INTRODUCTION
Many fungi from various environments on earth have been identified to produce an extensive variety of chemical compounds which possess bioactivity. The formation of these chemical compounds can be through primary and secondary metabolism. Chemical compounds generated by fungi cannot be thoroughly demonstrated by their function in the metabolism of these fungi. Generally, these chemical compounds can function as a detoxification mechanism for fungi and have the ability to communicate between fungi and the surrounding environment (Cometto-Muñiz et al., 2005; Humphris et al., 2002). One kind of metabolites discovered in fungi is a volatile compound. Terpenes, alcohols, hydrocarbons, ketones, esters, carboxylic acids, and some volatile sulfur-containing compounds are among the diverse chemical classes represented in the secondary metabolites of fungi. Fungi from oceanic regions are also able to produce these volatile chemical compounds, in addition to those produced by fungi from lands. Researchers are currently interested in studying marine fungi because they produce a variety of chemical compounds that are distinct from those produced by their terrestrial counterparts. An example of a fungus revealed in marine environments is one connected to marine sponges. The marine sponge is a marine biota whose body is almost 60% in symbiosis with other microorganisms; the one incorporating them is a fungus. Marine sponge-associated fungi contribute significantly to the metabolic chain in the sponge body (Brinkmann et al., 2017; Taylor et al., 2007; Zhang et al., 2005). There are several factors causing fungi originating from marine areas, particularly marine sponge-associated fungi, to own different chemical compounds from their relatives that grow in land areas, one of which is extreme environmental conditions or experiencing biological stress such as limited carbon supply and insufficient sunlight, and the osmotic pressure is relatively high. It causes marine-derived fungi to develop their own metabolic pathways to generate new secondary metabolites with various biological activities (Debbab et al., 2011; Huang et al., 2011; Samirana et al., 2021a).
One of the marine sponges with the species Stylissa flabelliformis has been understood to possess several quite diverse marine sponge-associated functions. Sponge S. flabelliformis was obtained from the waters of Menjangan Island, West Bali National Park (Indonesia). The marine sponge-derived fungi that have been isolated from the S. flabelliformis sponge incorporate with Aspergillus flavus UPMZ02 (SAL 1), Aspergillus fumigatus CD1621 (SAL 2), Trichoderma reesei JCM 2267 (SAL 3), Aspergillus nomius KUB105 (SAL 4), Aspergillus sp. TLWK-09 (SAL 5), A. flavus MC-10-L (SAL 6), Penicillium sp. RMA-2 (SAL 7), Aspergillus sp. TLWK-09 (SAL 9), A. fumigatus (SAL 9), and T. reesei TV221 (SAL 10). The marine sponge-associated fungus S. flabelliformis with codes SAL 5, SAL 6, SAL 7, SAL 9, and SAL 10 owned antimicrobial activity on Staphylococcus aureus American type culture collection (ATCC) 29213, Escherichia coli (EC) ATCC 25922, and methicillin-resistant S. aureus (MRSA) which were assessed from their zone of inhibition. In accordance with the further research, among the five marine sponge-associated fungi of S. flabelliformis that possess antimicrobial bioactivity, T. reesei TV221 owns the most antimicrobial potential (Samirana et al., 2021b; Setyowati et al., 2018, 2017). In accordance with the results of this study, the fungus T. reesei TV221 associated with S. flabelliformis sponges is tremendously potential to be advanced as an antimicrobial agent in the future and can be detected to have bioactive compounds that act as antimicrobial agents.
The search for antimicrobial bioactive compounds from the fungus T. reesei TV221 can be performed by utilizing a metabolomics approach. The metabolomics approach is a process of discovering bioactive compounds by employing the chemical profile of natural products. The combination of chemical profiles with bioactivity data produces big data that will lead to bioactive compounds from these natural products (Xu et al., 2006; Yuliana et al., 2011). As previously identified, most of the secondary metabolites of fungi are volatile chemical compounds. A reliable chemical profile which is able to represent the active components and their chemical characteristics are acquired through appropriate instrumentation techniques. Precise instrumentation enhances separation capability, measurement precision, and selectivity and reduces instrument interference. The most appropriate instrumentation for the separation of volatile compounds is by employing the gas chromatography-mass spectroscopy (GC-MS) technique. The GC-MS technique in research associated with metabolomics requires the separated metabolites to be volatile and thermostable (Han et al., 2009; Samirana et al., 2022).
In this study, to obtain an adequate variety of chemical profiles, variations were conducted on the fermentation medium of the fungus T. reesei TV221 in the form of variations in the Dextrose content and salinity of the fermentation medium. Furthermore, fermented products such as supernatant and biomass were extracted with ethyl acetate to acquire different types of extracts. It is expected that with numerous factors of variation, the chemical profile and antimicrobial activity produced can also vary which will later be able to lead to bioactive compounds that function as antimicrobials.
MATERIAL AND METHOD
Materials
Marine sponge-derived fungus T. reesei TV221 from the marine sponge S. flabelliformis was obtained from the waters of Menjangan Island, West Bali National Park (Indonesia). The test microbes employed in the antimicrobial study were S. aureus (SA) ATCC 25923 and EC ATCC 25922, Sabouraud Dextrose Agar (SDA) (Oxoid, UK), Dextrose (Oxoid, UK), Tryptone (Oxoid, UK), Peptone P (Oxoid, UK), nutrient agar (Oxoid, UK), nutrient broth (Oxoid, UK), ethyl acetate (Merck, German), methanol (Merck, German).
Fermentation and extraction of secondary metabolites from fungi
The T. reesei TV221 fungal isolate associated with the S. flabelliformis sponge was used in a previous study and had been stored at the Indonesian Center for Biodiversity and Biotechnology Bogor (ICBB) for recultivation (Setyowati et al., 2018). Identification and authentication have also been carried out at ICBB. The fungus T. reesei TV211 associated with sponge S. flabelliformis was recultured on SDA with 30 parts per trillion (ppt) salinity seawater as a solvent to enhance the number of fungi. Fungal cultures were incubated for 7 days at room temperature. Fermentation was administered with medium variations in Dextrose and salinity levels in accordance with Table 1 for 12 days at 25°C and placed on a rotary shaker (120 rpm). After 12 days of fermentation, the supernatant and biomass were separated. The supernatant and biomass extraction processes followed the procedures performed in previous studies (Samirana et al., 2021b).
Antimicrobial activity
Antimicrobial testing in this study utilized the determination of the zone of inhibition. The bacteria employed in the antimicrobial test were EC ATCC 25922 and S. aureus ATCC 25923. The antimicrobial testing procedure in this study followed the procedures administered in previous studies with minor adjustments (Samirana et al., 2021b; Syukri et al., 2021).
GC-MS chemical profiling
The supernatant and biomass extract samples were dissolved in methanol at a rate of 1 mg/ml. GC-MS preparation for chemical profile determination was administered in accordance with the procedure in a previous study (Setyowati et al., 2017). The profile data obtained are compared with the WILEY7.LIB library.
Statistical analysis
After obtaining the GC-MS chromatogram profile data, the mass spectrum data were correlated with the WILEY7.LIB library. The matching factor or Similarity Index (SI) is implemented at least 80%. The metabolomics approach in this study was examined by chemometric methods such as principal component analysis (PCA) and cluster analysis with a hierarchical approach employed to assess the differences between the chromatogram profiles.
