INTRODUCTION
Clavulanic acid (CA) is an antibiotic of the β-lactam class with moderate antimicrobial activity but a high inhibitory effect on β-lactamase enzymes. CA is one of the most important specialized metabolites produced by Streptomyces clavuligerus; indeed, this strain is still the only industrial source for CA production (López-Agudelo et al., 2021). The CA’s importance lies in its wide use as a medical treatment for infections caused by antibiotic-resistant bacteria, including some of those listed in the World Health Organization’s priority list, such as the case of Streptococcus pneumoniae, Escherichia coli, Neisseria gonorrhoeae, Staphylococcus aureus, Enterobacteriaceae, and Klebsiella species (Gómez-Ríos and Ramírez-Malule, 2019).
The nutritional restriction in S. clavuligerus activates the secondary metabolism, leading to the secretion of penicillin and clavaminic acid derivatives, with cephamicyn C (CephC) and CA being the most important of them. The low CA productivity in the submerged cultivation of S. clavuligerus and the complexity of the downstream process have a direct impact on the production cost of this pharmaceutical compound, which is commonly coformulated with wide-spectrum β-lactam antibiotics. Strategies like nutritional and environmental perturbation appear as feasible alternatives to enhance the strain’s productivity (López-Agudelo et al., 2021; Ramirez-Malule, 2018). Metabolic engineering emerges as an alternative for the rational enhancement of CA in S. clavuligerus cultures through the perturbation of metabolic and regulatory features (Liras and Martín, 2021).
In this sense, the systems biology approaches have contributed to the understanding of S. clavuligerus metabolism under different nutritional and environmental conditions (Gómez-Rios et al., 2022; Ramirez-Malule et al., 2021). Nevertheless, the classical steady-state approaches like the flux balance analysis (FBA) present limitations like the prediction of cellular growth and product secretion rates only for fixed values of substrate uptake rates. In addition, it has strict applicability to the balanced growth phase in batch cultures or steady-state growth in continuous cultures. The intrinsic dynamic condition of batch and fed-batch cultivations has repercussions on the prediction of cell phenotypical states, product yield, and bioreactor control. Thus, modeling tools like FBA must be adapted to cover dynamic states that allow us to get a more accurate description of cellular physiology during such nonsteady processes. Indeed, the highest CA titers for wild-type S. clavuligerus cultivation have been reported in batch and fed-batch operation; thus, high CA productivity necessarily must consider this operation mode. In this work, steady-state FBA was used for the in silico identification of potential gene targets exerting a positive effect on the CA biosynthesis. Afterward, a dynamic FBA (dFBA) framework was used for the in silico evaluation of fed-batch production scenarios, potentially leading to CA overproduction considering the identified gene targets as promissory alternatives for further S. clavuligerus strain engineering.
MATERIALS AND METHODS
Genome-scale metabolic model and sensitivity analysis
The validated genome-scale metabolic model iDG1237 of S. clavuligerus metabolism was used in both FBA and dFBA methodologies (Gómez-Ríos et al., 2020). The iDG1237 biomodel consists of 2,177 reactions, 543 internal/external metabolites, 1,237 annotated genes, and it has been checked for thermodynamic and topological consistency (Gómez-Ríos et al., 2020). Our experimental studies have previously determined suitable environmental and nutritional conditions favoring CA production in wild-type S. clavuligerus (Gómez-Ríos et al., 2019; Gómez-Rios et al., 2022). In this regard, glycerol, glutamate, and oxygen have been identified as key nutrients in the CA biosynthetic pathway, which is activated under phosphate limitation. Previous studies have not considered CA production as a physiological objective in the FBA formulation. In this study, the FBA simulations were performed for exploring the effect of glycerol, glutamate, oxygen, ammonium, phosphate, CephC, and clavam-2-carboxylate (a clavam metabolite) on both physiological objectives, biomass synthesis (Z) and CA secretion flux (vCA,ex). The FBA formulations are described by Equations (1) and (2), respectively.
where S is the stoichiometric matrix; v is the reaction fluxes; and lb and ub are the lower and upper bounds for the metabolic fluxes, respectively.
