INTRODUCTION
In agricultural farming, plant diseases are one of the major causes of crop yield losses. During the past decades, chemical pesticides have been widely used to control and prevent infections. However, due to the growing resistance of phytopathogens, several chemical pesticides have markedly become less effective, and higher doses are needed to obtain the same results (Tirado et al., 2008). Moreover, many of these pesticide residues may pose potential risks to the health of farmers and consumers and serious environmental hazards. In Thailand, chemical fungicides are among the top three imported pesticides, with amounts of 12,000–15,000 tons per year (Office of Agricultural Economics, 2021). These chemicals, including dinocap, maneb, ferbam, benomyl, carbendazim, captafol, and tridemorph, are used to prevent and inhibit the growth of fungi, both before and after harvesting. The phytopathogenic resistance to these chemical fungicides has been continuously reported (Kongtragoul et al., 2021), and this problem has resulted in poor disease control. In addition, most of these fungicides are known to be hazardous to people, and their long-term usage may harm both the economy and the environment. There is an urgent need to promote organic farming and search for new biological agents to control plant diseases and replace these chemicals.
Actinomycetes are a promising source of agricultural biocontrol agents since they produce a vast array of secondary metabolites that can inhibit the growth of various bacterial and fungal phytopathogens without toxicity to humans and harm to the environment (Chitraselvi, 2018). Numerous reports revealed that endophytic actinomycetes could promote the growth of host plants and reduce disease symptoms in various environmental stresses through the activities of their secondary metabolites. For example, 3-methyl-3,5-amino-4-vinyl-2-pyrone, a broad-spectrum antifungal compound from Streptomyces sp. N2, was effective against anthracnose of grapefruits, a disease caused by the fungus Colletotrichum gloeosporioides (Xu et al., 2015). 2,5-Bis(hydroxymethyl)furan monoacetate, a furan derivative obtained from Streptomyces sp. CEN26, an endophytic actinomycete found in the root nodes of Centella asiatica (L.) Urban, inhibited and deformed the conidial germination of Alternaria brassicicola, a fungal pathogen of cabbage (Phuakjaiphaeo et al., 2016). Natamycin, a polyene macrolide isolated from the fermentation broth of Streptomyces lydicus AZ-55, inhibited the growth of the filamentous fungi Fusarium oxysporum, Alternaria alternata, and Rhizoctonia solani (Atta et al., 2015). Blasticidin-S, an antibiotic from Streptomyces griseochromogenes, has been used commercially in Japan to reduce rice blasts (Tapadar and Jha, 2013). In a recent study, the crude extracts from the culture filtrate of Streptomyces sp. SCA2-4T destroyed the cell structure and inhibited mycelial growth and spore germination of the banana wilt fungus F. oxysporum f. sp. cubense tropical race 4 (Foc TR4) (Qi et al., 2021). However, it should be noted that if these biological control agents are to be employed to treat several kinds of phytopathogenic infections, new and potent secondary metabolites with a broad-spectrum activity must be obtained. In this regard, whole-genome sequencing analysis has been a useful tool for identifying important biocontrol agents (Zerikly and Challis, 2009).
Endophytes are ubiquitous and remain in specific association with the host plants, for example, mutualism or antagonism but not parasitism (Nair and Padmavathy, 2014). Endophytic actinobacteria have been reported from various plants, including dominant Streptomyces, along with other genera Micromonospora, Microbispora, Nocardia, Nocardioides, and Streptosporangium (Shimizu, 2011). It has been demonstrated that plant-associated microorganisms create compounds of therapeutic potential (Wu et al., 2021). However, there have been few investigations on the endophytic microorganisms in orchids. For example, two new cytotoxic and antifungal chemicals isolated from the endophytic fungus Pestalotiopsis sp. DO14 of Dendrobium officinale are excellent resources for preventing or treating pathogens (Wu et al., 2015). Fungal endophytes from seven Dendrobium species demonstrated a powerful and diverse antibacterial range (Cui et al., 2012) and from D. devonianum and D. thyrsiflorum exhibited both antibacterial and antifungal activity against pathogenic organisms (Xing et al., 2011). This study deals with the identification and evaluation of the antifungal activity of endophytic Streptomyces from three Dendrobium orchids. Four isolates are recognized based on their phenotypic, chemotaxonomic, and genotypic characteristics. The identification of secondary metabolites from the most promising strain and its genome analysis are discussed.
MATERIALS AND METHODS
Isolation, cultivation, and maintenance of actinomycetes
Actinomycetes were isolated from the roots of three Dendrobium orchids: D. kentrophyllum, D. findlayanum, and D. chrysanthum. The roots were washed with running tap water and dried at room temperature and prepared according to our previously reported protocols (Tedsree et al., 2021). The suspension was serially diluted 10 times. Each diluted suspension was spread on starch casein gellan gum (Küster and Williams, 1964) supplemented with nalidixic acid (25 μg/ml) and cycloheximide (50 μg/ml) (Kuncharoen et al., 2019). Bacterial colonies were picked up and purified after being incubated at 30°C for three weeks. The purified strains were preserved on ISP 2 slants and freeze-dried for long-term storage.
Phenotypic characteristics
Cell morphologies of the isolates were investigated by light microscopy (CX41, Olympus) and scanning electron microscopy (JSM-IT500HR, JEOL) after being cultivated on ISP 2 agar plates at 30°C for 14 days. The cultural characteristics were observed on ISP 2 agar as described previously (Shirling and Gottlieb, 1966). The NBS/IBCC color system was used to determine the colors of aerial mycelia, substrate mycelia, and diffusible pigment (Kelly, 1964). Physiological characteristics, including the growth at different temperatures (20°C–45°C), NaCl concentrations (0%–10%, w/v), and pH range of 4–12 (at intervals of 1 pH unit), were evaluated in ISP 2 broth at 30°C for 14 days. Carbon utilization on ISP 9 supplemented with 1% (w/v) carbon sources, starch hydrolysis, nitrate reduction, milk coagulation, peptonization, and gelatin liquefaction was examined as described by Arai (1975). The isomer of diaminopimelic acid in cell wall peptidoglycan was determined as described by Staneck and Roberts (1974).