RESULTS AND DISCUSSION
Antimicrobial activity
Trichoderma reesei TV221 marine sponge-associated fungi from S. flabelliformis were obtained from the waters of Menjangan Island, West Bali National Park, Indonesia. The fungi T. reesei have been understood to embody the highest antimicrobial activity under fermentation by employing Sabouraud Dextrose Broth medium (1l SDB containing 2% b/v Dextrose, 0.5% b/v Peptone P, and 0.5% b/v Tryptone), salinity of 30 ppt, and fermentation time of 12 days. It was also affected by the growth curve of the fungus T. reesei TV221 which acquired its peak growth on day 12 (Samirana et al., 2021b; Sibero et al., 2018). The production of secondary metabolites that possess bioactivity as antimicrobial is affected by nutrition during the fermentation process. Carbon sources and medium salinity influence the production of secondary metabolites from marine sponge-associated fungi that own bioactivity (Samirana et al., 2021a; Stanbury et al., 1995; Ukhty et al., 2017).
The fungus T. reesei TV221 in previous studies has been identified to possess antimicrobial activity on several bacteria which are S. aureus ATCC 29213, EC ATCC 25922, and MRSA. The fermented component that owns the highest antimicrobial activity is the supernatant portion compared to the biomass portion (Samirana et al., 2021b; Setyowati et al., 2018). In this study, antimicrobial tests were administered on S. aureus ATCC 29213 and EC ATCC 25922 by varying the components of the fermentation medium. Variations in the components of the fermentation medium are hoped to produce diverse amounts of secondary metabolites so that various antimicrobial activities are obtained for the objective of tracing antimicrobial bioactive compounds with a metabolomic approach by administering the GC-MS technique. The antimicrobial activity test results of the supernatant ethyl acetate extract and fermented biomass with various medium components are demonstrated in Figure 1.
The most potent antimicrobial activity of supernatant extracts and biomass was displayed in the culture of S. aureus ATCC 25923. Staphylococcus aureus ATCC 25923 bacteria were employed in this study to represent Gram-positive bacteria. The highest antimicrobial activity in S. aureus ATCC 25923 was revealed in the ethyl acetate supernatant extract with variations in the numbers 8 and 9 media, that is, with 2% Dextrose concentration and 30 and 15 ppt salinities. It is in accordance with the previous study, in which the ethyl acetate supernatant extract from the fermentation of the fungus T. reesei TV221 for 12 days with Sabouraud Dextrose Broth medium (2% Dextrose) and 30 ppt salinity presented the most potent antimicrobial activity against S. aureus bacteria (Samirana et al., 2021b; Setyowati et al., 2017). The antimicrobial activity of the supernatant extract and fermented biomass with various medium variations (Dextrose and salinity) on EC ATCC 25922 presented lower results than the antimicrobial activity of S. aureus ATCC 25923 bacteria. This result is in accordance with previous studies conducted by previous researchers (Samirana et al., 2021b; Setyowati et al., 2017). The ethyl acetate extract of biomass fermented with various medium variations did not present antimicrobial activity on EC ATCC 25922. EC ATCC 25922 bacteria were administered in this study to represent Gram-negative bacteria. It can be displayed here that the antimicrobial activity of the ethyl acetate extract, both supernatant, and biomass is robustly affected by the variation of the medium. The concentration of Dextrose is revealed to possess an effect on antimicrobial activity, particularly when it was perceived against the bacteria S. aureus ATCC 25923, in which the higher the concentration of Dextrose is, and the more potent antimicrobial activity is. Dextrose is one of the carbon sources required for fungi in their growth cycle. Fungi require large amounts of carbon sources as these carbon sources will later be administered in the formation of fungal cell walls. Simple carbon sources such as Dextrose are more easily processed by fungi in their metabolism; thus, their levels in the fermentation medium possess an effect on fungi in generating bioactive compounds, particularly here as antimicrobials (Kjer et al., 2010; Moore-Landecker, 1996; Muthukumar and Venkatesh, 2013; Samirana et al., 2021a). The level of salinity in the fermenting medium seems to possess little effect on the metabolism of the fungi T. reesei TV221. It can be perceived in the antimicrobial activity of S. aureus ATCC 25923 and EC ATCC 25922 which did not own a significant effect. Although the fungi T. reesei TV221 originated from marine sponges, it is uncovered that the salinity of the medium did not significantly influence the production of secondary metabolites that function as antimicrobials. Several Trichoderma genera originating from the marine environment possess good growth with salinity levels ranging from 0.5% to 2% (5–20 ppt). Salinity levels exceeding 3% (30 ppt) are able to decrease the growth of fungal colonies of the genus Trichoderma (Mishra et al., 2016; Sánchez-Montesinos et al., 2019).
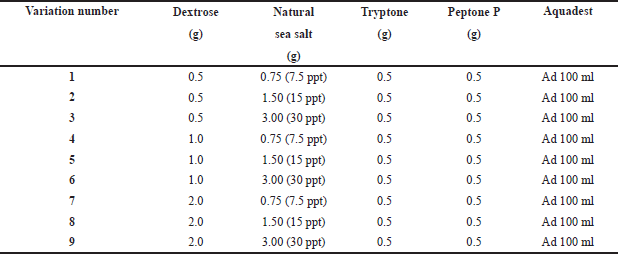 | Table 1. Variation of fermentation medium (Dextrose and salinity levels). [Click here to view] |
In this antimicrobial study, measurements were made of the diameter of the inhibition zone against the bacteria S. aureus ATCC 25923 and EC ATCC 25922. The diameter of the inhibition zone was able to illustrate the strength of the sample in inhibiting microbial growth. The category of antimicrobial power strength, an inhibition zone diameter of 5 mm or less, was categorized as weak; an inhibition zone of 5–10 mm was classified as moderate; an inhibition zone of 10–20 mm was classified as strong; an inhibition zone of 20 mm or more was categorized as tremendously strong (Davis and Stout, 1971; Samirana et al., 2021b). Figure 2 depicts the inhibition zone in broad strokes. The ethyl acetate supernatant extract with a medium variation of eighth (14.33 mm) and the ethyl acetate supernatant extract with a variation of the ninth medium (13.67 mm) both discovered the diameter of the inhibition zone in the antimicrobial study of the bacteria EC ATCC 25922, while the medium variations 3 and 4 displayed the narrowest zone of inhibition (6.33 mm). While this was progressing on, the ethyl acetate extract of biomass with different medium variations did not demonstrate that bacteria possessed an inhibition zone. Based on the inhibition zone strength category, the supernatant extract with medium variations 8 and 9 owned robust antimicrobial activity, while the supernatant ethyl acetate extract in medium variations 3–7 possessed moderate antimicrobial activity. It implies that the supernatant ethyl acetate extract with variations 8 and 9 owns the potential as an antimicrobial agent in EC ATCC 25922 representing Gram-negative bacteria. In the antimicrobial study of S. aureus ATCC 25923, the diameter of the widest inhibition zone was implied by the ethyl acetate supernatant extract with a medium variation of 8 (17.33 mm) followed by ethyl acetate supernatant extract with a medium variation of 9 (16.67 mm); then for the lowest inhibition zone narrow was unveiled by the ethyl acetate extract of biomass with a medium variation of 3 (6.33 mm). The biomass ethyl acetate extract with medium variations 1 and 2 did not present any inhibition zones. In accordance with the strength category of the zone of inhibition, the ethyl acetate supernatant extract with a medium variation of 4–9 possessed a category of robust antimicrobial activity, while the ethyl acetate supernatant extract with a medium variation of 1–3 and biomass ethyl acetate extract with a medium variation of 3–9 owned moderate antimicrobial activity. It implies that the supernatant ethyl acetate extract with variations 8 and 9 possesses the potential as an antimicrobial agent in S. aureus ATCC 25923 which represents Gram-positive bacteria. The results displayed that the antimicrobial activity was more active in S. aureus ATCC 25923 compared to EC ATCC 25922; it could be due to the fact that EC ATCC 25922, which are Gram-negative bacteria, possess a more complex cell wall structure that owns more lipid composition and is thicker than the bacteria in S. aureus ATCC 25923 which are categorized as Gram-positive bacteria (Pelczar and Chan, 2008a, 2008b). In this study, the negative control (CO−) administered was the solvent implemented to dissolve the extract, that is, methanol. The utilization of negative control aims to ensure that the solvent utilized is inert and does not possess antimicrobial activity. The positive control (CO+) used in this study was chloramphenicol. Chloramphenicol is a broad-spectrum antibiotic that is tremendously effective against Gram-negative and Gram-positive bacteria (Tjay and Rahardja, 2015).