After the sensitivity analysis, the genome-scale model was used for the identification of potential metabolic targets for gene knockout, dampening, and/or overexpression, which might favor the carbon flux toward the CA biosynthetic pathway. Initially, a gene essentiality analysis in a synthetic medium according to the usual production conditions was performed. Then, the application of the (robust, overexpression, knockout, and dampening) pipeline allowed the determination of the potential gene candidates for strain engineering (Stanford et al., 2015).
dFBA implementation
The dFBA was formulated according to the static optimization approach, which assumes a pseudo-steady-state condition in each time interval, and the optimization FBA problem is solved. The FBA problem is constrained dynamically for each time interval by the solution of a set of ordinary differential equations (ODE), whose parameters correspond to experimentally measured rates in S. clavuligerus wild-type cultivations. The set of ordinary equations [Eqs. (3)–(5)] accounts for the dynamics of growth rate, glycerol, glutamate, phosphate, and ammonia depletions, as well as gas transfer rates for oxygen and carbon dioxide. Based on the simplified stoichiometry of biomass synthesis, a kinetic Monod model was proposed for growth, as shown in Equation (6).
where Ci is the concentration of i species, X is the biomass concentration, F is the constant feed rate during fed-batch operation (from t = 34 hours onwards), and V is the volume. For the Monod growth rate expression [Eq. (6)], μD is the specific growth rate for the dynamic model, Kd is the specific death constant, Ks is the affinity constant, and Cs is the concentration of the carbon source. The values of the kinetic used are currently available in the literature for wild-type S. clavuligerus (Sanchez et al., 2012). dFBA simulations for the in silico engineered strains were conducted under the same kinetic macroscopic parameters validated for wild-type fed-batch cultivation since the carbon flux toward secondary metabolism is very low in comparison with the central metabolism (Gómez-Ríos et al., 2022). The initial conditions are presented in Table 1.
dFBA was constrained by the solutions of the ODE set and steady-state constraints for exchange reactions of low-concentrated nutrients and metabolites. The growth rate estimated is conciliated through the error minimization between the growth rates predicted by solving the FBA and ODE set, as observed in Equation (7). The dFBA definition is presented in Equation (8).
s?t
where μF is the growth rate estimated for the pseudo-steady-state model, vm is the fluxes calculated by integration of the ODE set, and vn is the fluxes determined from the optimization of the FBA problem [Eq. (14)]. The FBA was implemented in COBRA Toolbox 3.0 for MATLAB. The optimization steps were solved with Gurobi 7.5 MATLAB API.
RESULTS AND DISCUSSION
Sensitivity analysis of nutrients uptake and antibiotics secretion
The effects of key nutrients variation on biomass synthesis and CA secretion were assessed via FBA. Additionally, the effects of CephC and clavam-2-carboxylate biosynthesis as the respective products of cephalosporins and clavams 5S pathways were also considered since the synthesis of these compounds were expected to compete with CA biosynthesis. The results of the effect of the abovementioned variables on the specific growth rate are shown in Figure 1.
The increase in the uptake of carbon and nitrogen, i.e., glycerol, glutamate, and ammonium, entails a positive effect on the growth rate since those are the main nutrients during the cultures. The increase in glycerol influx supports biomass synthesis up to 0.095 hour−1, as shown in Figure 1a. Moreover, glutamate may further enhance the growth rate (up to 0.25 hour−1) due to its content of carbon and nitrogen and its direct incorporation into the urea cycle as an amino acid precursor. Ammonium as the main nitrogen source is essential for growth and promotes growth as long as enough carbon source, either glycerol or glutamate, is provided (Fig. 1b). In the case of phosphate, there exists a threshold in the phosphate influx to support the biomass accumulation and CA biosynthesis (Saudagar and Singhal, 2007). As shown in Figure 1b, phosphate acts as a biomass precursor but uptake rates over 0.2 mmol.gram dry cell weight (gDCW.hour)−1 might inhibit growth. Interestingly, the phosphate limitation slightly decreases the growth rate but does not cause inhibition, suggesting that metabolic mechanisms of phosphate compensation and energetic regulation might compensate for the phosphate scarcity, as indeed occurs in other Streptomyces species, in which this correlates with antibiotics production (Esnault et al., 2017; Millan-Oropeza et al., 2020).
In S. clavuligerus, secondary metabolism demands for substrates that are simultaneously required for growth as in the case of amino acids from the urea cycle. Specifically, CA biosynthesis requires glyceraldehyde-3-phosphate (GAP), arginine, and tricarboxylic acid (TCA) cycle intermediates. As shown in Figure 1c, the increase in CA synthesis beyond 0.067 mmol. (gDCW.hour)−1 in wild-type S. clavuligerus is expected to be detrimental to biomass synthesis. The CephC biosynthesis requires valine and cysteine, also biomass precursors. Similarly, CephC secretion affects biomass production but to a lesser extent. These results suggest that feasible phenotypes producing either CA or CephC can be attained at specific growth rates higher than 0.04 hour−1 and lower than 0.9 hour−1.
