mechanisms of action on colon cancer.
MATERIALS AND METHODS
Target identification
The canonical SMILE of Chrysin was retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and its targets were predicted using the SwissTargetPrediction online server (http://www.swisstargetprediction.ch/). The server predicts molecular protein targets based on similarity measures depending on 2D and 3D combinations with known ligands. Furthermore, the protein molecules involved in colorectal cancer were identified using the Open Targets Platform (https://platform.opentargets.org/) and Therapeutic Target Database (http://db.idrblab.net/ttd/).
Gene set enrichment and network analysis
A set of gene IDs of probable protein target for Chrysin were submitted as input into the STRING database (https://string-db.org/) to study protein–protein interactions. Furthermore, molecular pathways modulated by a set of genes were identified using the KEGG pathway. The network among Chrysin, probable protein targets, and its pathways were constructed with the aid of Cytoscape 3.6.1 version. The constructed network was analyzed by edge count topological parameter by treating it as direct. The node size of the network was fixed to “low values to small size” with color to “low values to bright colors” (Patil et al., 2020, 2021).
Docking studies
Selection, preparation, and quality of protein target
Based on the network analysis, protein targets having larger node sizes and highest edge counts, and based on the clinical success full targets, we selected epidermal growth factor receptor (EGFR), a major potential therapeutic target of colon cancer. The 3D structure with X-ray crystallographic of EGFR having PDB ID: 1M17 was retrieved from Research Collaboratory for Structural Bioinformatics Protein Data Bank (https://www.rcsb.org). The distribution of amino acids and the quality of proteins were determined by PROCHECK and ERRAT (https://servicesn.mbi.ucla.edu/ERRAT) online servers, respectively. The active site residues that are involved in the ligand-binding were noted from the selected PDB file via Discovery Studio Visualizer 2019v (DSV v2019) and further, the listed amino acid residues were confirmed from P2RANK (https://prankweb.cz/analyze?database=v2-conservation&code=1M17) and Galaxy site web servers (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=SITE).
Preparation of ligand
The 3D structure of Chrysin and the reference drug molecule, i.e., Erlotinib, was taken from the PubChem chemical database. To avoid interference during docking simulation, the initial free energy of the ligand molecules was minimized by Marvin Sketch using the MMFF94 force field.
Docking validation
Before initiating the docking studies, the grid box and docking conformation are validated via the pre-docking step. The structure of Erlotinib from the crystal structure of EGFR (PDB ID 1M17) was retrieved in .pdb format using DSV v2019. Docking of EGFR with both Erlotinib extracted from the 1M17 and PubChem database was carried out using PyRx 0.8v software. The .pdb format protein molecule and the ligand were imported into the software and converted into AutoDock Vina molecules, i.e., .pdbqt files. The grid box was set to maximum for size and center [center x = 26.6909, y = 9.667, and z = 59.1236; size x = 58.124, y = 66.546, and z = 51.512] and the system exhaustiveness was set to 8. Intermolecular interactions and post-docking ligand orientation (conformation) root mean square deviation (RMSD) of both Erlotinib extracted from the 1M17 and PubChem was analyzed by DSV v2019 software and DockRMSD online server (https://zhanglab.ccmb.med.umich.edu/DockRMSD/).
Chrysin–EGFR docking
The affinity between Chrysin and EGFR protein target was determined using AutoDock Vina by using PyRx 0.8v software. The .pdb format protein molecule and the ligand were imported into the software and converted into AutoDock Vina molecule (i.e., Chrysin.pdbqt) file. As mentioned above, the grid box was set to maximum for both size and center [center x = 26.6909, y = 9.667, and z = 59.1236; size x = 58.124, y = 66.546, and z = 51.512] and the system exhaustiveness was set to eight. The ligand–protein interactions within the site-specific domains along with a transformation in the orientation of ligand after docking were envisioned with the aid of DSV v2019.