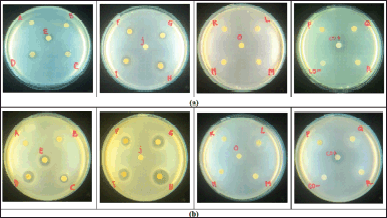 | Figure 1. Antimicrobial activity of supernatant extract (sample code A–I) and biomass extract (sample code J–R); CO− = negative control; CO+ = positive control. (a) EC ATCC 25922; (b) S. aureus ATCC 25923. [Click here to view] |
In an antimicrobial assay employing the disc method, the type of inhibition zone produced signifies the sensitivity of bacteria to antimicrobial or antibiotic agents from the sample. The radical and irradical zone are two types of inhibition zones of antibacterial activities. The radical zone is the uncontaminated area that is free of bacterial growth and has a distinct boundary from the bacterial colony. The inhibition zone is referred to as the irradical zone if it appears faint and there is no discernible boundary between the bacterial colony and the inhibition zone (Davis and Stout, 1971; Pelczar and Chan, 2008b; Samirana et al., 2021b). Table 2 presents the types of inhibition zones from antimicrobial studies on bacteria S. aureus ATCC 25923 and EC ATCC 25922 from ethyl acetate supernatant extracts and fermented biomass with various medium variations (Dextrose and salinity). The type of inhibition zone in the antimicrobial test of S. aureus bacteria ATCC 25923 possesses a radical zone type for ethyl acetate supernatant extract and an irradical zone type for biomass ethyl acetate extract. It implies that the supernatant ethyl acetate extract was more effective in killing bacterial colonies of S. aureus ATCC 25923 than the biomass ethyl acetate extract which was merely able to inhibit the growth of bacterial colonies. For the type of inhibition zone in the study of antimicrobial bacteria, EC ATCC 25922 owned a radical zone type for the supernatant ethyl acetate extract, while the biomass ethyl acetate extract did not possess antimicrobial activity on EC ATCC 25922. It implied that the ethyl acetate supernatant extract was effective in eradicating colonies of bacteria EC ATCC 25922 although it owns a diameter that is not extensive enough. Due to the type of inhibition zone from antimicrobial studies on EC ATCC 25922 and S. aureus ATCC 25923, it can be indicated that the supernatant ethyl acetate extract possesses the potential to inhibit bacterial colonies. In the future, to ensure its antibacterial strength, it is necessary to conduct minimum inhibitory concentration and minimum bactericidal concentration tests.
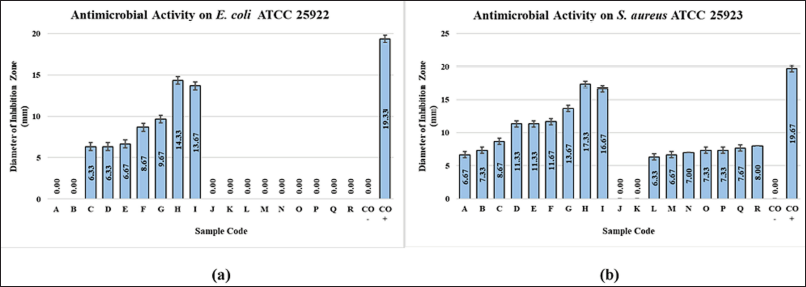 | Figure 2. Value of antibacterial inhibition zone diameter at (a) EC ATCC 25922 and (b) S. aureus ATCC 25923. [Click here to view] |
GC-MS chemical profile
The working principle of GC-MS is separation by employing the gas chromatography (GC) technique, which employs differences in the vapor points of volatile compounds. This GC is correlated with a Quadrupole Mass Analyzer and an ionizing electron (EI) source. EI is one of the ionization techniques functioning by striking electron ions into molecules resulting from GC elution so as to produce pieces of ions from the molecular mass. The mass spectrum of EI was acquired under standard conditions (electron energy of 70 eV and ion source temperature of 200°C–250°C); thus, compounds, particularly volatile compounds, could be identified easily and could compare the obtained mass spectra with the reference mass spectra of pure compounds collected through specialized libraries (Schauer et al., 2005).
In this study, there were 18 samples, the GC-MS chemical profile of which was identified. In Figure 3, there are five GC-MS chemical profiles from the sample, which are the ethyl acetate supernatant extract with medium variations of 3 (C), 6 (F), 7 (G), 8 (H), and 9 (I). All GC-MS chemical profiles of all extracts from both the supernatant and biomass were documented on the area under the curve and retention time (Rt) data, which were then normalized to the Rt data. In Figure 3, it is displayed that there are similarities in the patterns formulated in the three GC-MS chemical profiles; only there are differences in the area of the produced peaks. This difference in peak area is because of variations in the fermentation medium causing different amounts of secondary metabolites to be generated. It is what causes differences in antimicrobial activity resulting from each of these extracts. This difference in area and activity can later be scrutinized with a metabolomic approach to be able to tell which peaks possess an essential role in antimicrobial activity (Yuliana et al., 2011; Yulianto et al., 2016).