According to previous experimental observations in batch and fed-batch cultures of wild-type S. clavuligerus, a minimum specific growth rate of 0.05 hour−1 was defined for FBA with CA secretion flux as the objective function (Gómez-Rios et al., 2022; Gómez-Ríos et al., 2019). CA production is not possible without providing glycerol due to its incorporation as GAP into the glycolytic pathway; thus, an increase in glycerol uptake would favor the CA secretion up to a maximum of 0.09 mmol. (gDCW.hour)−1, as shown in Figure 1d. The simultaneous availability of glutamate enhances CA production (Fig. 1d) by increasing the flux of arginine from the urea cycle, which is an early precursor of CA. Similarly, the assimilation of ammonium is essential for secondary metabolism; as can be seen in Figure 1e, an increase in the ammonium uptake is expected to impact positively the CA production. Figure 1e also shows that the maximum CA production is attained when the phosphate limitation is reached, and no production occurs at phosphate uptake higher than 0.28 mmol. (gDCW.hour)−1. Indeed, the phosphate limitation has been reported as a condition triggering the biosynthesis of antibiotics in several species of Streptomyces (Barreiro and Martínez-Castro, 2019).
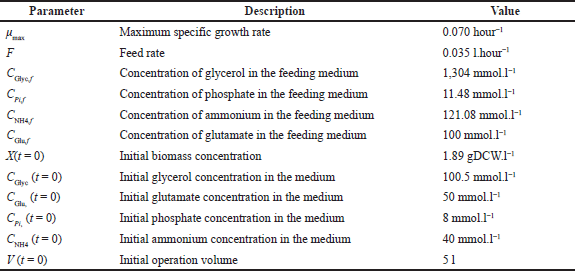 | Table 1. Initial conditions for the kinetic model of S. clavuligerus. [Click here to view] |
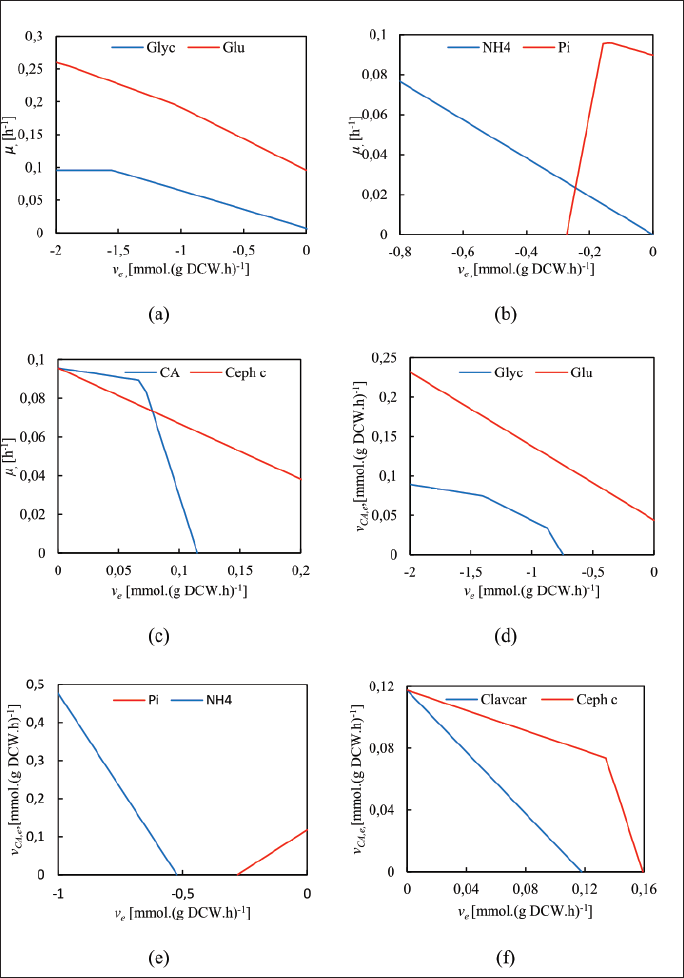 | Figure 1. Effect of nutrient uptake and secondary metabolites on the specific growth rate (μ) and CA secretion (vCA,e). [Click here to view] |
Figure 1f shows that the higher the CephC biosynthesis flux, the lower the CA production. CephC biosynthesis has been reported as a competitor pathway of CA biosynthesis, although both can be secreted under appropriate nutritional conditions (Bellão et al., 2013). This is probably linked to the consumption of 2-oxoglutarate and oxygen in the oxidation steps of the pathway which also occurs in CA biosynthesis. Moreover, the synthesis of CephC amino acid precursors (valine, cysteine, and lysine) requires the flux of glutamate as also occurs with arginine in the urea cycle and further CA synthesis. The clavam-2-carboxylate is a product of the 5S clavam pathway. Figure 1f also suggests that the higher the clavam 2-carboxylate production, the lower the CA secretion. The 5S clavams pathway is a direct competitor of CA biosynthesis since both pathways use the same precursor, clavaminic acid, whose synthesis acts as a bifurcation point between both pathways (Tahlan et al., 2007).