Molecular dynamics
Molecular dynamics of Chrysin with EGFR complex was carried out by Desmond software (version 6.1). The MD system was solvated by Simple Point Charge water model in a cubic box [dimension 10Å × 10Å × 10Å]. Chlorine ions (–5) were added to neutralize the system. Furthermore, the complex system was minimized for a 100ps production run. The Isothermal-Isobaric ensemble: moles (N), pressure (P) and temperature (T) was applied, and 1.01325bar atmospheric pressure and temperature 300K were set. Furthermore, 2,000 frames were set for 20 ns MD production run at 10 ps sampling analysis time interval. Finally, the RMSD and residue-wise interaction fluctuations were analyzed.
Druglikeness characteristics and ADMET profile
Chrysin and Erlotinib canonical SMILES were retrieved to envisage the characteristics of druglikeness which includes the molecular weight of the drugs (MW), the total number of hydrogen bond donor (NHBD), the total number of hydrogen bond acceptor (NHBA), its lipid/water partition coefficient (Log P), and its druglikeness score (DLS) by employing MolSoft online server. With the aid of ADVERPred and ADMET SAR2.0 by the online server, numerous side effects along with the probable inactivity (Pi) and probable activity (Pa) and ADMET profile were anticipated, respectively.
RESULTS
Target prediction
A total of 40 protein targets were identified for the Chrysin from SwissTargetPrediction. Among them, 13 protein targets were significantly targeted by Chrysin and found to involve in colon cancer, namely calmodulin 1, EGFR, insulin-like growth factor 1, insulin receptor, myosin light chain kinase, glycogen synthase kinase 3 beta, neurotrophic receptor tyrosine kinase 2, cyclin-dependent kinase 6, nitric oxide synthase 2, monoamino oxidase A, arachidonate 15-lipoxygenase, arachidonate 12-lipoxygenase, and arachidonate 15-lipoxygenase.
Gene set enrichment and network analysis
The enrichment analysis identified 13 targets to play a major role in the 40 molecular pathways [false discovery rate (FDR) < 0.05]. Among them, pathways in cancer, metabolic pathways, Ras and PI3K–Akt signaling pathways, and nitrogen metabolism were significantly modulated via scoring the lowest FDR value. Table 1 summarizes the modulated pathways with their respected modulated genes by Chrysin and Figure 1 shows the network of interacted phytoconstituents with molecules of proteins and the pathways involved in colon cancer.
Active site residues of EGFR protein
The active site residues of EGFR present in the .pdb file 1M17 are Lys721, Val702, Ala719, Leu764, Met742, Leu694, Glu738, Asp831, Met769, and Asp831 (confirmed via P2RANK and GalaxySite web server).
Docking studies
Docking validation
The binding energy (BE) of Erlotinib extracted from the 1M17 and PubChem was –7.1 and –7.5 kcal/mol, respectively. Erlotinib extracted from the 1M17 formed one hydrogen bond interaction, i.e., Asp831, with NH group of Erlotinib and formed five non-hydrogen bond interactions with Gly695, Val702, Leu764, Ala719, and Met742. However, Erlotinib extracted from PubChem formed two hydrogen bond interactions, i.e., Met769…O- and Asp831…NH, and formed six non-hydrogen bond interactions with Leu694, Val702, Lys721 (2), Met742, and Leu764. Among the total interactions of Erlotinib extracted from 1M17 and PubChem with EGFR, Asp831, Val702, Met742, and Leu764 were found to be the common interactive residues. Furthermore, the Erlotinib docked orientation (conformation) was analyzed by the DockRMSD server and the RMSD was found to be 1.657 Å. Figure 2 shows the interactions and ligand orientation RMSD of both Erlotinib extracted from 1M17 and PubChem.
Chrysin and EGFR docking studies
Docking was carried out for Chrysin and compared its affinity and interactions with the docked conformation of Erlotinib extracted from PubChem due to its lowest BE and maximum intermolecular interactions. Chrysin scored the lowest BE of –8.8 kcal/mol with EGFR by forming two hydrogen bond interactions with site-specific residues that is Glu738…OH and Asp831…=O. However, Erlotinib scored the lowest BE of –7.5 kcal/mol with EGFR by forming two hydrogen bonds with active site residues, i.e., Met769…O- and Asp831…NH. Among these interactions, Asp831 amino acid residue was found to be common for Chrysin and Erlotinib. After docking, the BE and the swap in the orientation of ligand along with the hydrogen bond interactions are displayed in Table 2 and the interactivity of Chrysin and Erlotinib concerning EGFR is shown in Figure 3.