16S rRNA gene sequence analysis
Genomic DNAs of the isolates were generated using the technique earlier described (Kudo et al., 1998). The 16S rRNA gene sequence was amplified according to a well-established method (Suriyachadkun et al., 2009) and then sequenced on a DNA sequencer (Macrogen) using universal primers, 27F forward (5?-AGAGTTTGATCMTGGCTCAG-3?) and 1492R reverse (5?-TACGGYTACCTTGTTACGACTT-3?). The sequence similarity values between the isolates and their related neighbors were calculated using the EzBioCloud service (Yoon et al., 2017).
Evaluation of antifungal activity
The isolates were cultivated in the broth of medium no. 57 (0.5% peptone, 2% glucose, 0.5% meat extract, 0.3% dry yeast, 0.5% NaCl, and 0.3% CaCO3) at 180 rpm, 30°C. After 14 days, the metabolites were extracted with 95% ethanol, and the cell suspension was centrifuged. The supernatant was used for testing the antifungal activity. The medium without the culture was used as a negative control. Antifungal activity was determined using the agar disc diffusion method (Mearns-Spragg et al., 1998). The tested filamentous fungi, F. oxysporum SA01, Fusarium solani SA02, Alternaria alternata SA01, C. gloeosporioides SA03, and Curvularia oryzae SA04, were cultivated on potato dextrose agar (PDA) plates. The paper disc was added with the supernatant amount of 50 μl and dried at room temperature. The actinomycete discs and phytopathogenic fungal disc were placed at a position 2.0 cm from the edge and center of the plate, respectively. The plates were incubated at 30°C until the fungal mycelium in the control plate grew to the edge of the plate. The inhibition zone was measured as the distance between the actinomycete discs and fungal mycelium edge. The percentage of inhibition is [(C − T)/C] × 100, where C is the fungal radius in the control plate and T is the fungal radius in the treatment plate. All experiments were carried out in triplicate.
Fermentation and extraction
The seed strain was cultivated in ISP 2 broth on a shaker (180 rpm), at 30°C, for 3 days and then was transferred into the broth of medium no. 57 on a shaker (180 rpm), at 30°C for 14 days. The culture broth of DR7-3 was extracted with ethyl acetate three times. The ethyl acetate layer was evaporated to dryness under a vacuum. The crude extract was stored in a dark and cold place for further tests.
Antifungal activity on the mycelial growth
The crude extract was determined for antifungal activity against the phytopathogenic fungus C. oryzae SA04. The percentage of mycelial inhibition was obtained using the poisoned food technique. The fungi were cultured on a PDA medium. The crude extract was prepared as a solution in dimethyl sulfoxide (DMSO) in different concentrations. The warm PDA medium of 45°C–50°C was added with different concentrations of the crude extract and then poured into sterilized Petri dishes. The control plate was a PDA medium with DMSO. The phytopathogenic fungal disc was placed at the center of the plate, for both control and treatment plates. The plates were incubated at 30°C until the fungal mycelia in the control plate reached the edge of the plate. The percentage of mycelial inhibition was calculated from the formula [(C − T)/C] × 100, where C is the colony diameter in the control plate and T is the colony diameter in the treatment plate. The results were compared with the antifungal activity of benomyl, a commercial chemical fungicide. To calculate the IC50 value, an online tool “Quest Graph™ IC50 Calculator” (by AAT Bioquest) was used (https://www.aatbio.com/tools/ic50-calculator).
The antifungal effect of crude extract DR7-3 on C. oryzae SA04 mycelia was determined using scanning electron microscopy (SEM, JSM-IT500HR, JOEL). The fungal mycelia samples (direct contact with crude of strain DR7-3 at IC50) and control (without the crude) were cut and examined by SEM.
Chemical profile of the crude extract
The chemical profile of the crude extract of the strain DR7-3 was investigated using gas chromatography (GC-MS). The analysis was carried out with GC Agilent 6890/MS Hewlett 5973. The injection port temperature was maintained at 250°C, and the column oven temperature program was set at 40°C for 2 minutes and then increased to 250°C (5°C/minute), ending with a 20 minutes isothermal process at 250°C. The carrier gas was helium (1 ml/minute), HP-5MS (30 m × 0.32 mm × 0.25 μm) was the stationary phase, and an injection volume of 1 μl was used. The chemical components were identified by comparison of their mass fragmentation patterns to those of the standard reference data of NIST libraries.
Genomic analyses of DR7-3
Whole-genome sequence analysis of the selected strain was performed with the Illumina MiSeq platform (Illumina, Inc., San Diego, CA) using 2 × 250 bp paired-end reads. The assembling of the reads to contigs was managed using SPAdes 3.12 (Bankevich et al., 2012). The draft genome of strain DR7-3 was determined using the antiSMASH server (Blin et al., 2019) to detect putative biosynthetic gene clusters (BGCs). All genomes were annotated on Prokka software 1.13 (Seemann, 2014) in line with the NCBI Prokaryotic Genome Annotation Pipeline. A phylogenomic tree of strain DR7-3 and its closest type strains was constructed using the TYGS web server (https://tygs.dsmz.de/) (Meier-Kolthoff and Göker, 2019). Average nucleotide identity (ANI), ANI-BLAST (ANIb), and ANI-MUMmer (ANIm) values between strain DR7-3 and closely related type strains were calculated pairwise using the JSpeciesWS web service (Richter et al., 2016). The digital DNA–DNA hybridization (dDDH) was evaluated using the Genome-to-Genome Distance Calculator (GGDC 2.1) with the BLAST+ method (Meier-Kolthoff et al., 2013), and the results were dependent on recommended formula 2 (identities/HSP length), which is proposed for use with incomplete whole-genome sequences.
Statistical analysis
Data were statistically analyzed by analysis of variance using the SPSS software package (SPSS 28 for Windows). The grouping was performed by Duncan’s multiple range tests at p = 0.05 on each of the significant variables measured. The data were expressed as mean values of triplicates ± standard deviation.