Potential gene targets for enhancing CA production
The reaction essentiality analysis performed with the iDG1237 biomodel showed that 216 reactions (9.7%) are essential for growth in the synthetic medium; these reactions are associated with 109 genes, which is 8.8% of the genes annotated. In the context of the pseudo-steady-state condition, 96 reactions were identified initially as potential targets for a single or double knockout and 72 for overexpression or dampening linked to the objective of increasing CA production. The ratio of growth rate-CA flux to glycerol uptake (μ.vCA,e/vGlyc,e) was used as an indicator for selecting the best potential candidates for the strain design. For the case of wild-type S. clavuligerus, this ratio was 0.0024; on the contrary, for the best candidate, i.e., overexpression of genes encoding (CEAS) enzyme N2-(2-carboxyethyl)-arginine synthase, this ratio was 0.028. A summary of the best potential target candidates for strain engineering is presented in Table 2.
Some studies have focused on the genetic engineering of expression levels of genes directly associated with the biosynthetic pathway of CA or its regulation. Consistent with what has been reported by other authors, the overexpression of the regulatory genes associated with the CA synthesis would generate significant increases in its production (Jnawali et al., 2011; Kurt-Kizildo?an et al., 2017; Song et al., 2010). In our study, the best ranking in increasing the CA secretion was obtained for the overexpression of the enzymes in the early steps of the CA biosynthesis pathway. Particularly, the overexpression of CEAS leads to an increase in the carbon flux along the complete CA pathway, leading to an increase in the CA secretion. In our FBA simulations, no significant fluxes through the 5S clavams pathway were observed under phosphate limitation and adequate carbon, nitrogen, and oxygen supply. Therefore, all the carbon fluxes directed to CEAS might directly impact the CA biosynthesis. As suggested by the results shown in Table 2, the dampening of phosphoglycerate kinase (PGK) would improve the flux of GAP as the C-3 precursor of CA, while the flux of aspartate synthesized from glutamate would provide the flux of the C-5 precursor required for CA production under such condition.
The analysis of potential strain modifications indicates that a lower increase in CA production could be achieved by overexpressing the pyridoxal phosphate-dependent aminotransferase catalyzing the production of clavulanate-1-aldehyde, either by single overexpression or in combination with other modifications. Nevertheless, the synthesis of (3R, 5R) clavulanate-1-aldehyde has not been completely elucidated. However, this step might be related to the orf17 cluster encoding the N-glycyl-clavaminate synthetase, which has been proposed as one of the enzymes catalyzing the reactions leading to (3R, 5R) clavaldehyde (Gómez et al., 2020; Ramirez-Malule et al., 2016). Moreover, the clavaminic acid synthesis is the bifurcation point between 5S clavams and CA biosynthesis; therefore, the overexpression of regulatory or encoding genes linked to clavulanate-1-aldehyde synthesis would drive the carbon flux preferably toward CA instead of 5S clavams. Similarly, the single overexpression of clavulanate-1-aldehyde dehydrogenase (CAR) would also potentially increase the CA secretion as the last step in biosynthesis. Since the demand for reducing cofactors increases when enhancing the CA production, these modifications might be accompanied by attenuations that would reduce the cost of these cofactors in other reactions without compromising cell growth. Likewise, to preserve the intracellular pool of intermediates required for the biosynthesis of CA, some decarboxylases can also be attenuated, such as pyruvate decarboxylase (PYRDC) and ornithine decarboxylase (ORNDC).