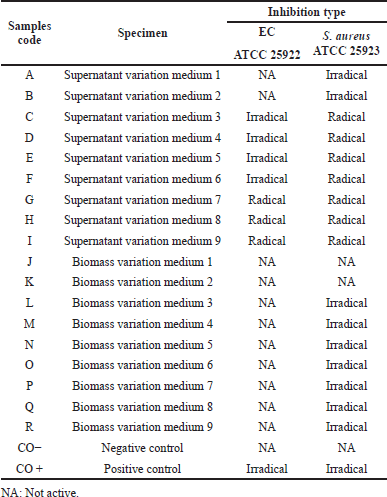 | Table 2. Inhibition type of antimicrobial activity on EC ATCC 25922 and S. aureus ATCC 25923. [Click here to view] |
Figure 3 displays that there are several peaks that possess increase in height and breadth. The changes in peak area and height were associated with the number of secondary metabolites of the fungus T. reesei TV221 generated during the fermentation process. In addition to the ability of the bioactivity of the extract, as previously mentioned, it is affected by the nutrients and conditions of the fungal fermentation medium. In the variation of the fermentation media 7 (G), 8 (H), and 9 (I), in which there is only variation in the salinity of the medium, there are variations in the spectrum pattern that occurs. It can be performed for instance at the peak with Rt 27,423 minutes, in which the peak looks wider and higher as the salinity increases from 7.5 to 30 ppt. Meanwhile, in Figure 3, the spectrum pattern of the supernatant extract from the variation of the fermentation media 3 (C), 6 (F), and 9 (I), in which there is a difference in the variation in the Dextrose content of the medium, presents a significant variation of the medium compared to the spectral pattern of the supernatant extract with variations in the salinity of the medium. It implies that the level of carbon source, which in this study is dextrose, in the fermentation medium of the T. reesei TV221 fungus tremendously affects the formation of volatile secondary metabolites compared to the level of salinity in the medium. It is directly proportional to the antimicrobial activity test results in this study. Secondary metabolites derived from marine sponge-associated fungi can be of various types and possess various kinds of bioactivity. In order to isolate bioactive metabolites from marine sponge-associated fungi, these fungi were fermented in a liquid medium the nutrients of which were formulated as close as possible to the sponge host. It is intended that marine sponge-associated fungi be able to generate secondary metabolites which are as similar as possible to those in their natural habitat (Kjer et al., 2010; Stanbury et al., 1995). Several genera of Trichoderma fungi, such as Trichoderma lignorum and Trichoderma viride, possess a tremendous significant response to the levels of carbon sources in their growth, in which escalating levels of carbon sources in their growth media increase the dry biomass weight and maximize the biological activity of these fungi (Fuentes et al., 2015; Gautam et al., 2010; Seto and Tazaki, 1975; Sguros and Simms, 1963). Fungi T. reesei TV221 are one of the fungi isolated from marine biota, which is sponge. Therefore, salinity greatly influences its life. The increase in salinity levels conducted in this study did not significantly influence the secondary metabolites produced by the fungus T. reesei TV221. In this study, salinity variations were administered at levels of 7.5, 15, and 30 ppt; each of these levels did not significantly affect the production of secondary metabolites of the fungus T. reesei TV221. There is a study which explains that an increase in salinity levels that exceed the salinity of seawater resulting in some associated fungal species from the marine environment shows a decrease in growth rate and a decrease in the production of active secondary metabolites. On the other hand, the absence of salinity in the growing medium of associated fungi from the marine environment inhibits the growth and production of secondary metabolites (Bheemaraya et al., 2013; Huang et al., 2011).
Metabolomics approach
The utilization of a metabolomics approach in this study was aimed to correlate the relationship between antimicrobial activity and the chemical profile of GC-MS, the results of the ethyl acetate supernatant extract, and biomass from fermentation with various medium composition variations (Dextrose and salinity). The metabolomics approach cannot be separated from the chemometric analysis. Chemometrics is a measurement of a chemical system or process in special or certain circumstances by implementing mathematical or statistical methods. One of the objectives of utilizing chemometrics is qualitative, which aims to recognize chemical patterns (without supervision) (Brereton, 2003; Gemperline, 2006). The chemical data employed in this study are the Rt and the area under the peak on the chromatogram. These chemical data are presented in variables (multivariate data) which are arranged in the same number of dimensions as the number of existing variables. Modeling utilizing chemometrics requires instruments and software to interpret the data. The chemometrics method administered in this study is cluster analysis with a hierarchical approach and PCA (Berrueta et al., 2007; Hanrahan and Gomez, 2010; Rohman, 2014).
The metabolomics approach by employing the chemometric analysis method hierarchical cluster analysis (HCA) is in accordance with the creation of branched structures, which are acknowledged as dendrogram clusters. This dendrogram demonstrates multivariate data qualitatively and provides cluster visualization and correlations between samples. The prior objective of the HCA method is to visualize data in a cluster which is in a two-dimensional space (Beebe et al., 1998; Brereton, 2003). In this study, the HCA method was administered to cluster the GC-MS chemical profile pattern which would later be associated with its antimicrobial activity. In Figure 4, a cluster dendrogram of the GC-MS chemical profile of each sample in this study is displayed. It can be perceived from the dendrogram that the clusters are extensively divided into two major groups; the two groups encompass ethyl acetate supernatant extract and biomass ethyl acetate extract. It presents that the GC-MS chemical profile of the supernatant extract and biomass possesses a different pattern. These different GC-MS chemical profile patterns can represent that the secondary metabolites extracted from the supernatant and biomass possess different amounts and types; hence, they can be clustered as a group of supernatant and biomass extracts. For clustering associated with antimicrobial bioactivity, it can be perceived that, in general, it has been clustered in accordance with the existing antimicrobial bioactivity. On the antimicrobial activity of S. aureus ATCC 25923, the dendrogram is clustered according to the activity that occurs. The ethyl acetate supernatant extract with medium variations of 6 (F), 7 (G), 8 (H), and 9 (I) was revealed to be clustered in one group; this extract owned a robust category of antimicrobial activity for S. aureus ATCC 25923. For supernatant extracts with medium variations of 1 (A), 2 (B), 3 (C), 4 (D), and 5 (E), this group of extracts possessed moderate to strong antimicrobial activity for S. aureus ATCC 25923 bacteria. For the ethyl acetate biomass extract with medium variations of 5 (N), 6 (O), 7 (P), 8 (Q), and 9 (R), these five biomass extracts were clustered in one group with antimicrobial activity category, medium on S. aureus ATCC 25923. For ethyl acetate biomass extract with medium variations of 1 (J), 2 (K), 3 (L), and 4 (M), these biomass extract were grouped in one with the same antimicrobial activity and it showed to moderate activity. In the ethyl acetate supernatant extract with medium variations of 6 (F), 7 (G), 8 (H), and 9 (I), it can be identified that the four extracts are clustered in one group, where this extract possesses an antimicrobial activity of moderate to robust category for bacteria EC ATCC 25922. For the ethyl acetate extract supernatant variation of media 1 (A), 2 (B), 3 (C), 4 (D), and 5 (E), these five extracts were classified into one group and acknowledged to possess antimicrobial activity category, with no to moderate antimicrobial activity for EC ATCC 25922. Meanwhile, for all biomass ethyl acetate extracts with medium variations of 1 (J) to 9 (R), it is identified that they do not possess antimicrobial activity for bacteria EC ATCC 25922, and these nine extracts were clustered in one group. Therefore, what can be identified from the metabolomic approach by employing the HCA method is that it is able to cluster extracts that possess similar antimicrobial activity in accordance with the GC-MS chemical profile. Hence, the metabolomics approach by utilizing the HCA method is appropriate for antimicrobial studies of the fungi T. reesei TV221 as it is able to cluster extracts which possess antimicrobial potential and those that do not own antimicrobial potential.