RESULTS
Isolation and identification of isolates
Four endophytic actinomycetes were recovered from the roots of the Thai orchid. Strains DR5-1 and DR7-3 were isolated from D. kentrophyllum and D. findlayanum, respectively, whereas strains DR8-5 and DR8-8 were obtained from D. chrysanthum. Results from the 16S rRNA gene sequence and phenotypic characteristic data indicated that all the isolates were Streptomyces spp. The pairwise alignment of the 16S rRNA gene sequence among all had 99.15%-99.93% similarity. Strains DR5-1, DR7-3, DR8-5, and DR8-8 were closely related to S. pravulus NBRC 13193T (99.40%), S. solisilvae HNM0141T (99.15%), S. daghestanicus NRRL B-5418T (99.93%), and S. malaysiense MUSC 136T (99.85%), based on 16S rRNA gene sequence similarity. The 16S rRNA gene sequences of these strains have been deposited in the NCBI database, and their accession numbers are listed in Table 1. The strains produced spiral spore chains with pale blue to gray color on ISP 2 agar after seven-day incubation. The scanning electron micrograph of strain DR7-3 revealed spiral spore chains on aerial hyphae (Fig. 1). All strains contained LL-diaminopimelic acid (Williams and Cross, 1971). The optimum temperature of all strains was 25°C–30°C, and the pH range was 5–10. All strains utilized various sugars for growth and grew in a range of 5%–10% NaCl concentrations. The cultural, physiological, and biochemical characteristics of the isolates are shown in Table 1.
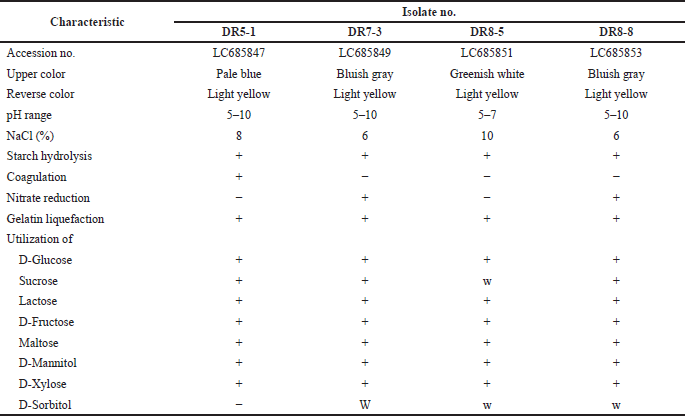 | Table 1. Phenotypic characteristics of isolates. [Click here to view] |
 | Figure 1. Scanning electron micrograph of strain DR7-3 grown on ISP 2 agar at 30°C for 14 days. [Click here to view] |
Orchids are known to be a rich source of endophytic microorganisms. Studies on the tissues of several terrestrial orchids indicated that the number and type of endophytes follow the seasonal rhythm of the year (Chutima et al., 2011). In this study of Dendrobium orchids, Streptomyces was found to be a dominant genus, similar to a previous report (Tsavkelova et al., 2007). In addition, Streptomyces sp. viji10 was reported from the velamen roots of the Vanda spathulata orchid (Senthilmurugan et al., 2013), while Actinomycetospora endophytica was identified as a novel species from the wild orchid Podochilus microphyllus (Sakdapetsiri et al., 2018).
Evaluation of antifungal activity
The antagonistic activity of the endophytic Streptomyces in this study was investigated in vitro against a wide range of phytopathogens using the dual culture technique. The inhibitory activities of strains DR5-1, DR7-3, DR8-5, and DR8-8 against five phytopathogenic fungi are shown in Table 2.
Among the four Streptomyces isolates in this study, DR7-3 presented the most interesting result, showing the highest antifungal potential, with 35.05 ± 0.95% inhibition against C. oryzae SA04, and thus was selected for further detailed studies (see below). Strain DR5-1 also exhibited remarkable antifungal activity, with the highest effects observed in three fungi, including against A. alternata SA01, F. solani SA02, and C. gloeosporioides SA03, whereas DR8-5 was the strongest isolate when tested against F. oxysporum SA01.
Endophytic actinomycetes from several sources have been studied earlier for their inhibitory activity against phytopathogenic fungi. For example, Streptomyces sp. CMUAc130 isolated from Zingiber officinale could inhibit Colletotrichum musae and F. oxysporum (Taechowisan and Lumyong, 2003), Streptomyces sp. S12-10 from rice showed high percentages of inhibition against Fusarium moniliforme, Helminthosporium oryzae, and R. solani, whereas Streptomyces strain CEN26 isolated from C. asiatica (L.) displayed significant antifungal activity against A. brassicicola (Phuakjaiphaeo and Kunasakdakul, 2015). In a more recent study, wetland-derived Streptomyces sp. ActiF450 exhibited a broad-spectrum antifungal activity against Aspergillus niger MA2, F. oxysporum F15, Penicillium chrysogenum ICF59, and Scopulariopsis candida ICF53 (Benhadj et al., 2020). The inhibitory potential of soilborne Streptomyces hygroscopicus against the fungus C. gloeosporioides was described earlier (Prapagdee et al., 2008). This study constitutes the first report of the antifungal activity of endophytic actinomycetes isolated from Dendrobium orchids.
Antifungal activity on the mycelial growth
The extract from the strain DR7-3 inhibited the mycelial growth of C. oryzae SA04 in a dose-dependent manner (Table 3). The colony growth was suppressed (100% inhibition) when the concentration of the extract reached 3,000 ppm. The IC50 value of the extract against C. oryzae SA04 was 25.75 ppm, significantly lower than that of the chemical fungicide benomyl (178.5 ppm). Benomyl has been widely used against a variety of phytopathogenic fungi (Tobih et al., 2015), and its fungicide action was related to its capacity to be absorbed by phytopathogen cells (Summerbell, 1993).
Scanning micrograph analyses revealed that the C. oryzae SA04 mycelia taken from the colony’s edge differed from the control. The control has a typical structural feature, such as a smooth outer surface on the cylindrically formed mycelium (Fig. 2A), whereas mycelia treated with DR7-3 extract were severely deformed, with uneven shrinkages, roughness, loss of smoothness, and a swollen mycelium surface (Fig. 2B). The unusual morphology of fungal hyphae can be taken as evidence of the antifungal activity of the test sample (Hashem et al., 2016). In a previous report, the fungicidal activity of a butanol extract of Streptomyces blastmyceticus 12-6 on Colletotrichum acutatum and F. oxysporum was studied using the SEM technique, which showed the abnormal morphology of hyphae, such as swelling and a reduction in cytoplasmic content, with apparent separation of the cytoplasm from the cell wall (Kim et al., 2019). These phenomena were also observed in C. oryzae PSUNK1012 when treated with the culture filtrate of S. angustmyceticus (Pithakkit et al., 2015). Similar observations were also reported for F. oxysporum race 4 upon adding the extract of Streptomyces sp. CB-75 (Chen et al., 2018).