Molecular dynamics
Chrysin combined with EGFR exhibited a very stable RMSD (Å) at 20 ns production run. Initially, from 0 to 6 ns the ligand RMSD (Å) was found stable (0.3Å) and suddenly slight fluctuation was seen (0.3–0.4Å) at 6 ns and further from 6 to 20 ns again RMSD was found to be very stable at 0.4Å. Furthermore, Chrysin formed a very stable bond throughout the 20 ns production run. It formed stable interactions with Asp831 (56%), Thr766 (52%), Ala719 (17%), Glu738 (94%), Lys721 (32%), and Thr830 (12%). Figure 4 shows the Chrysin–EGFR RMSD and residue-wise interactions.
Druglikeness and side effects
Erlotinib, a clinically approved molecule was found to obey the rule of Lipinski via MW ≤500 g/mol (i.e., 393.17 g/mol), HBA ≤10 (i.e., 6), HBA ≤5 (i.e., 1), LogP ≤5 (i.e., 2.21), and scored positive DLS of 0.90. Similarly, Chrysin was also found to obey the rule of Lipinski via MW ≤500 g/mol (i.e., 254.06 g/mol), HBA ≤10 (i.e., 4), HBA ≤5 (i.e., 2), LogP ≤5 (i.e., 3.61), and scored negative DLS of –0.21. On looking at the side effects, Erlotinib was predicted to exert hepatotoxicity, myocardial infarction, nephrotoxicity, and cardiac failure via scoring Pa values of 0.932, 0.665, 0.594, and 0.362, respectively. Similarly, Chrysin was predicted to exert only hepatotoxicity via a scoring Pa value of 0.516. The results are shown in Table 3.
DISCUSSION
The present study was carried out to recognize the molecular mechanism of anticancer activity of phytoconstituent “Chrysin” which belongs to the flavonoid category with the analysis of gene set enrichment, network pharmacology, and in silico molecular docking analysis. Network pharmacology is a novel innovative method to forecast the association between the phytoconstituent and protein targets (Chandran et al., 2017). Primarily, taking into account, the fundamentals of “similar compounds target similar proteins” forecasted the Chrysin targets with the help of SwissTargetPrediction and the protein molecules involved in targeting colon cancer concerning the approved targets which are available in the Therapeutic Target database. Later, we executed enrichment analysis of the compound gene set with the help of KEGG to recognize the biological pathways modulated by the Chrysin. Consequently, for colon cancer, 40 molecular pathways were notably clarified. The network was built displaying the interactivity and interconnection among Chrysin and its protein molecules, the protein molecules along with its associated pathways, and also with the phytoconstituents–targets pathways. Based on the polypharmacology perspective, it is important to assume that one pathway which consisted of several molecules of protein regulated by a single compound is commendable than the function of a single molecule of proteins that requires numerous pathways regulated by a single drug molecule (Patil et al., 2019). The influence of a single target in several routes can be fewer and the influence for that single route consisting of many protein targets regulated by that single compound could be vast. Our study disclosed that phytoconstituent Chrysin revealed the excessive gene count for various routes or pathways which are entangled in colon cancer and ultimately Chrysin falls under the polypharmacology therapy via multiple proteins and multiple pathways’ mechanism. The constructed network linking phytoconstituents–proteins–pathways discloses the role of Chrysin (flavonoid) for the treatment of colon cancer by targeting significant molecules of protein. Flavonoids are polyphenolic compounds that are found drastically in plant sources. It contains two aromatic rings that are A and B which form a C ring by attaching a 3-carbon chain (Kumar and Pandey, 2013). Considering various in vitro and in vivo research studies, it can be seen that flavonoids had potential activity as an anticancer agent against innumerable types of cancer models which is progressed through modulation of key signaling pathways that are responsible for the transfer and conquering of cancer cells and also by arresting the metastatic progression like regulatory molecules like MMPs, TGF-β, uPA/uPAR, and other contributors of the complicated process which are involved in metastatic spread (Liskova et al., 2020). In one study, the growth of epidermoid carcinoma A431 cells and upregulation of EGFR was inhibited significantly by tyrosine kinase inhibitors (Huang et al., 2009). The present study recognized Chrysin to potentially