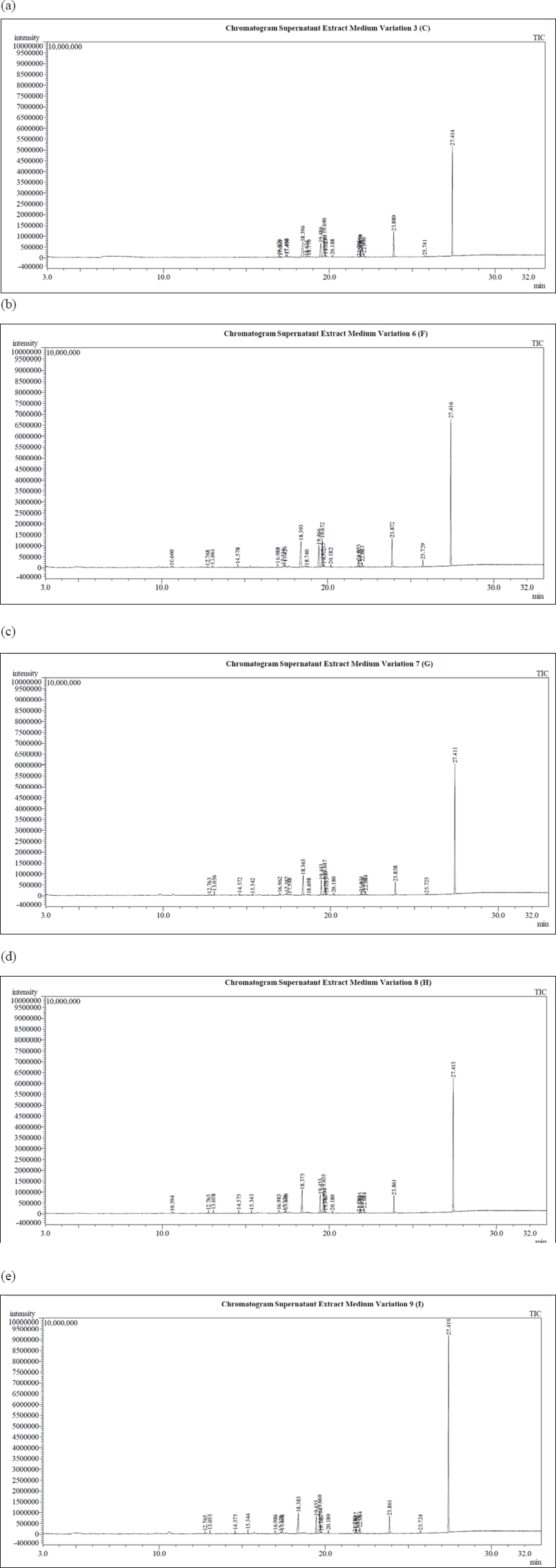 | Figure 3. (a) GC-MS chemical profile of supernatant extract variation medium 3 (C). (b) GC-MS chemical profile of supernatant extract variation medium 6 (F). (c) GC-MS chemical profile of supernatant extract variation medium 7 (G). (d) GC-MS chemical profile of supernatant extract variation medium 8 (H). (d) GC-MS chemical profile of supernatant extract variation medium 9 (I). [Click here to view] |
For the metabolomics approach, the PCA method aims to decrease the large dimensions of the data space into small dimensions the data space. Thus, it can be demonstrated in a simple way. PCA is able to reduce the number of variables in a matrix to generate new variable data while maintaining the information possessed by the data before it is simplified. This method is able to produce advantage of subtle differences from chemical profile spectral data and reduce the influence of noise on the spectrum (Che Man et al., 2011; Pratiwi and Harjoko, 2013). Data from the PCA method can be interpreted through loading analysis. Loading is the relationship between the actual variable and the newly formed variable; hence, it provides data indicating the original variable that is tremendously essential or influences the formation of new variables, in which the higher the loading value is, the more influential the original variable is in the formation of new variables. Furthermore, the interpretation of PCA data can be perceived from the biplot. There is some crucial information perceived from the biplot display, that is, the diversity of variables, the value of the variable on an object, the closeness between the observed objects, and the correlation between variables (Che Man et al., 2011; Sartono et al., 2003; Sharma, 1996). The scatter plot data from PCA in this study is illustrated in Figure 5. Meanwhile, the loading plot data PC1 and PC2 are presented in Figure 6. In the scatter plot data from PCA, it can be perceived that the extracts are clustered and this grouping of extracts is in accordance with the results of the metabolomics approach by employing the HCA method, as presented above that the extract which owns the potential as an antimicrobial agent is the ethyl acetate supernatant extract with a variety of media 6 (F), 7 (G), 8 (H), and 9 (I). These results certainly strengthen the metabolomics approach for extracts that own potential as antimicrobials which possess similar chemical profiles to GC-MS; hence, peaks can be estimated that own a major influence on antimicrobial activity.
To be able to predict peaks that possess a large influence on antimicrobial activity, further identification is administered on the loading plot data from PCA. In Figure 6, it can be perceived that there are two loading plots of PCA. Figure 6a is the loading plot of PC1 and Figure 6b is the loading plot of PC2. In accordance with scatter plot data from PCA (Fig. 5), the position of the points representing extracts which own the most potent antimicrobial activity is placed in quadrant I, where quadrant I possess positive eigenvalues of PC1 and PC2. Referring back to the loading plots of PC1 and PC2, the peaks affecting the extract points that possess the most potent antimicrobial activity are peaks with positive eigenvalues both from PC1 and from PC2. After identification, the peaks were at Rts as follows: 10,600, 12,768, 13,069, 14,577, 15,346, 17,325, 19,740, 21,934, 27,423 minutes. The eight peaks with Rt own positive eigenvalues for both PC1 and PC2. The eight peaks were then examined by employing mass spectroscopy to acquire their mass spectrum. The mass spectra of the eight peaks were then synchronized with the WILEY7.LIB library. After being compared with the WILEY7.LIB library, the predictions of compounds that are similar are acquired in accordance with the adjusted library and the SI. The eight compounds were successfully estimated by the WILEY7.LIB library and sorted associated with their Rts as follows: mevalonolactone (Rt 10,600 minutes); methyl (2-hydroxyphenyl) acetate (Rt 12,768 minutes); methyl 2-hydroxy-3-phenylpropionate (Rt 13,069 minutes); methyl (p-hydroxyphenyl) acetate (Rt 14,577 minutes); methyl ester of N-isovaleryl-L-isoleucine (Rt 15,346 minutes); methyl ester of N-isovaleryl-phenylalanine (Rt 17,325 minutes); 1,4-diaza-2,5-dioxobicyclo[4.3.0]nonane (Rt 19,740 minutes); 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo[4.3.0]nonane (Rt 21,934 minutes); 3-benzyl-1,4-diaza-2,5-dioxobicyclo[4.3.0]nonane (Rt 27,423 minutes). Looking at the SI values of all these compounds, there are two compounds that do not require the SI criteria expected from this study (minimum SI 80), methyl ester of N-isovaleryl-L-isoleucine (Rt 15.346 minutes) and methyl ester of N-isovaleryl-phenylalanine (Rt 17,325 minutes), which possess SI values of 72 and 74, respectively; thus, it cannot be incorporated in the compounds predicted to own antimicrobial activity in this study. Therefore, the remaining six compounds were estimated to possess antimicrobial activity. After further searching the literature, it was discovered that there were three compounds that had been previously examined and were identified to possess antimicrobial activity. The three compounds were 1,4-diaza-2,5-dioxobicyclo[4.3.0]nonane (Rt 19,740 minutes); 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo[4.3.0]nonane (Rt 21,934 minutes); 3-benzyl-1,4-diaza-2,5-dioxobicyclo[4.3.0]nonane (Rt 27,423 minutes). The mass spectra of the three compounds are demonstrated in Figure 7.