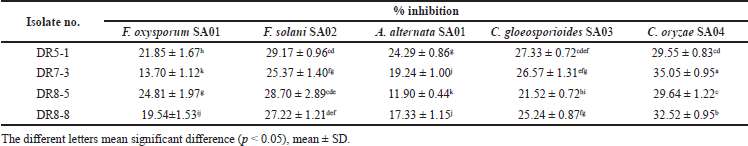 | Table 2. Antifungal activity of isolates. [Click here to view] |
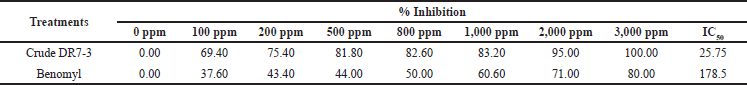 | Table 3. Antifungal activity of ethyl acetate extract against C. oryzae SA04 of strain DR7-3. [Click here to view] |
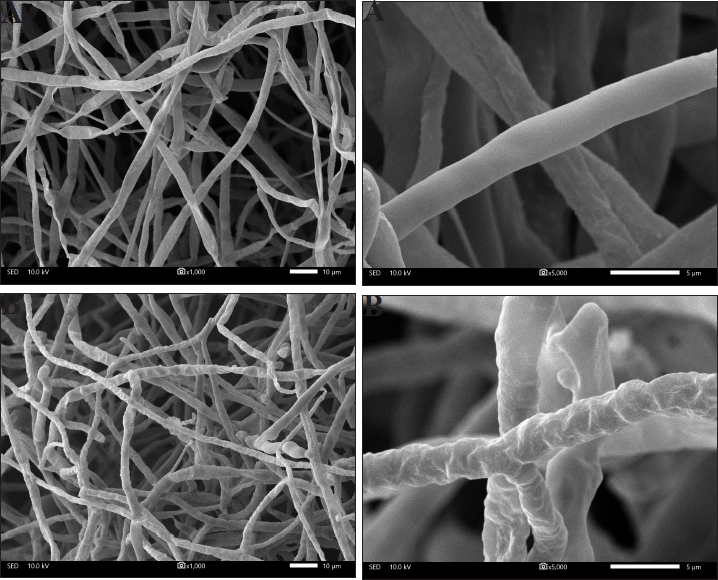 | Figure 2. Scanning electron micrograph of mycelia of C. oryzae SA04 without treatment (A and magnified A); treatment with ethyl acetate extract from strain DR7-3 (B and magnified B). [Click here to view] |
Genomic sequencing analysis
Genome analysis of strain DR7-3 revealed the size of 11,331,527 bp distributed in 159 contigs with a G+C content of 71.12% and 9,582 protein-coding sequences (CDSs). The phylogenetic analysis based on whole-genome sequences (Fig. 3) indicated that strain DR7-3 was phylogenetically closed to S. solisilvae HNM0141T. The ANIb and ANIm of the genomes DR7-3 and S. solisilvae HNM0141T were 98.49% and 98.71%, respectively. The dDDH values were the highest, 88.40% with S. solisilvae HNM0141T. For genome comparison, ANI and dDDH values are considered well correlated when the values were ≥95% (ANI) and ≥70% (dDDH), respectively (Fitch, 1971; Seemann, 2014). Since the dDDH (90.90%) and the ANI (98.59%–99.03%) values between strain DR7-3 and S. solisilvae HNM0141T were higher than the species cut-off, DR7-3 was identified as S. solisilvae. The GenBank accession number for the draft genome sequence of strain DR7-3 is JAMQOH000000000.
Gene function annotation and secondary metabolism gene clusters
The draft genome of strain DR7-3 was determined using the antiSMASH server to detect putative BGCs. More than 70 gene clusters were observed on DR7-3 genomes related to various BGCs, mainly type I polyketide synthase (T1PKS), nonribosomal peptide synthetase (NRPS), terpene, and siderophore (Table 4). The secondary metabolite biosynthetic gene clusters (smBGCs) exhibited 100% similarity in genetic relatedness to the known clusters producing geosmin, desferrioxamine B, ectoine, coelichelin, pristinol, and echoside. Interestingly, desferrioxamine B, echoside A, and echoside B have a potential as SARS-CoV-2 inhibitors (Bellotti and Remelli, 2021; Melinda et al., 2021). Thus, strain DR7-3 might be one of the sources of natural anti-COVID-19 compounds. Strain DR7-3 is predicted to produce anticancer agents such as geldanamycin (Fukuyo et al., 2010), salinomycin (Antoszczak and Huczy?ski, 2015), and hygrocin A/hygrocin B (Yin et al., 2017). The predicted secondary metabolites meilingmycin and herboxidiene can be used as insecticides (Sun et al., 2003) and herbicides (Pokhrel et al., 2015; Wideman, 1992), respectively, in agricultural farming.
Comparing the BGCs of DR7-3 to those of the close strain S. solisilvae HNM0141T revealed similar main gene clusters. However, the numbers of some gene clusters, such as T1PKS and NRPS-like, were different. T1PKS and NRPS-like in DR7-3 predicted secondary metabolites not found in S. solisilvae HNM0141T (Table 4). The strain DR7-3 exhibited genetic relatedness to five known smBGCs that are associated with antifungal activity, rustmicin, elaiophylin, coelichelin, cyphomycin, and rapamycin, and found in S. solisilvae HNM0141T. However, eight smBGCs, heronamide, niphimycins, fluvirucin B2, primycin, sceliphrolactam, niphimycins, pentamycin, and mediomycin A, were present only in strain DR7-3. In addition, the genome of strain DR7-3 contained nine smBGCs that displayed no similarity to any known smBGCs in antiSMASH (Table 4). These results suggested that strain DR7-3 might be a source of novel secondary metabolites with antifungal activity.