target EGFR as a vital protein molecule, which is inherently entangled in colon cancer, respectively. Previous studies reported that drugs like Erlotinib or Gefitinib produced a strong synergistic effect with serine/threonine-protein kinase B-Raf (BRAF) (V600E) inhibition which in turn leads to the inhibition of EGFR and thus such drugs are recommended as an inherent anticancer compound (Prahallad et al., 2012). In one study, it has been found that the therapeutic effect of anti-EGFR therapy is overruled by activated Kirsten rat sarcoma viral oncogene homolog protein mutations, which further lead to the transduction of the activation signal from EGFR. Apart from these, supplementary factors may be the portion for different signaling routes or pathways which are triggered by EGFR which include EGFR itself, the phosphatase, BRAF, and phosphatase and tensin homolog (PTEN). These are the genes that are the major targets of genetic modifications for colon cancer and they have been related to the effectiveness of therapies regulated against EGFR or the members of the EGFR-activated pathways (Laurent-Puig et al., 2009). Inhibition of EGFR is one of the restorative approaches in the management of colon cancer. In previous reports, it has been found that the subjects with KRAS wild type tumors and the administration of EGFR MAb drugs to standard treatment, there was progression of the disease which was reduced by 30%, and the rate of death was also shrunk by 12% and the tumor shrinkage rate was seen to be improved, i.e., from 31% to 46%. For subjects having both KRAS and neuroblastoma RAS viral oncogene homolog (extended Rat sarcoma) wild type, the risk of disease progression was found to be lowered, i.e., 40% and the risk of death was lowered by 23% and the rate of tumor shrinkage increased from 21% to 48% (Chan et al., 2017). In the present study, the phytoconstituent Chrysin is predicted for the inhibition of EGFR. It is suggested that Chrysin reduced the proliferation of cancer cells by downregulation of the EGFR tyrosine kinase enzyme (Liu et al., 2019). Additionally, this phytoconstituent is also envisaged to interconnect with numerous target proteins and regulate the pathways which are entangled in colon cancer. In the present study, we recognized Chrysin as a potent EGFR tyrosine kinase inhibitor. Considering, the small molecules function only on the active site domain on specific targets, according to the theory of the “lock and key” model. The present study employed molecular docking simulation to delineate the affinity of the standard drug and phytoconstituent toward envisaged target molecules. For the docking study, we selected the phytoconstituent Chrysin, which is predicted to target one inherent therapeutic protein target of colon cancer, i.e., EGFR. Furthermore, the binding affinity of the phytoconstituent Chrysin was compared with the therapeutically approved drug, i.e., Erlotinib. Among the selected drug, Chrysin expressed the highest binding affinity to EGFR. The results revealed that the phytoconstituent Chrysin, when coupled with long-chain, can obtain a better anticancer effect and those compounds with 16 carbon atoms chain were found to show inherent anti-proliferative activity (Lv et al., 2010). Observing the hydrogen bond interactions, the selected standard drugs, i.e., Erlotinib and Chrysin, had hydrogen bond interactions with the predicted protein targets. The residues amino acids of EGFR having links with Chrysin were Lys721, Val702, and Ala719; and with Erlotinib are Ala719, Leu764, Lys721, Met742, and Leu69. The interactions of these residues were found within its active site domain. Likewise, Erlotinib and Chrysin shared Asp831 as the common residue of amino acids for EGFR. Furthermore, the interactions of Chrysin with EGFR were confirmed via a 20 ns MD production run. Chrysin was found to 56% stable with Asp831 and also it formed stable interactions with Thr766, Ala719, Glu738, Lys721, and Thr830. The study revealed that Chrysin was a better inhibitor of EGFR when compared to Erlotinib. Considering these findings from in silico molecular docking and MD analysis, Chrysin interacts with the active site domain and possibly plays a major role in the inhibition of EGFR as well as numerous protein molecules as observed in the Chrysin network-based mechanisms. Thus, employing phytoconstituents formulations for the management of complicated diseases could be the most favorable approach.