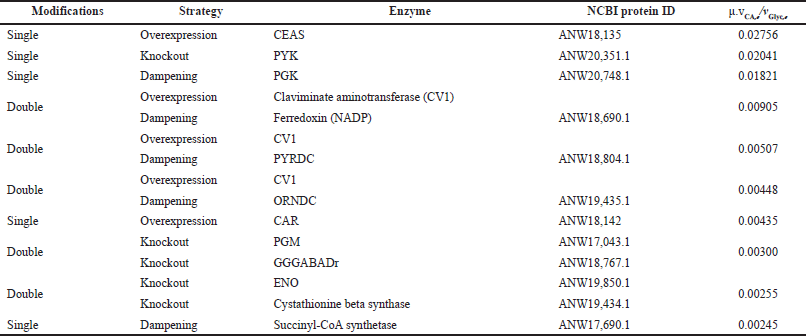 | Table 2. Potential reaction targets for the enhancement of CA production by strain engineering of S. clavuligerus. [Click here to view] |
Previous experimental reports have shown that a significant increase in the production of CA can be achieved by disrupting the glyceraldehyde 3-phosphate dehydrogenase (GAPD), directing part of the flux of GAP to CA biosynthesis (Jnawali et al., 2010; Li and Townsend, 2006). Our in silico analysis showed that the single deletion of the gene encoding the reaction catalyzed by pyruvate kinase (PYK) would also improve the flux of GAP toward CA biosynthesis. This modification implies the regulation of the flux passing from glycolysis to TCA cycle through the phosphoenolpyruvate carboxylase (PPC) to maintain the oxidative metabolism in the TCA cycle. Nevertheless, the kinase regulation in the Streptomyces genus is complex and it is still not completely elucidated, given its direct relationship with phosphate utilization and the response to environmental conditions. Phosphate limitation affects the adenosine triphosphate (ATP) availability and abundant generation of reduced cofactors and ATP may favor antibiotics synthesis to adjust the ATP generation to low phosphate availability (Gómez-Ríos et al., 2020). In the genus, more than 80 sensor kinases and more than 60 response regulators have been identified in the genome (Paradkar, 2013). Although the model shows insights into the stoichiometric effect of phosphate depletion on CA biosynthesis, the regulatory features are not captured by the FBA and dFBA approaches. Some reports have indicated that the expression levels of genes downstream from the CA gene cluster (orf-22 and orf-23) with sensor kinase activity affect the CA production levels (Paradkar, 2013). This suggests that the expression of kinases would play a role in the carbon flux regulation toward the CA biosynthetic pathway and those conditions might arise during the cultivation under specific conditions and not only via gene manipulation.
The potential knockout of phosphoglycerate mutase (PGM) would cut the conversion of 3-phosphoglycerate to 2-phosphoglycerate by alternatively activating the alcohol dehydrogenase for the use of glycerol as the sole source of carbon. This pathway is conducive to the formation of 2-phosphoglycerate via glycerate in three reactions, which is also a shorter route than glycolysis itself, so the flow of carbon to the TCA cycle is not compromised. The consequent accumulation of 2-phosphoglycerate and 3-phosphoglycerate favors the synthesis of amino acids and 2-oxoglutatarate, both widely required by secondary metabolism. Similarly, the formation of 3-phosphoglycerate is favored by the deletion of the cystathionine beta-synthase, avoiding the supply of 3-phosphoglycerate as a precursor in the serine–cysteine pathway. The gamma-glutamyl-gamma aminobutyric acid dehydrogenase (GGGABADr) connects succinate from the TCA cycle with glutamate metabolism; its dampening would reduce the carbon flux from glutamate to the TCA cycle, favoring the flux toward the urea cycle. The deletion of enolase would cause the repression of PPC from 2-phosphoglycerate, generating the accumulation of this intermediate, then increasing the production of CA. The in silico modifications with the highest ratio of CA to glycerol usage were further assessed under a dFBA approach.
dFBA assessment of potential targets for strain engineering
Considering that fed-batch operation using a medium with glycerol, ammonium, and glutamate is suitable for the enhancement of CA production, an in silico analysis of cultivation was performed considering the most promissory gene modifications exerting the highest CA flux. In silico single overexpression of ceas2 and single deletion of pyk genes showed the best ratio of glycerol usage and CA secretion. Therefore, fed-batch cultivation of overexpressed ceas2 (twofold) and deleted pyk were simulated to assess the potential CA yield. As shown in Figure 2, the time course of CA secretion was different in all cases indicating a flux redistribution that positively impacts the CA biosynthesis. The highest in silico CA concentration was observed for S. clavuligerus ?ceas2, whose maximum concentration was 1.9-fold calculated for the wild-type strain. The knockout of the pyk would also increase the CA production but to a lesser extent, showing a maximum increase of 1.1-fold for the concentration of the wild-type cultivation.