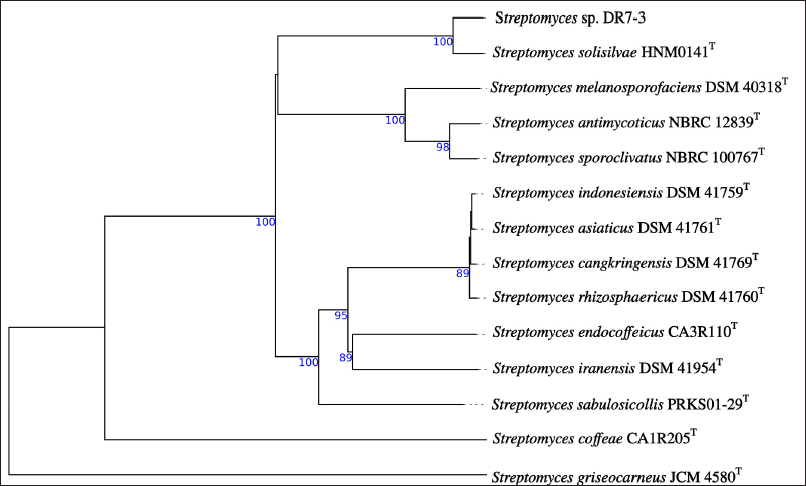 | Figure 3. Phylogenomic tree of strain DR7-3 and related Streptomyces species obtained from TYGS. The numbers above branches are GBDP pseudo-bootstrap support values from 100 replications. [Click here to view] |
Identification of bioactive compounds of strain DR7-3 by GC-MS
The secondary metabolites of the strain DR7-3 extract were analyzed by GC-MS. A total of 15 chemical compounds were identified by alignment of the NIST library based on retention time, molecular mass, molecular formula, and their biological activity (Table 5). These compounds were identified as 1) 2,3-butanediol, 2) 4-hydroxy-4-methyl-2-pentanone, 3) phenyl ethanol, 4) phenyl propanoic acid, 5) 2-phenylethyl ester of acetic acid, 6) N-tetradecane, 7) 2,4-bis(1,1-dimethylethyl) phenol, 8) hexadecane, 9) 1-heptadecane, 10) n-octadecane, 11) 2-phenylethyl ester of phenyl-acetic acid, 12) methyl ester of hexadecanoic acid, 13) 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo, 14) eicosane, and 15) bis(2-ethylhexyl) ester of hexanedioic acid ester of hexanedioic acid. Among these compounds, eicosane, 2,4-bis(1,1-dimethylethyl) phenol, hexadecane, and methyl ester of hexadecanoic acid have been reported earlier to possess antifungal activity (Table 5).
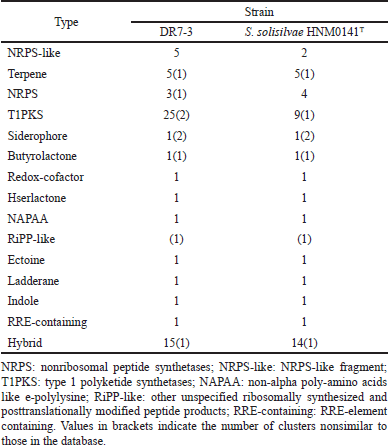 | Table 4. Comparison of BGCs composition of strain DR7-3 and S. solisilvae HNM0141T. [Click here to view] |
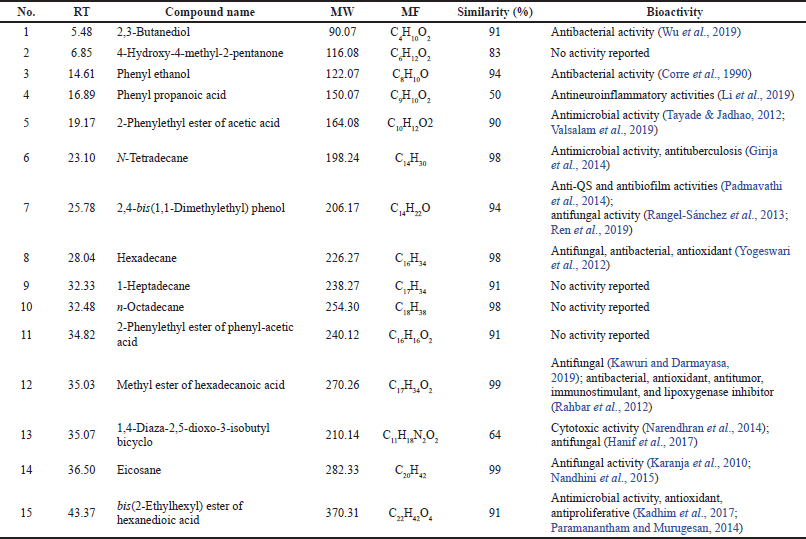 | Table 5. Chemical profile and bioactivity of ethyl acetate extract of strain DR7-3. [Click here to view] |
Eicosane, a long-chain fatty acid, was detected in the crude extract of Streptomyces sp. KX852460 showed antifungal activity against R. solani AG-3 KX852461, the cause of leaf spot disease (Ahsan et al., 2017). This compound is also present in the flower of Allium atroviolaceum, contributing to the antimicrobial activity of the plant extract (Dehpour et al., 2012). 2,4-Bis(1,1-dimethylethyl)-phenol from the ethyl acetate extract of Kutzneria sp. TSII inhibited the pathogenic fungus Pithomyces atro-olivaceous (Devi et al., 2021). This compound was produced by Pseudomonas fluorescens TL-1 and showed antifungal activity against Curvularia lunata (Ren et al., 2019). The long-chain hydrocarbon hexadecane from Jatropha curcas leaf extracts was exhibited against groundnut late leaf spot disease caused by Phaeoisariopsis personata (Francis et al., 2021). Hexadecanoic acid-methyl ester, a long-chain fatty ester produced from Streptomyces galbus TP2 and Streptomyces humidus, has been identified as an antifungal constituent (Kawuri and Darmayasa, 2019). It should be noted that bis(2-ethylhexyl)-hexanedioic acid, the major compound in the DR7-3 extract, possessed antimicrobial, antioxidant, and antiproliferative activities (Kadhim et al., 2017; Paramanantham and Murugesan, 2014).