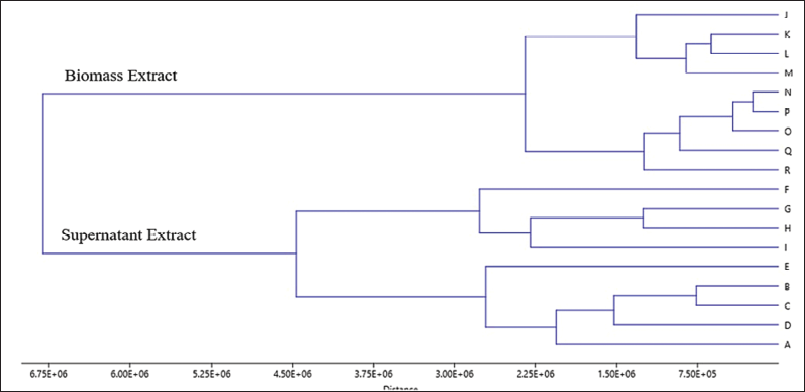 | Figure 4. Dendogram of HCA from the sample extracts. [Click here to view] |
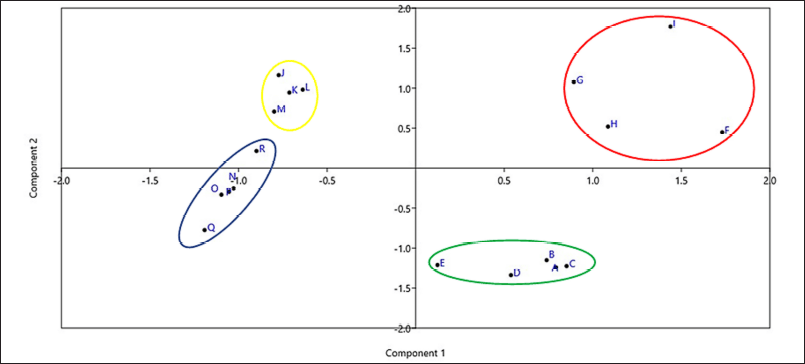 | Figure 5. Scatter plot of PCA. [Click here to view] |
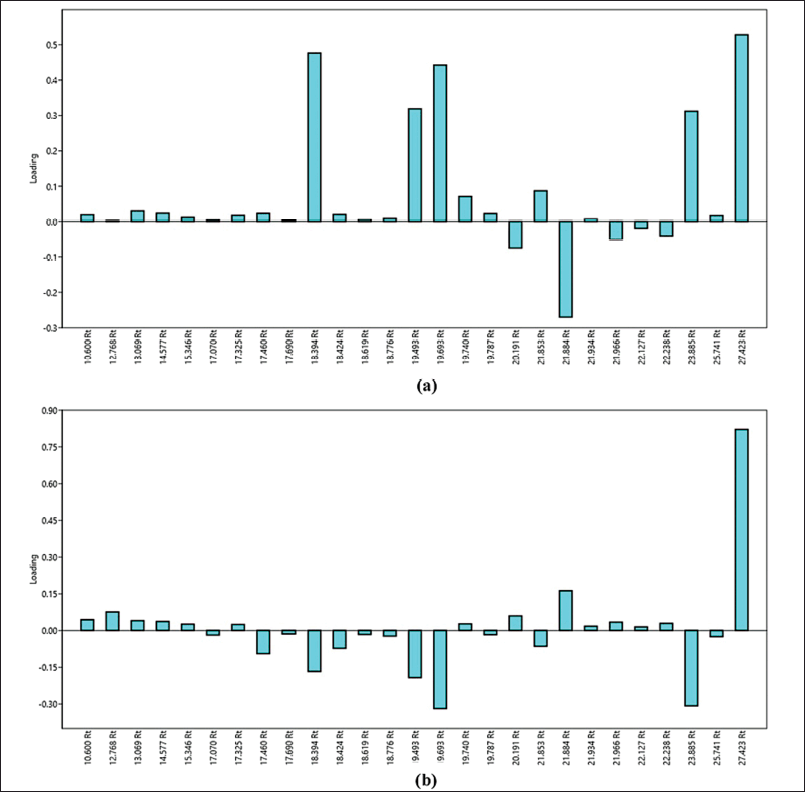 | Figure 6. Loading plot of PCA from PC1 (a) and PC2 (b). [Click here to view] |
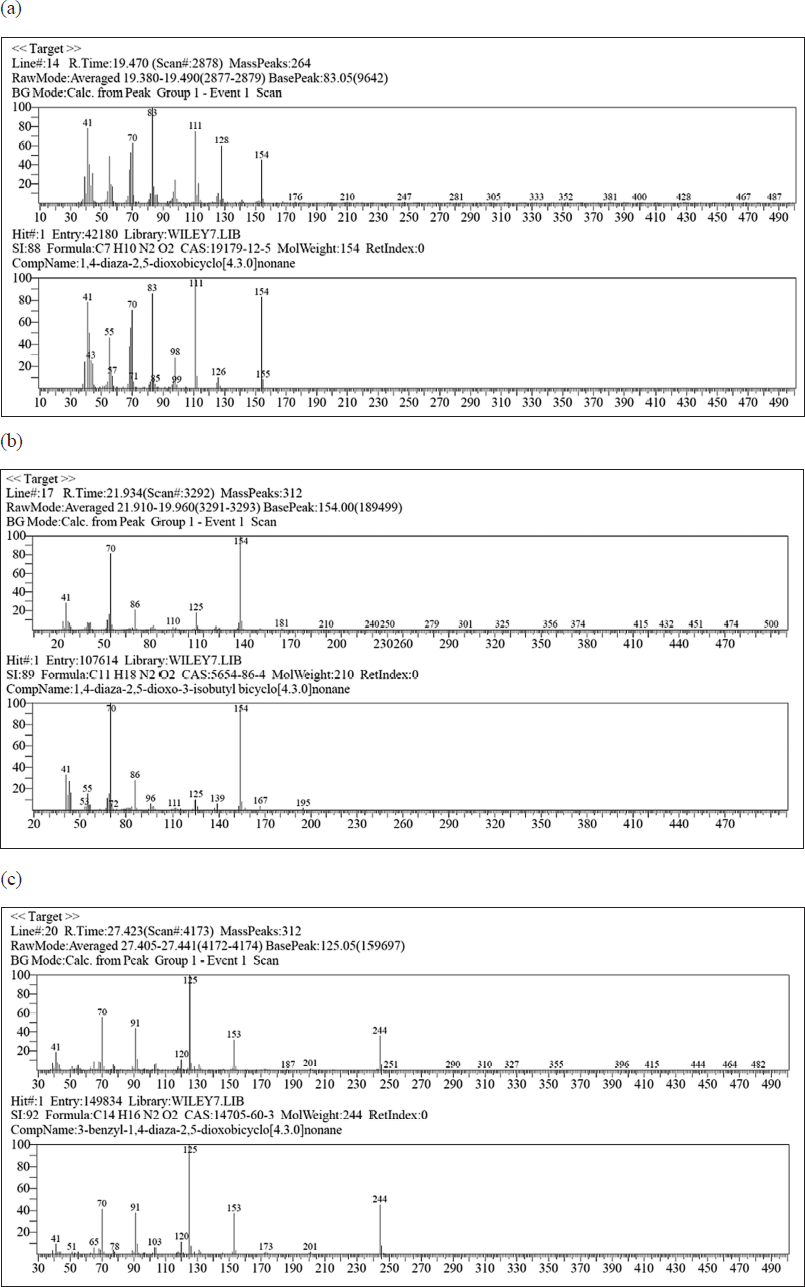 | Figure 7. (a) Mass spectrum from the peak with Rt 19,740 minutes. (b) Mass spectrum from the peak with Rt 21,934 minutes. (c) Mass spectrum from the peak with Rt 27,423 minutes. [Click here to view] |
The chemical compound 1,4-diaza-2,5-dioxobicyclo[4.3.0]nonane whose peak appeared at Rt 19,740 minutes and the chemical compound 3-benzyl-1,4-diaza-2,5-dioxobicyclo[4.3.0]nonane whose peak appeared at Rt 27,423 minutes were identified to possess antimicrobial activity in previous studies. Both compounds have been acknowledged to possess antimicrobial activity on Aeromonas hydrophila, Edwardsiella tarda, and Vibrio ordalii bacteria, all of which are Gram-negative bacteria. Furthermore, other studies also uncovered that these two compounds possess antimicrobial activity on S. aureus and Bacillus subtilis bacteria which are encompassed in Gram-positive bacteria. In addition to possessing antimicrobial activity, these two compounds are also discovered to possess antioxidant activity (Gohar et al., 2010; Graz et al., 1999; Kusumawati et al., 2021). The 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo[4.3.0]nonane compound in the previous study was also revealed to possess antimicrobial activity. These compounds are identified to possess antimicrobial activity on several bacteria, encompassing S. aureus, B. subtilis, Pseudomonas aeruginosa, Vibrio parahaemolyticus, and EC Moreover, 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo[4.3.0]nonane is also understood to possess antioxidant activity (Azman et al., 2017; Raharja et al., 2019). A compound that is identified to own antioxidant activity is also likely to possess antimicrobial activity (Samirana et al., 2017). Based on the literature search, it can be indicated that the compounds 1,4-diaza-2,5-dioxobicyclo[4.3.0]nonane; 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo[4.3.0]nonane; 3-benzyl-1,4-diaza-2,5-dioxobicyclo[4.3.0]nonane are antimicrobial compounds in bacteria S. aureus ATCC 25923 and EC ATCC 25922 which are potentially present in the ethyl acetate supernatant extract in various media as 6 (F), 7 (G), 8 (H), and 9 (I).