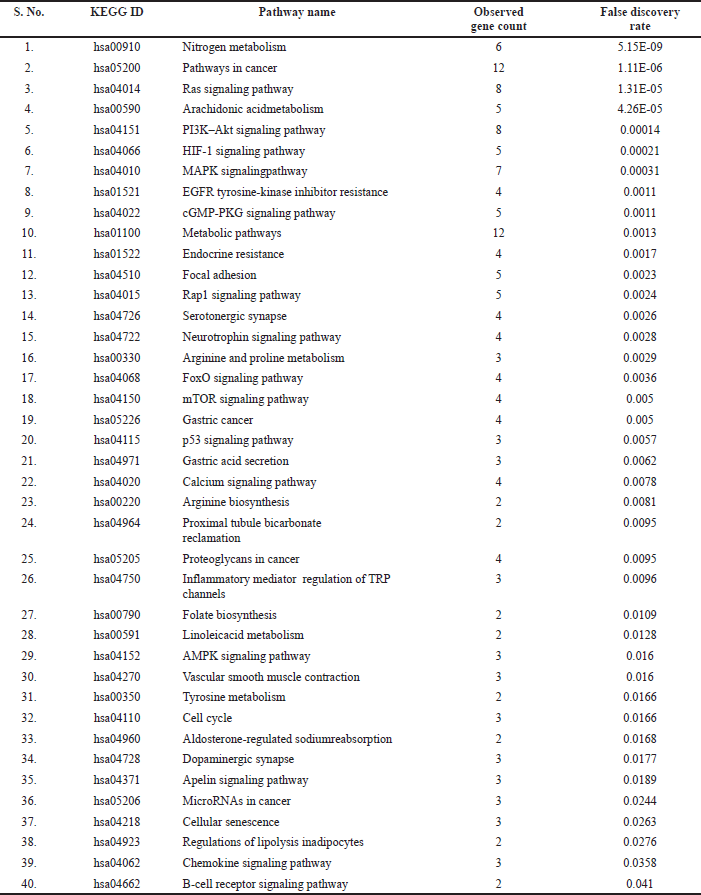 | Table 1. Gene set enrichment analysis of probable protein targets of Chrysin. [Click here to view] |
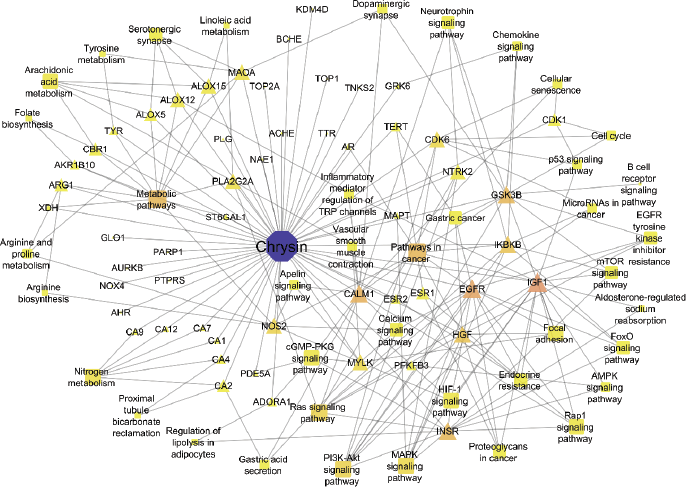 | Figure 1. Network relationship between Chrysin (blue color node), probable protein targets, and pathways (yellow color nodes) involved in colon cancer. [Click here to view] |
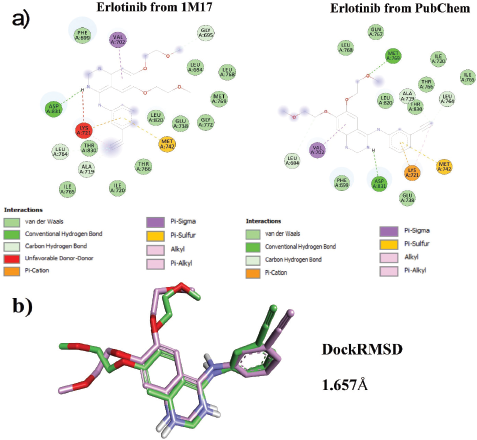 | Figure 2. Docking validation. (a) Interactions of both Erlotinib extracted from 1M17 and PubChem. (b) Ligand orientation RMSD. [Click here to view] |
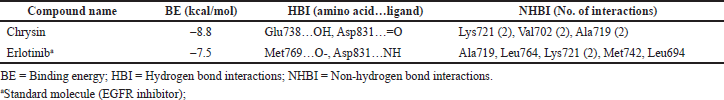 | Table 2. Affinity and interactions of Chrysin and Erlotinib with EGFR. [Click here to view] |
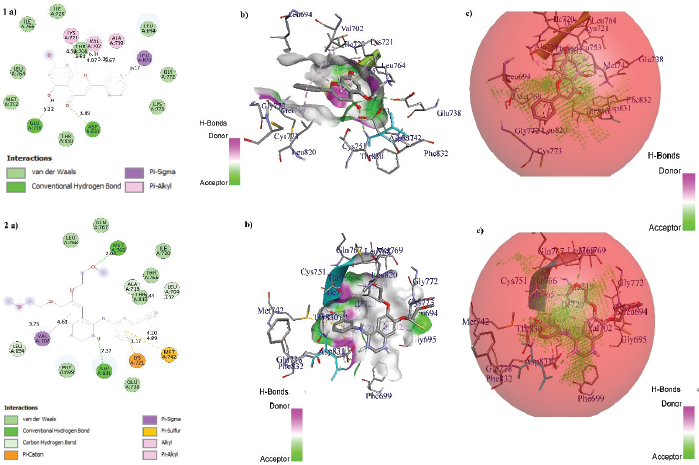 | Figure 3. Interaction of (1). Chrysin and (2) Erlotinib with EGFR. (a) 2D representation, (b) Ligand within binding pockets, and (c) Ligands within active site domains, respectively. [Click here to view] |
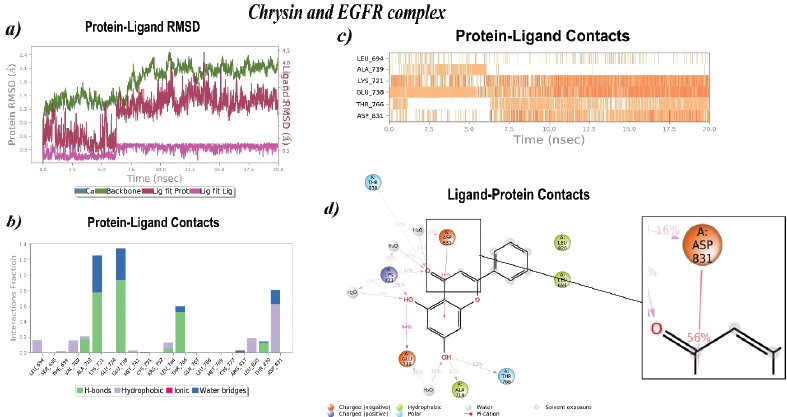 | Figure 4. Stability of intramolecular interactions of Chrysin with EGFR. (a) RMSD fluctuations, (b–d) Residue-wise Chrysin–EGFR contacts. [Click here to view] |
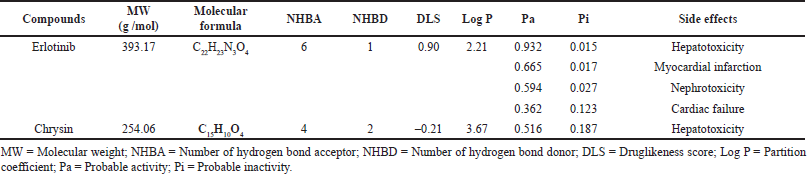 | Table 3. Druglikeness characteristics and its probable side effects of Erlotinib and Chrysin. [Click here to view] |
CONCLUSION
In this study, a gene set enrichment and network analysis of Chrysin possible protein targets coupled with its molecular interactions with EGFR via molecular docking and dynamics study were examined. We identified the mechanisms by which Chrysin modulates various signaling pathways which led to the inhibition of tumor progression. The study provided Chrysin as a major lead molecule as an anticancer agent against colon cancer. Chrysin majorly acts via pathways in cancer, metabolic pathways, and Ras and PI3K–Akt signaling pathways. However, it has major action on an EGFR as an inhibitor compared to standard molecule Erlotinib via molecular docking studies. The results which are obtained in the currents study will therefore narrow down the inherent protein targets of Chrysin which will direct a better pathway in future experiments. The major hurdle to deploy Chrysin as an anticancer agent for clinical use is its inadequate bioavailability and its reduced chemical stability under physiological conditions. However, we propound that novel drug delivery systems like nanoparticles might be intended to increase the bioavailability and stability of Chrysin. Thus, in the drug-development process, molecular docking analysis provides a rapid and cost-effective method to recognize the inherent protein targets for different anticancer compounds emerging from several natural sources along with the potential toxic side effects which occur through the interactions with proteins.