Figure 2 shows a summary of the dynamic fluxes distribution along primary metabolism and CA biosynthesis. The implementation of a fed-batch operation at 34 hours before the carbon sources starvation promotes higher and more stable metabolic activity in most of the enzymes, contributing to maintaining higher fluxes along the CA biosynthesis (Gómez-Ríos et al., 2022). The simulation of cultivation with ?ceas2 strain shows less carbon flux along the glycolysis in comparison with the ?pyk and wild-type strains, shown in the lower reaction fluxes of GAPD and pyruvate dehydrogenase (PDH) in Figure 2. This is due to considerable demand for GAP directly from CEAS enzyme. Additionally, this modification would also cause an increase in the demand for TCA cycle products 2-oxoglutarate and arginine, leading to an increase in the reaction fluxes of most of the enzymes in the TCA cycle, and downstream in the urea cycle. Indeed, transcriptomic evidence has shown upregulation of CA pathway enzymes when synthesized under extensive amino acid supply (Pinilla et al., 2019). The demand for carbon from central metabolism and CA biosynthesis also reduces the fluxes considerably toward the pentose–phosphate pathway. Therefore, the highest fluxes for the whole TCA cycle and CA biosynthesis are expected for the ?ceas2 strain that showed the highest in silico CA production.
The deletion of pyk causes a slight redistribution of the metabolic fluxes by cutting the pyruvate synthesis from phosphoenolpyruvate. Thus, the carbon flux is redistributed toward the pentose–phosphate pathway, CA biosynthesis, and PPC. The flux through the Glucose 6-phosphate dehydrogenase as the first reaction on the pentose–phosphate pathway was the highest observed among all the simulations, as shown in Figure 2. In case of glycolysis, the PPC must act as the auxiliary connection point between the glycolysis and TCA cycle maintaining the carbon flux to the oxidative metabolism and the intermediates supply to the urea cycle and CA biosynthesis. Additionally, the demand for pyruvate from PDH and TCA cycle is compensated by the malic enzyme, increasing the demand for malate, and then causing the flux inversion of the malate dehydrogenase. Thus, the flux throughout the TCA cycle does not decrease, despite the deletion of the last reaction of the glycolysis. The lower CA biosynthesis in comparison with the ?ceas2 strain is concordant with the lower activity of Argininosuccinate synthase, Aspartate dehydrogenase, and CEAS in connection with a lower demand for C-5 precursors.
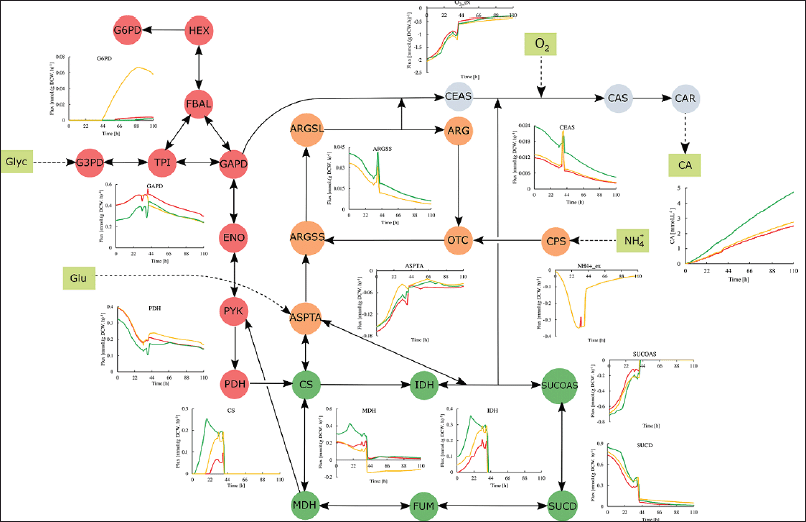 | Figure 2. In silico dynamic flux distributions in S. clavuligerus for fed-batch cultures with wild-type (red line), ?ceas2 (green line), and ?pyk (orange line) strains. The color in the metabolic network indicates enzymes related to central carbon pathways; red: glycolysis; green: TCA cycle; orange: urea cycle; and blue: CA biosynthesis. [Click here to view] |
CONCLUSION
Pseudo-steady-state simulations allowed us to analyze the sensitivity of the genome-scale metabolic model to constraints associated with the environmental cultivation conditions with biomass and CA as physiological objectives. Potential metabolic targets were determined via FBA based on the CA flux ratio. The single perturbations of CEAS and PYK reactions would cause a redistribution of metabolic fluxes during the culture, potentially favoring the CA accumulation. The dynamic simulations indicated that an increase in final antibiotic titers up to 1.9-fold for ?ceas2 and 1.1-fold for ?pyk might be possible in fed-batch submerged cultivation. Our approach applied to the genome-scale under dynamic conditions provided complementary perspectives for genetic perturbation of S. clavuligerus metabolism, which aimed at improving antibiotic production levels. The proposed framework constitutes a valuable tool for the exploration of production scenarios under different environmental conditions, genetic perturbations, and bioreactor operations (for instance, cell retention or product separation during the culture) as feasible strategies for redirecting the metabolism toward products of interest.
AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit it to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
LIST OF ABBREVIATIONS
ATP: Adenosine triphosphate; CA: Clavulanic acid; CAR: clavulanate-1-aldehyde dehydrogenase; CEAS: N2-(2-carboxyethyl)-arginine synthase; CephC: cephamycin C; CV1: Claviminate aminotransferase; dFBA: dynamic flux balance analysis; FBA: flux balance analysis; GAP: Glyceraldehyde 3-phosphate; GAPD: Glyceraldehyde 3-phosphate dehydrogenase; gDCW grams dry cell weight; GGGABADr: Gamma-glutamyl-gamma aminobutyric acid dehydrogenase; ODE: ordinary differential equation; ORNDC: Ornithine decarboxylase; PDH: Pyruvate dehydrogenase; PGK: Phosphoglycerate kinase; PGM: Phosphoglycerate mutase; PYK: Pyruvate kinase; PYRDC: Pyruvate decarboxylase; TCA cycle: Tricarboxylic acid cycle.
CONFLICTS OF INTEREST
The authors report no financial or other conflicts of interest in this work.
FUNDING
There is no funding to report.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Barreiro C, Martínez-Castro M. Regulation of the phosphate metabolism in Streptomyces genus: impact on the secondary metabolites. Appl Microbiol Biot, 2019; 103(4):1643–58. CrossRef
Bellão C, Antonio T, Araujo MLGC, Badino AC. Production of clavulanic acid and cephamycin c by streptomyces clavuligerus under different fed-batch conditions. Braz J Chem Eng, 2013; 30(2):257–66. CrossRef
Esnault C, Dulermo T, Smirnov A, Askora A, David M, Deniset-Besseau A, Holland IB, Virolle MJ. Strong antibiotic production is correlated with highly active oxidative metabolism in Streptomyces coelicolor M145. Sci Rep, 2017; 7(1):1–10. CrossRef
Gómez S, Ramírez-Malule H, Cardona-G W, Osorio E, Restrepo A. Double-ring epimerization in the biosynthesis of clavulanic acid. J Phys Chem A, 2020; 124(45):9413–26. CrossRef
Gómez-Ríos D, Junne S, Neubauer P, Ochoa S, Ríos-Estepa R, Ramírez-Malule H. Characterization of the metabolic response of Streptomyces clavuligerus to shear stress in stirred tanks and single-use 2D rocking motion bioreactors for clavulanic acid production. Antibiotics, 2019; 8(4):1–16. CrossRef
Gómez-Ríos D, López-Agudelo VA, Ramírez-Malule H, Neubauer P, Junne S, Ochoa S, Ríos-Estepa R. A genome-scale insight into the effect of shear stress during the fed-batch production of clavulanic acid by streptomyces clavuligerus. Microorganisms, 2020; 8(9):1–19. CrossRef
Gómez-Ríos D, Ramírez-Malule H. Bibliometric analysis of recent research on multidrug and antibiotics resistance (2017–2018). J Appl Pharm Sci, 2019; 9(5):112–6. CrossRef
Gómez-Rios D, Ramírez-Malule H, Ochoa S, Ríos-Estepa R. Rational selection of culture medium for clavulanic acid production by Streptomyces clavuligerus based on a metabolic modeling approach. Agric Nat Resour, 2022; 56(2):267–76. CrossRef
Gómez-Ríos D, Ramírez-Malule H, Neubauer P, Junne S, Ríos-Estepa R, Ochoa S. Tuning of fed-batch cultivation of Streptomyces clavuligerus for enhanced clavulanic acid production based on genome-scale dynamic modeling. Biochem Eng J, 2022; 185(1):1–10. CrossRef
Jnawali HN, Lee HC, Sohng JK. Enhancement of clavulanic acid production by expressing regulatory genes in gap gene deletion mutant of Streptomyces clavuligerus NRRL3585. J Microbiol Biotechnol, 2010; 20(1):146–52. CrossRef