CONCLUSION
The fungi T. reesei TV221 which were cultured on nine variations of the fermentation medium with various variations of Dextrose and salinity levels produced 18 different types of extracts taken from the supernatant and biomass. All of these extracts were examined for antimicrobials on S. aureus ATCC 25923 and EC ATCC 25922, and it was discovered that the ethyl acetate supernatant extract from various media as 6 (F), 7 (G), 8 (H), and 9 (I) owned potential antimicrobial activity. The GC-MS chemical profiles of the extracts with different salinity and Dextrose concentrations differ from one another. There was a difference in the chemical profile of GC-MS between the extracts with salinity variation and the extracts with Dextrose variation, with the chemical profile of GC-MS in the extracts with Dextrose variation being more obvious. As a result, the formation of secondary metabolites and the antimicrobial activity of the fungus T. reesei TV221 are more dependent on the role of Dextrose as a carbon source in the fermentation medium than they are on salinity. Three compounds with antimicrobial activity are predicted by the metabolomic approach using the HCA and PCA methods. The three compounds were 1,4-diaza-2,5-dioxobicyclo[4.3.0]nonane; 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo[4.3.0]nonane; 3-benzyl-1,4-diaza-2,5-dioxobicyclo[4.3.0]nonane. To be able to determine which compounds possess the highest potential as antimicrobials, it is necessary to isolate the three compounds and administer antimicrobial tests on every single compound.
ACKNOWLEDGMENTS
The authors would like to thank the Minster of Research and Higher Education of Republic of Indonesia for funding this work (1620/UNl/DITLIT /Dit-Lit/PT .01.03/2022).
LIST OF ABBREVIATION
ATCC: American type culture collection; GC-MS: Gas chromatography-mass spectroscopy; ICBB: Indonesian Center for Biodiversity and Biotechnology Bogor; PCA: Principle component analysis; HCA: Hierarchical cluster analysis ; MRSA: Methicillin-resistant Staphylococcus aureus.
AUTHOR CONTRIBUTIONS
Conceptualization, E. P. S. and P. O. S.; investigation, E. P. S., P. O. S., Y. B. M. and R. I. J.; writing—preparation of original draft, E. P. S., P. O. S. and Y. B. M.; writing—review and editing, E. P. S., P. O. S., Y. B. M. and R. I. J.; visualization, P. O. S. and Y. B. M.; surveillance, E. P. S.; project administration, E. P. S. All authors have read and approved the published version of the manuscript.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Azman AS, Othman I, Fang CM, Chan KG, Goh BH, Lee LH. Antibacterial, anticancer and neuroprotective activities of rare Actinobacteria from mangrove forest soils. Indian J Microbiol, 2017; 57:177–87; https://doi.org/10.1007/s12088-016-0627-z
Beebe KR, Pell RJ, Seasholtz MB. Chemometrics: a practical guide. Wiley, New York, NY, 1998.
Berrueta LA, Alonso-Salces RM, Héberger K. Supervised pattern recognition in food analysis. J Chromatogr A, 2007; 1158:196–214; https://doi.org/10.1016/j.chroma.2007.05.024
Bheemaraya PMB, Ramesh YST, Amaresh YS, Naik MK. Salinity stress tolerance in native Trichoderma isolates. Environ Ecol, 2013; 31:727–9.
Brereton RG. Chemometrics: data analysis for laboratory and chemical plant. John Wiley and Sons Ltd, Chichester, UK, 2003.
Brinkmann CM, Marker A, Kurtböke DI. An overview on marine sponge-symbiotic bacteria as unexhausted sources for natural product discovery. Diversity, 2017; 9:1–31; https://doi.org/10.3390/d9040040
Che Man YB, Rohman A, Mansor TST. Differentiation of lard from other edible fats and oils by means of Fourier transform infrared spectroscopy and chemometrics. J Am Oil Chem Soc, 2011; 88:187–92; https://doi.org/10.1007/s11746-010-1659-x
Cometto-Muñiz JE, Cain WS, Abraham MH. Determinants for nasal trigeminal detection of volatile organic compounds. Chem Senses, 2005; 30:627–42; https://doi.org/10.1093/chemse/bji056
Davis WW, Stout TR. Disc plate method of microbiological antibiotic assay. II. Novel procedure offering improved accuracy. Appl Microbiol, 1971; 22:666–70; https://doi.org/10.1128/aem.22.4.666-670.1971
Debbab A, Aly AH, Proksch P. Bioactive secondary metabolites from endophytes and associated marine derived fungi. Fungal Divers, 2011; 49:1–12; https://doi.org/10.1007/s13225-011-0114-0
Fuentes ME, Quiñones RA, Gutiérrez MH, Pantoja S. Effects of temperature and glucose concentration on the growth and respiration of fungal species isolated from a highly productive coastal upwelling ecosystem. Fungal Ecol, 2015; 13:135–49; https://doi.org/10.1016/j.funeco.2014.09.006
Gautam SP, Bundela PS, Pandey AK, Jamaluddin, Awasthi MK, Sarsaiya S. Optimization of the medium for the production of cellulase by the Trichoderma viride using submerged fermentation. Int J Environ Sci, 2010; 1:656–65.
Gemperline P. Practical guide to chemometrics. Second. Taylor & Francis, Boca Raton, FL, 2006.
Gohar YM, El-Naggar MMA, Soliman MK, Barakat KM. Characterization of marine Burkholderia cepacia antibacterial agents. J Nat Prod, 2010; 74:86–94.
Graz M, Hunt A, Jamie H, Grant G, Milne P. Antimicrobial activity of selected cyclic dipeptides. Pharmazie, 1999; 54:772–5.
Han J, Datla R, Chan S, Borchers CH. Mass spectrometry-based technologies for high-throughput metabolomics. Bioanalysis, 2009; 1:1665–84; https://doi.org/10.4155/bio.09.158
Hanrahan G, Gomez FA. Chemometric methods in capillary electrophoresis. J. Wiley & Sons, Hoboken, NJ, 2010.
Huang J, Lu C, Qian X, Huang Y, Zheng Z, Shen Y. Effect of salinity on the growth, biological activity and secondary metabolites of some marine fungi. Acta Oceanol Sin, 2011; 30:118–23; https://doi.org/10.1007/s13131-011-0126-3
Humphris SN, Bruce A, Buultjens E, Wheatley RE. The effects of volatile microbial secondary metabolites on protein synthesis in Serpula lacrymans. FEMS Microbiol Lett, 2002; 210:215–9; https://doi.org/10.1016/S0378-1097(02)00604-3
Kjer J, Debbab A, Aly AH, Proksch P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat Protoc, 2010; 5:479–90; https://doi.org/10.1038/nprot.2009.233
Kusumawati DE, Purwanto UMS, Bintang M, Pasaribu FH. Analysis of metabolite compound profiles of miana leaves endophytic bacteria (Coleus scutellariodes) using GC-MS. Curr Biochem, 2021; 8:63–7.