CONCLUSION
The endophytic actinomycetes DR5-1, DR7-3, DR8-5, and DR8-8 isolated from three Dendrobium orchids belong to the genus Streptomyces. They showed inhibitory activity against several phytopathogenic fungi. Strain DR7-3 was identified as S. solisilvae and exhibited a broad-spectrum antifungal activity against five fungi that are causal agents of plant diseases. The ethyl acetate extract from this strain showed a high level of inhibition against C. oryzae SA04 compared with a standard chemical fungicide. Moreover, it suppressed mycelial growth and damaged the cell structure of the fungi. Four chemical components with antifungal activity were identified from the extract using the gas chromatography-mass spectrometric (GC-MS) technique. The draft genome sequence analysis of strain DR7-3 indicated that 13 gene clusters are involved in the biosynthesis of these antifungal metabolites. In our investigation, S. solisilvae DR7-3 appears to be a promising source for developing new antifungal agents against phytopathogenic fungi.
ACKNOWLEDGMENT
The authors would like to thank the Pharmaceutical Research Instrument Center, Faculty of Pharmaceutical Sciences, Chulalongkorn University, for providing research facilities.
CONFLICTS ON INTEREST
The authors declare that there are no conflicts of interest.
FUNDING
This research was supported by the Ministry of Higher Education, Science, Research and Innovation as a Ph.D. scholarship to Nisachon Tedsree and the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund), Graduate School, Chulalongkorn University.
ETHICAL APPROVAL
This article does not contain any studies with human participants and/or animals performed by any authors. Formal consent was not required in this study.
AUTHORS’ CONTRIBUTIONS
All authors have made significant contributions to the conceptualization and design, data acquisition, data analysis and interpretation, and revision of the manuscript.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Ahsan T, Chen J, Zhao X, Irfan M, Wu Y. Extraction and identification of bioactive compounds (eicosane and dibutyl phthalate) produced by Streptomyces strain KX852460 for the biological control of Rhizoctonia solani AG-3 strain KX852461 to control target spot disease in tobacco leaf. AMB Express, 2017; 7(1):1–9. CrossRef
Antoszczak M, Huczy?ski, A. Anticancer activity of polyether ionophore-salinomycin. Anti-Cancer Agents Med Chem, 2015; 15(5):575–91. CrossRef
Arai T. Culture media for actinomycetes. The Society for Actinomycetes Japan, Tokyo, Japan, 1975.
Atta H, El-Sayed A, El-Desoukey M, Hassan M, El-Gazar,M. Biochemical studies on the natamycin antibiotic produced by Streptomyces lydicus: fermentation, extraction and biological activities. J Saudi Chem Soc, 2015; 19(4):360–71. CrossRef
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol, 2012; 19(5):455–77. CrossRef
Bellotti D, Remelli M. Deferoxamine B: a natural, excellent and versatile metal chelator. Molecules, 2021; 26(11):3255. CrossRef
Benhadj M, Metrouh R, Menasria T, Gacemi-Kirane D, Slim FZ, Ranque S. Broad-spectrum antimicrobial activity of wetland-derived Streptomyces sp. ActiF450. EXCLI J, 2020; 19:360.
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res, 2019; 47(W1):W81–7. CrossRef
Chen Y, Zhou D, Qi D, Gao Z, Xie J, Luo Y. Growth promotion and disease suppression ability of a Streptomyces sp. CB-75 from banana rhizosphere soil. Front Microbiol, 2018; 8:2704. CrossRef
Chitraselvi R. Actinomycetes: dependable tool for sustainable agriculture. Curr Investig Agric Curr Res, 2018; 1:128–30. CrossRef
Chutima R, Dell B, Vessabutr S, Bussaban B, Lumyong S. Endophytic fungi from Pecteilis susannae (L.) Rafin (Orchidaceae), a threatened terrestrial orchid in Thailand. Mycorrhiza, 2011; 21(3):221–9. CrossRef
Corre J, Lucchini J, Mercier G, Cremieux A. Antibacterial activity of phenethyl alcohol and resulting membrane alterations. Res Microbiol, 1990; 141(4):483–97. CrossRef
Cui J, Wang Y, Xing Y, Guo S, Xiao P, Wang M. Antimicrobial activity of endophytic fungi isolated from Dendrobium species in southwestern China. China J Chin Mater Med, 2012; 37: 764–70.
Dehpour A, Yousefian M, Jafary Kelarijani S, Koshmoo M, Mirzanegad S, Mahdavi V, Javad Bayani M. Antibacterial activity and composition of essential oils of flower Allium rotundum. Adv Environ Biol, 2012; 6(3):1020–25.
Devi TS, Vijay K, Vidhyavathi R, Kumar P, Govarthanan M, Kavitha T. Antifungal activity and molecular docking of phenol, 2, 4-bis (1, 1-dimethylethyl) produced by plant growth-promoting actinobacterium Kutzneria sp. strain TSII from mangrove sediments. Arch Microbiol, 2021; 203(7):4051–64. CrossRef
Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol, 1971; 20(4):406–16. CrossRef
Francis M, Chacha M, Ndakidemi PA, Mbega E. Phytochemical analysis and in vitro antifungal evaluation of Jatropha curcas against late leaf spot disease on groundnut. J Anim Plant Sci, 2021; 47(1):8358–71
Fukuyo Y, Hunt CR, Horikoshi N. Geldanamycin and its anti-cancer activities. Cancer Lett, 2010; 290(1):24–35. CrossRef
Girija S, Duraipandiyan V, Kuppusamy PS, Gajendran H, Rajagopal R. Chromatographic characterization and GC-MS evaluation of the bioactive constituents with antimicrobial potential from the pigmented ink of Loligo duvauceli. Int Sch Res Notices, 2014; 2014:820745. CrossRef
Hanif A, Soekarno BPW, Munif A. Selection of endophytic bacteria producing metabolite compound to control seedborne fungal pathogen of maize. J Fitopatol Indones, 2017; 12(5):149. CrossRef
Hashem M, Alamri SA, Alrumman SA, Moustafa MF. Suppression of phytopathogenic fungi by plant extract of some weeds and the possible mode of action. Br Microbiol Res J, 2016; 15:1–13. CrossRef
Kadhim MJ, Al-Rubaye AF, Hameed IH. Determination of bioactive compounds of methanolic extract of Vitis Vinifera using GC-MS. Int J Toxicol Pharmacol, 2017; 9(2):113–26. CrossRef
Karanja E, Boga H, Muigai A, Wamunyokoli F, Kinyua J, Nonoh J. Growth characteristics and production of secondary metabolites from selected novel Streptomyces species isolated from selected Kenyan national parks. In: Jkuat Annual Scientific Conference Proceedings, 2010.