Jnawali HN, Liou K, Sohng JK. Role of σ-factor (orf21) in clavulanic acid production in Streptomyces clavuligerus NRRL3585. Microbiol Res, 2011; 166(5):369–79. CrossRef
Kurt-Kizildo?an A, Vanli-Jaccard G, Mutlu A, Sertdemir I, Özcengiz G. Genetic engineering of an industrial strain of Streptomyces clavuligerus for further enhancement of clavulanic acid production. Turk J Biol, 2017; 41(2):342–53. CrossRef
Li R, Townsend CA. Rational strain improvement for enhanced clavulanic acid production by genetic engineering of the glycolytic pathway in Streptomyces clavuligerus. Metab Eng, 2006; 8(3):240–52. CrossRef
Liras P, Martín JF. Streptomyces clavuligerus: the omics era. J Ind Microbiol Biotechnol, 2021; 48(9–10):1–18. CrossRef
López-Agudelo VA, Gómez-Ríos D, Ramirez-Malule H. Clavulanic acid production by Streptomyces clavuligerus: insights from systems biology, strain engineering, and downstream processing. Antibiotics, 2021; 10(1):1–26. CrossRef
Millan-Oropeza A, Henry C, Lejeune C, David M, Virolle MJ. Expression of genes of the Pho regulon is altered in Streptomyces coelicolor. Sci Rep, 2020; 10(1):1–21. CrossRef
Paradkar A. Clavulanic acid production by Streptomyces clavuligerus: biogenesis, regulation and strain improvement. J Antibiot (Tokyo), 2013; 66(2):411–20. CrossRef
Pinilla L, Toro LF, Laing E, Alzate JF, Ríos-Estepa R. Comparative transcriptome analysis of Streptomyces clavuligerus in response to favorable and restrictive nutritional conditions. Antibiotics, 2019; 8(3),96:1–18. CrossRef
Ramirez-Malule, H. Bibliometric analysis of global research on clavulanic acid. Antibiotics, 2018; 7(4):1–14. CrossRef
Ramirez-Malule H, López-Agudelo VA, Gómez-Ríos D, Ochoa S, Ríos-Estepa R, Junne S, Neubauer P. TCA cycle and its relationship with clavulanic acid production: a further interpretation by using a reduced genome-scale metabolic model of Streptomyces clavuligerus. Bioengineering, 2021; 8(8):1–16. CrossRef
Ramirez-Malule H, Restrepo A, Cardona W, Junne S, Neubauer P, Rios-Estepa R. Inversion of the stereochemical configuration (3S, 5S)-clavaminic acid into (3R, 5R)-clavulanic acid: a computationally-assisted approach based on experimental evidence. J Theor Biol, 2016; 395:40–50. CrossRef
Sanchez C, Gomez N, Quintero JC. Producción de Ácido Clavulánico por fermentación de Streptomyces clavuligerus?: evaluación de diferentes medios de cultivo y modelado matemático. Dyna, 2012; 79:158–65.
Saudagar PS, Singhal RS. Optimization of nutritional requirements and feeding strategies for clavulanic acid production by Streptomyces clavuligerus. Bioresour Technol, 2007; 98(10):2010–7. CrossRef
Song JY, Jensen SE, Lee KJ. Clavulanic acid biosynthesis and genetic manipulation for its overproduction. Appl Microbiol Biot. 2010; 88(3):659–69. CrossRef
Stanford NJ, Millard P, Swainston N. RobOKoD: microbial strain design for (over)production of target compounds. Front Cell Dev Biol, 2015; 3(17):1–12. CrossRef
Tahlan K, Anders C, Wong A, Mosher RH, Beatty PH, Brumlik MJ, Griffin A, Hughes C, Griffin J, Barton B, Jensen SE. 5S clavam biosynthetic genes are located in both the clavam and paralog gene clusters in Streptomyces clavuligerus. Chem Biol, 2007; 14(2):131–42. CrossRef