ACKNOWLEDGMENTS
The authors would like to thank KLE College of Pharmacy, Belagavi, KLE Academy of Higher Education and Research (KAHER) Belagavi, for providing support to carry out the research.
AUTHORS’ CONTRIBUTIONS
All the authors have equally contributed to study design, collection, analysis, and interpretation of the data, writing, and drafting of the manuscript. All the authors read and approved the final version.
ETHICAL APPROVAL
This study does not involve any animals or human subjects.
CONFLICT OF INTEREST
The authors declare no conflict of interests for this manuscript.
FUNDING
None.
REFERENCES
Bahadori M, Baharara J, Amini E. Anticancer properties of chrysin on colon cancer cells, in vitro and in vivo with modulation of caspase-3, -9, bax and sall4. Iran J Biotechnol, 2016; 14(3):117–24. CrossRef
Benarba B, Pandiella A. Colorectal cancer and medicinal plants: principle findings from recent studies. Biomed Pharmacother, 2018; 107:408–23. CrossRef
Bray F, Ferlay J, Soerjomataram I, Siege R; Torre L, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of Incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2018; 68(6):394–424. CrossRef
Chan DLH, Segelov E, Wong RS, Smith A, Herbertson RA, Li BT, Tebbutt N, Price T, Pavlakis N. Epidermal growth factor receptor (EGFR) inhibitors for metastatic colorectal cancer. Cochrane Database Syst Rev, 2017; 6(6):CD007047. CrossRef
Chandran U, Mehendale N, Patil S, Chaguturu R, Patwardhan B. Network pharmacology. Innovative Approaches Drug Discov, 2017; 2017:127–64. CrossRef
Filho CB, Jesse CR, Donato F, Giacomeli R, Del Fabbro L, da Silva Antunes M, de Gomes MG, Goes ATR, Boeira SP, Prigol M, Souza LC. Chronic unpredictable mild stress decreases BDNF and NGF levels and Na+, K+-ATPase activity in the hippocampus and prefrontal cortex of mice: antidepressant effect of Chrysin. Neuroscience, 2015; 289:367–80. CrossRef
Hellinger MD, Santiago CA. Reoperation for recurrent colorectal cancer. Clin Colon Rectal Surg, 2006; 19(4):228–36. CrossRef
Huang YT, Hwang JJ, Lee PP, Ke FC, Huang JH, Huang CJ, Kandaswami C, Middleton E, Lee MT. Effects of luteolin and quercetin, inhibitors of tyrosine kinase, on cell growth and metastasis-associated properties in A431 cells overexpressing epidermal growth factor receptor. Br J Pharmacol, 1999; 128(5):999–1010. CrossRef
Khan R, Khan AQ, Qamar W, Lateef A, Tahir M, Rehman MU, Ali F, Sultana S. Chrysin protects against cisplatin-induced colon. toxicity via amelioration of oxidative stress and apoptosis: probable role of p38MAPK and p53. Toxicol Appl Pharmacol, 2012; 258(3):315–29. CrossRef
Kumar S, Pandey AK. Chemistry and biological activities of flavonoids: an overview. ScientificWorldJournal, 2013; 2013:162750. CrossRef
Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, Ducreux M, Ychou M, Bibeau F, Bouché O, Reid J, Stone S, Penault-Llorca F. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol, 2009; 27(35):5924–30. CrossRef