Mishra N, Khan SS, Sundari SK. Native isolate of Trichoderma: a biocontrol agent with unique stress tolerance properties. World J Microbiol Biotechnol, 2016; 32:1–23; https://doi.org/10.1007/s11274-016-2086-4
Moore-Landecker E. Fundamentals of the fungi. Fourth. Prentice Hall, Hoboken, NJ, 1996.
Muthukumar A, Venkatesh A. Physiological studies of Sclerotium rolfsii Sacc. causing collar rot of peppermint. African J Biotechnol, 2013; 12:6837–42; https://doi.org/10.5897/AJB2013.13201
Pelczar MJ, Chan ECS. Dasar-Dasar Mikrobiologi I. Jilid 1. UI-Press, Jakarta, Indonesia, 2008a.
Pelczar MJ, Chan ECS. Dasar-Dasar Mikrobiologi II. Jilid 2. UI-Press, Jakarta, Indonesia, 2008b.
Pratiwi DE, Harjoko A. Implementasi pengenalan wajah menggunakan PCA (principal component analysis). IJEIS, 2013; 3:175–84; https://doi.org/10.1002/jlac.19335020105
Raharja NI, Widanarni, Wahyudi AT. Marine sponge-associated bacteria as biocontrol agents of vibriosis on whiteleg shrimp caused by Vibrio parahaemolyticus. Biodiversitas, 2019; 20:3164–9; https://doi.org/10.13057/biodiv/d201108
Rohman A. Statistika dan Kemometrika dasar dalam analisis farmasi. Pustaka Pelajar, Yogyakarta, Indonesia, 2014.
Samirana PO, Jenie RI, Murti YB, Setyowati EP. Application of metabolomics on marine sponges and sponge-associated microorganisms: a review. J Appl Pharm Sci, 2022; 12:18–33; https://doi.org/10.7324/JAPS.2022.120702
Samirana PO, Murti YB, Jenie RI, Setyowati EP. Marine sponge-derived fungi: fermentation and cytotoxic activity. J Appl Pharm Sci, 2021a; 11:21–39; https://doi.org/10.7324/japs.2021.110103
Samirana PO, Murti YB, Istighfari Jenie R, Prawita Setyowati E. Antibacterial and cytotoxic activities of supernatant and mycelium extracts from fermentation of fungal symbiont Trichoderma reesei TV221. J Appl Pharm Sci, 2021b; 11:90–9; https://doi.org/10.7324/japs.2021.1101207
Samirana PO, Susidarti RA, Rohman A. Isolation and 2,2’-diphenyl-1-picrylhydrazyl radical scavenging activity of active compound from Jujube tree (Zizyphus mauritiana Auct. non Lamk.). Int J Food Prop, 2017; 20:1523–9; https://doi.org/10.1080/10942912.2016.1233427
Sánchez-Montesinos B, Diánez F, Moreno-Gavira A, Gea FJ, Santos M. Plant growth promotion and biocontrol of Pythium ultimum by saline tolerant Trichoderma isolates under salinity stress. Int J Environ Res Public Health, 2019; 16:1–11; https://doi.org/10.3390/ijerph16112053
Sartono B, Affendi FM, Syafitri UD, Sumertajaya I, Angraeni Y. Analisis Peubah Ganda. IPB Press, Bogor, Indonesia, 2003.
Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Tunali UR, Forbes MG, Willmitzer L, Fernie AR, Kopka J. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett, 2005; 579:1332–7; https://doi.org/10.1016/j.febslet.2005.01.029
Seto M, Tazaki T. Growth and respiratory activity of mold fungus (Trichoderma lignorum). Bot Mag Tokyo, 1975; 88:255–66; https://doi.org/10.1007/BF02488368
Setyowati EP, Pratiwi SUT, Hertiani T, Samirana O. Bioactivity of fungi Trichoderma reesei, Associated with sponges (Stylissa flabelliformis) collected from National ark West Bali, Indonesia. J Biol Sci, 2017; 17:362–8; https://doi.org/10.3923/jbs.2017.362.368
Setyowati EP, Pratiwi SUT, Purwantiningsih, Samirana PO. Antimicrobial activity and identification of fungus associated (Stylissa flabelliformis) sponge collected from Menjangan Island West Bali National Park, Indonesia. Indones J Pharm, 2018; 29:66–73; https://doi.org/10.14499/indonesianjpharm29iss2pp66
Sguros P, Simms J. Role of marine fungi in the biochemistry of the oceans. II. Effect of glucose, inorganic nitrogen, and tris (Hydroxymethyl) aminomethane on growth and pH changes in synthetic media. Mycologia, 1963; 55:728–41; https://doi.org/10.1080/00275514.1963.12018064
Sharma S. Applied multivariate techniques. John Wiley and Sons, New York, NY, 1996.
Sibero MT, Radjasa OK, Sabdono A, Trianto A, Triningsih DW, Hutagaol ID. Antibacterial activity of Indonesian sponge associated fungi against clinical pathogenic multidrug resistant bacteria. J Appl Pharm Sci, 2018; 8:088–94; https://doi.org/10.7324/JAPS.2018.8214
Stanbury PF, Whitaker A, Hall SJ. Principles of fermentation technology. 2nd edition, Elsevier, London, UK, 1995; https://doi.org/10.1016/0167-7799(85)90016-2
Syukri Y, Fitria A, Hanifah S, Idrati M. Development of new indonesian propolis extract-loaded self-emulsifying: characterization, stability and antibacterial activity. Adv Pharm Bull, 2021; 11:120–9; https://doi.org/10.34172/apb.2021.013
Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev, 2007; 71:295–347; https://doi.org/10.1128/mmbr.00040-06
Tjay TH, Rahardja K. Obat-Obat Penting?: Khasiat, Penggunaan, dan Efek-Efek Sampingnya. Edisi VIII. PT. Elex Media Komputindo, Jakarta, Indonesia, 2015.
Ukhty N, Tarman K, Setyaningsih I. Isolation of endophytic fungi from the coastal plant terong pungo (Solanum sp.) and its antibacterial activity against oral pathogenic bacteria. Biotropia (Bogor), 2017; 24:9–15; https://doi.org/10.11598/btb.2017.24.1.453
Xu CJ, Liang YZ, Chau FT, Heyden Y Vander. Pretreatments of chromatographic fingerprints for quality control of herbal medicines. J Chromatogr A, 2006; 1134:253–9; https://doi.org/10.1016/j.chroma.2006.08.060
Yuliana ND, Khatib A, Choi YH, Verpoorte R. Metabolomics for bioactivity assessment of natural products. Phyther Res, 2011; 25:157–69; https://doi.org/10.1002/ptr.3258
Yulianto W, Andarwulan N, Giriwono PE, Pamungkas J. HPLC-based metabolomics to identify cytotoxic compounds from Plectranthus amboinicus (Lour.) Spreng against human breast cancer MCF-7Cells. J Chromatogr B Anal Technol Biomed Life Sci, 2016; 1039:28–34; https://doi.org/10.1016/j.jchromb.2016.10.024
Zhang L, Parente J, Harris SM, Woods DE, Hancock REW, Falla TJ. Antimicrobial peptide therapeutics for cystic fibrosis. Antimicrob Agents Chemother, 2005; 49:2921–7; https://doi.org/10.1128/AAC.49.7.2921-2927.2005