Kawuri R, Darmayasa I. Bioactive compound of Streptomyces capoamus as biocontrol of bacterial wilt disease on banana plant. IOP Conf Ser: Earth Environ Sci, 2019; 347(1):012054.. CrossRef
Kelly K. Inter-society color council–national bureau of standards color-name charts illustrated with centroid colors. US Government Printing Office, Washington, DC, 1964.
Kim YJ, Kim, Jh, Rho JY. Antifungal activities of Streptomyces blastmyceticus strain 12-6 against plant pathogenic fungi. Mycobiology, 2019; 47(3):329–34. CrossRef
Kongtragoul P, Ishikawa K, Ishii H. Metalaxyl resistance of Phytophthora palmivora causing durian diseases in Thailand. Sci Hortic, 2021; 7(10):375. CrossRef
Kudo T, Matsushima K, Itoh T, Sasaki J, Suzuki KI. Description of four new species of the genus Kineosporia: Kineosporia succinea sp. nov., Kineosporia rhizophila sp. nov., Kineosporia mikuniensis sp. nov. and Kineosporia rhamnosa sp. nov., isolated from plant samples, and amended description of the genus Kineosporia. Int J Syst Evol Microbiol, 1998; 48(4):1245–55. CrossRef
Kuncharoen N, Fukasawa W, Mori M, Shiomi K, Tanasupawat S. Diversity and antimicrobial activity of endophytic actinomycetes isolated from plant roots in Thailand. Microbiology, 2019; 88(4):479–88. CrossRef
Küster E, Williams S. Selection of media for isolation of Streptomycetes. Nature, 1964; 202(4935):928–29. CrossRef
Li J, Duan M, Yao X, Tian D, Tang J. Prenylated benzenepropanoic acid analogues from the Citrus grandis (L.) Osbeck and their anti-neuroinflammatory activity. Fitoterapia, 2019; 139:104410. CrossRef
Mearns-Spragg A, Bregu M, Boyd K, Burgess J. Cross-species induction and enhancement of antimicrobial activity produced by epibiotic bacteria from marine algae and invertebrates, after exposure to terrestrial bacteria. Lett Appl Microbiol, 1998; 27(3):142–46. CrossRef
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC bioinformatics, 2013; 14(1):1–14. CrossRef
Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun, 2019; 10(1):1–10. CrossRef
Melinda YN, Widada J, Wahyuningsih TD, Febriansah R, Damayanti E, Mustofa M. Metabologenomics approach to the discovery of novel compounds from Streptomyces sp. GMR22 as anti-SARS-CoV-2 drugs. Heliyon, 2021; 7(11):e08308. CrossRef
Nair DN, Padmavathy S. Impact of endophytic microorganisms on plants, environment and humans. Sci World J, 2014; 2014(2):250693. CrossRef
Nandhini SU, Sangareshwari S, Lata K. Gas chromatography-mass spectrometry analysis of bioactive constituents from the marine Streptomyces. J Pharm Clin Res, 2015; 8(2):244–46.
Narendhran S, Rajiv P, Vanathi P, Sivaraj R. Spectroscopic analysis of bioactive compounds from Streptomyces cavouresis kuv39: evaluation of antioxidant and cytotoxicity activity. Int J Pharm Pharm Sci, 2014; 6:319–22.
Office of Agricultural Economics (OAE). Summary of imported pesticides, 2021 [ONLINE]. Available via http://www.oae.go.th (Accessed 20 Jan 2022).
Padmavathi AR, Abinaya B, Pandian SK. Phenol, 2, 4-bis (1, 1-dimethylethyl) of marine bacterial origin inhibits quorum sensing mediated biofilm formation in the uropathogen Serratia marcescens. Biofouling, 2014; 30(9):1111–22. CrossRef
Paramanantham M, Murugesan A. GC-MS analysis of Holarrhena antidysentrica Wall flower. Int J Sci Eng, 2014; 3(3):631–39.
Phuakjaiphaeo C, Chang C, Ruangwong O, Kunasakdakul K. Isolation and identification of an antifungal compound from endophytic Streptomyces sp. CEN 26 active against Alternaria brassicicola. Lett Appl Microbiol, 2016; 63(1):38–44. CrossRef
Phuakjaiphaeo C, Kunasakdakul K. Isolation and screening for inhibitory activity on Alternaria brassicicola of endophytic actinomycetes from Centella asiatica (L.) Urban. Int J Agric Technol, 2015; 11(4):903–12.
Pithakkit S, Petcharat V, Chuenchit S, Pornsuriya C, Sunpapao A. Isolation of antagonistic actinomycetes species from rhizosphere as effective biocontrol against oil palm fungal diseases. Walailak J Sci Technol, 2015; 12(5):481–90.
Pokhrel AR, Dhakal D, Jha AK, Sohng JK. Herboxidiene biosynthesis, production, and structural modifications: prospect for hybrids with related polyketide. Appl Microbiol Biotechnol, 2015; 99(20):8351–62. CrossRef
Prapagdee B, Akrapikulchart U, Mongkolsuk S, Prapagdee B, Mongkolsuk S. Potential of a soil-borne Streptomyces hygroscopicus for biocontrol of a anthracnose disease caused by Colletotrichum gloeosporioides in orchid. J Biol Sci, 2008; 8(7):1187–92. CrossRef
Qi DF, Zou L, Zhou D, Zhang M, Wei Y, Zhang L, Xie J, Wang W. Identification and antifungal mechanism of a novel actinobacterium Streptomyces huiliensis sp. nov. against Fusarium oxysporum f. sp. cubense tropical race 4 of banana. Front Microbiol, 2021; 12:1–14. CrossRef
Rahbar N, Shafaghat A, Salimi F. Antimicrobial activity and constituents of the hexane extracts from leaf and stem of Origanum vulgare L. ssp. viride (Boiss.) hayek growing wild in northwest Iran. J Med Plant Res, 2012; 6(13):2681–85. CrossRef
Rangel-Sánchez G, Castro-Mercado E, García-Pineda E. Avocado roots treated with salicylic acid produce phenol-2,4-bis (1,1-dimethylethyl), a compound with antifungal activity. J Plant Physiol, 2013; 171:189–98. CrossRef
Ren J, Wang J, Karthikeyan S, Liu H, Cai J. Natural anti-phytopathogenic fungi compound phenol, 2, 4-bis (1, 1-dimethylethyl) from Pseudomonas fluorescens TL-1. Indian J Biochem Biophys, 2019; 56(2):162–68.