Lin YM, Chen CI, Hsiang YP, Hsu YC, Cheng KC, Chien PH, Pan HL, Lu CC, Chen YJ. Chrysin attenuates cell viability of human colorectal cancer cells through autophagy induction unlike 5-fluorouracil/oxaliplatin. Int J Mol Sci, 2018; 19(6):1763. CrossRef
Liskova A, Koklesova L, Samec M, Smejkal K, Samuel SM, Varghese E, Abotaleb M, Biringer K, Kudela E, Danko J, Shakibaei M, Kwon TK, Büsselberg D, Kubatka P. Flavonoids in Cancer metastasis. Cancers, 2020; 12(6):1498. CrossRef
Liu YM, Li Y, Liu RF, Xiao J, Zhou BN, Zhang QZ, Song JX. Synthesis, characterization and preliminary biological evaluation of Chrysin amino acid derivatives that induce apoptosis and EGFR downregulation. J Asian Nat Prod Res, 2019; 23(1):39–54. CrossRef
Lv PC, Wang KR, Li QS, Chen J, Sun J, Zhu HL. Design, synthesis, and biological evaluation of Chrysin long-chain derivatives as potential anticancer agents. Bioorg Med Chem, 2010; 18(3):1117–23. CrossRef
Nunes CDR, Arantes MB, de Faria Pereira SM, da Cruz LL, de Souza Passos M, de Moraes LP, Vieira IJC, de Oliveira DB. Plants as sources of anti-inflammatory agents. Molecules, 2020; 25(16):3726. CrossRef
Patil RS, Khatib NA, Patil VS, Suryawanshi SS. Chlorogenic acid may be a potent inhibitor of dimeric SARS-CoV-2 main protease 3CLpro: an in silico study. Tradit Med Res, 2021; 6(2):20. CrossRef
Patil VS, Biradar PR, Attar V, Khanal P. In silico docking analysis of active biomolecules from Cissus quadrangularis L. against PPARG. Indian J Pharm Educ, 2019; 53(3s):s332–37. CrossRef
Patil VS, Deshpande SH, Harish DR, Patil AS, Virge R, Nandy S, Roy S. Gene set enrichment analysis, network pharmacology and in silico docking approach to understand the molecular mechanism of traditional medicines for the treatment of diabetes mellitus. J Proteins Proteom, 2020; 11:297–310. CrossRef
Prahallad A, Sun C, Huang S, Nicolantonio FD, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature, 2012; 483:101–3. CrossRef
Rani N, Bharti S, Bhatia J, Nag TC, Ray R, Arya DS. Chrysin, a PPAR-γ agonist improves myocardial injury in diabetic rats through inhibiting AGE-RAGE mediated oxidative stress and inflammation. Chem Biol Interact, 2016; 250: 59–67 CrossRef
Rehman MU, Ali N, Rashid S, Jain T, Nafees S, Tahir M, Khan AQ, Lateef A, Khan R, Hamiza OO, Kazim S, Qamar W, Sultana S. Alleviation of hepatic injury by Chrysin in cisplatin administered rats: probable role of oxidative and inflammatory markers. Pharmacol Rep, 2014; 66( 6):1050-59 CrossRef
Shi M, Li B, Yuan Q, Gan X, Ren X, Jiang S, Liu Z. Network pharmacology-based approach to investigate the mechanisms of mahai capsules in the treatment of cardiovascular diseases. Evid Based Complement Alternat Med, 2020; 2020:9180982. CrossRef
Zhang Z, Li G, Szeto SSW, Chong C, Quan Q, Huang C, Cui W, Guo B, Wang Y, Han Y, Michael Siu KW, Lee SMY, Chu IK. Examining the neuroprotective effects of protocatechuic acid and Chrysin on in vitro and in vivo models of Parkinson disease free radical. Biol Med, 2015; 84:331–43. CrossRef