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics, 2016; 32(6):929–31. CrossRef
Sakdapetsiri C, Ngaemthao W, Suriyachadkun C, Duangmal K, Kitpreechavanich V. Actinomycetospora endophytica sp. nov., isolated from wild orchid (Podochilus microphyllus Lindl.) in Thailand. Int J Syst Evol, 2018; 68(9): 3017–21. CrossRef
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics, 2014; 30(14):2068–69. CrossRef
Senthilmurugan G, Viji S, Sekar L, Suresh K. Enzyme analysis of endophytic new Streptomyces sp. viji10 isolated from velaman roots of orchid plant Vanda spathulata (L) spreng. Asian J Agri Biol, 2013; 1(13):149–54.
Shimizu M. Endophytic actinomycetes: biocontrol agents and growth promoters. In: Maheshwari DK (ed.). Bacteria in agrobiology: plant growth responses, Springer, Berlin, Heidelberg, pp 201–20, 2011. CrossRef
Shirling ET, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Evol, 1966; 16(3):313–40. CrossRef
Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol, 1974; 28:226–31. CrossRef
Summerbell RC. The benomyl test as a fundamental diagnostic method for medical mycology. J Clin Microbiol, 1993; 31(3):572–7. CrossRef
Sun Y, Zhou X, Tu G, Deng Z. Identification of a gene cluster encoding meilingmycin biosynthesis among multiple polyketide synthase contigs isolated from Streptomyces nanchangensis NS3226. Arch Microbiol, 2003; 180(2):101–7. CrossRef
Suriyachadkun C, Chunhametha S, Thawai C, Tamura T, Potacharoen W, Kirtikara K, Sanglier JJ. Planotetraspora thailandica sp. nov., isolated from soil in Thailand. Int J Syst Evol, 2009; 59(5):992–7. CrossRef
Taechowisan T, Lumyong S. Activity of endophytic actimomycetes from roots of Zingiber officinale and Alpinia galanga against phytopathogenic fungi. Ann Microbiol, 2003; 53(3):291–8.
Tapadar SA, Jha DK. 2013. Disease management in staple crops: a bacteriological approach. In: Maheshwari DK (ed.). Bacteria in agrobiology: disease management, Springer, Berlin, Heidelberg, pp 111–52. CrossRef
Tayade D, Jadhao N. Attempt in the synthesis of 2-[(2, 6 disubstitutedthiocarbamidophenyl) amino] benzeneacetic acid and their antimicrobial study. J Pure Appl Microbiol, 2012; 6:2025–8.
Tedsree N, Tanasupawat S, Sritularak B, Kuncharoen N, Likhitwitayawuid K. Amycolatopsis dendrobii sp. nov., an endophytic actinomycete isolated from Dendrobium heterocarpum Lindl. Int J Syst Evol, 2021; 71(7):004902. CrossRef
Tirado R, Englande AJ, Promakasikorn L, Novotny V.Use of agrochemicals in Thailand and its consequences for the environment. Greenpeace Research Laboratories Technical, Bangkok, Thailand, 2008.
Tobih F, Bosah B, Nweke F. Evaluation of the efficacy of radial growth, spore density of Curvularia lunata and Fusarium semitectum. Int J Agric Innov Res, 2015; 4(1):47–50.
Tsavkelova EA, Cherdyntseva TA, Botina SG, Netrusov AI. Bacteria associated with orchid roots and microbial production of auxin. Microbiol Res, 2007; 162(1):69–76. CrossRef
Valsalam S, Agastian P, Arasu MV, Al-Dhabi NA, Ghilan AKM, Kaviyarasu K, Ravindran B, Chang SW, Arokiyaraj S. Rapid biosynthesis and characterization of silver nanoparticles from the leaf extract of Tropaeolum majus L. and its enhanced in-vitro antibacterial, antifungal, antioxidant and anticancer properties. J Photochem Photobiol B Biol, 2019; 191:65–74. CrossRef
Wideman M. Herboxidiene, a new herbicidal substance from Streptomyces chromofuscus A7847. J Antibiotics, 1992; 45:914–21. CrossRef
Williams ST, Cross T. Actinomycetes. In: Booth C (ed.). Methods in microbiology. Academic Press, London, UK, pp 295–334, 1771. CrossRef
Wu LS, Jia M, Chen L, Zhu B, Dong HX, Si JP, Peng W, Han T. Cytotoxic and antifungal constituents isolated from the metabolites of endophytic fungus DO14 from Dendrobium officinale. Molecules, 2015; 21(1):0014. CrossRef
Wu W, Chen W, Liu S, Wu J, Zhu Y, Qin L, Zhu B. Beneficial relationships between endophytic bacteria and medicinal plants. Front Plant Sci, 2021; 12:758. CrossRef
Wu Y, Zhou J, Li C, Ma Y. Antifungal and plant growth promotion activity of volatile organic compounds produced by Bacillus amyloliquefaciens. Front Plant Sci, 2019; 8(8):e00813. CrossRef
Xing YM, Chen J, Cui JL, Chen XM, Guo, SX. Antimicrobial activity and biodiversity of endophytic fungi in Dendrobium devonianum and Dendrobium thyrsiflorum from Vietman. Curr Microbiol, 2011; 62(4):1218–24. CrossRef
Xu B, Chen W, Wu Zm, Long Y, Li K. A novel and effective Streptomyces sp. N2 against various phytopathogenic fungi. Appl Biochem Biotechnol, 2015; 177(6):1338–47. CrossRef
Yin M, Jiang M, Ren Z, Dong Y, Lu T. The complete genome sequence of Streptomyces autolyticus CGMCC 0516, the producer of geldanamycin, autolytimycin, reblastatin and elaiophylin. J Biotechnol, 2017; 252:27–31. CrossRef
Yogeswari S, Ramalakshmi S, Neelavathy R, Muthumary J. Identification and comparative studies of different volatile fractions from Monochaetia kansensis by GCMS. Glob J Pharmacol, 2012; 6(2):65–71.
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol, 2017; 67(5):1613. CrossRef
Zerikly M, Challis GL. Strategies for the discovery of new natural products by genome mining. ChemBioChem, 2009; 10(4):625–33. CrossRef