INTRODUCTION
Ever since the existence of humankind, infectious diseases have always been seen as a big threat. However, with the advancement of science and technology and the discovery of penicillin, people were hopeful to combat the threat posed by microorganisms. However, their joy did not last too long due to the emergence of resistance in microorganism against available antibiotics. Development of drug resistance is attributed to the excessive and over use of antibiotics (over the counter availability) (Demain, 1999). The evolution of resistant bacteria is also influenced by mutation and gene exchange between bacteria, therefore leaving most of the current antibiotics obsolete. Since last one and half decade, scientists have turned to natural products for the identification of novel compounds to combat the menace of drug resistance. Microorganisms are seen as the reservoir of natural and novel compounds due to their ability to produce diverse secondary metabolites with their environment (Demain, 1999; Schmidt, 2004). There are a variety of natural resources that can be used to find antibiotics. By studying marine-derived microorganisms, novel antibiotics can be discovered and developed as per several studies, as it is a home to a diverse range of biodiversity (Arumugam et al., 2015). The organisms of marine habitat evolve themselves not only to survive in harsh environment but also produce various metabolites which have been found biologically active (Kasanah and Hamann, 2004).
It is now widely acknowledged that marine is home to large and varied micro biomes. In addition to bacteria and archaea, fungi in deep-sea habitats have been extensively investigated (Wang et al., 2015). Fungi are known for their wide range of secondary metabolites, which include numerous life-saving medications as well as deadly poisonous mycotoxins. It has been found that screening and characterization of metabolites are key concepts in metabolomics (Roy and Banerjee, 2017). Many pharmacologically bioactive compounds have been identified from deep-sea fungi and tested for their anticancer, antifungal, antibacterial, antiviral, and anti-larval properties (Wang et al., 2015). Lots of fungi have been isolated from the deep sea or coastal seaside, like Rhodotorula, Aspergillus, Cladosporium, Penicillium, Alternaria, Fusarium, Engyodontium, Sistotrema, Schizophyllum, Tilletiopsis, etc. The most dominant species were found to be Penicillium and Aspergillus (Luo et al., 2020).
Novelty of marine-derived compounds and its wide applications have yet to find a way to solutions for many dreadful diseases caused by resistant microorganisms, which are difficult to treat today (Manimegalai et al., 2013). Due to a scarcity of knowledge, secondary metabolites of marine fungi are of special interest. Thus, the current study was carried out to isolate and identify marine fungi from the coastal region and to assess their antibacterial potential against several microorganisms.
MATERIALS AND METHODS
Sample collection
Water sample was collected in a sterile glass bottle from the coastal side of Gorai beach, Mumbai, India and stored in refrigerator at 4°C for further study.
Isolation and identification of fungi
Samples were diluted at 10 and 100 folds and spread onto potato dextrose agar (PDA) plate, and incubated for 3–4 days at room temperature. Media was supplemented with streptomycin (30 mcg/ml) for inhibition of bacterial growth. After incubation, hyphae growth was observed on plates, each fungal colony was separated on a new PDA plate for purification. Further identification was done by slide culturing method and lacto-phenol cotton blue staining (Shamly et al., 2014). All the experiments were repeated in triplicates.
Test bacteria
Test organisms such as Escherichia coli MTCC 64, Enterococcus faecalis MTCC 439, Enterococcus faecium MTCC 9728, Klebsiella pneumonia MTCC 432, Bacillus subtilis 441, and Staphylococcus aureus MTCC 96 were obtained from MTCC, Chandigarh, Punjab.
Fermentation and extraction of metabolites
Broth fermentation method and extraction
The fungal strains were cultivated on PDA. After 4–5 days of incubation little block of size 2 cm2 containing the mycelium was inoculated in 50 ml of potato dextrose broth in a 250 l flask and incubated for 14 days on a rotary shaker at 150 rpm under room temperature (VanderMolen et al., 2013).
Solvents (Chloroform and ethanol) in combination and individually were used for extraction of metabolites. 60 ml of ethanol and chloroform (1:1) were used in combination and individually and added in fermentation broth of Aspergillus fumigatus, Histoplasma capsulatum, Cladosporium cladosporioides, Cladosporium pseudocladosporioides, Trichophyton rubrum, Penicillium chrysogenum, Alternaria alternate, Neoscytalidium dimidiatum and Aspergillus terreus. Flasks were kept on a shaker at 150 rpm for 24 hours and filtered with Whatman paper. Mycelia were separated for further investigation and filtrate was allowed to evaporate (VanderMolen et al., 2013). Further, the sticky substance was collected in vials that were weighed earlier and again weighed with metabolite. Then vials were refrigerated at 4°C for further studies.
Dry mycelia extraction
Mycelia from broth fermentation were poured onto a muslin cloth and boiling water was added until the entire agar disappeared. Then mycelia was recovered by inverting the mesh on another cloth, pouring water again to release the mycelium from the mesh and weighing the final cloth. Further cloth was kept with mycelia in a hot air oven at 50°C till it dried out. Then mycelia were collected in a mortar and powdered with the pestle. Weight of powdered mycelia was calculated and stored in a cool place for further study (VanderMolen et al., 2013). Extraction was done by using three solvents: chloroform, ethyl acetate, and ethanol. 1 g of each dried mycelia was added to 10 ml of solvent separately and incubated at room temperature for 24 hours. The next day, it was filtered and the filtrate was store in refrigerator at 4°C. Mycelia was again extracted overnight by using more than 10 ml of solvent. That was the second cycle of extraction. Again, this was filtered and both the filtrate were mixed, allowed to evaporate, and the metabolite was collected in weighed vials and refrigerated at 4°C.
Antibacterial activity of crude extract by Well diffusion method
Bacterial cultures E. coli, E. faecalis, E. faecium, K. pneumonia, B. subtilis, and S. aureus were adjusted at 0.5 McFarland standard and spreaded on Mueller Hinton agar plate with a sterile swab moistened with bacterial suspension. Wells of 6 mm diameter were punched onto the agar medium and filled with a concentration that ranged between 2–10 g/ml (2, 4, 6, 8, 10 μg/ml) and 10–150 μg/ml (5, 10, 20, 50, 100, 150 g/ml). 100 μl of metabolite extracted from broth fermentation and dry mycelia was added to wells. Metabolite were allowed to diffuse at room temperature for 2 hours. Plates were incubated in the upright position at 37°C for 24 hours. Wells of positive (streptomycin 30 mcg/ml) and negative control (sterile saline) was also set. After incubation, the diameters of the inhibition zone were measured in millimeter and noted (Balouiri et al., 2016).
RESULT
Sample collected site
The sample was collected from the coastal waters of Gorai Beach, Mumbai, India. The pH was found to 5.9 and temperature of the sample was 34°C.
Isolated and identified fungi
Fungal identification was done by using the slide culture method and lacto-phenol cotton blue staining. Few fungal identification books were referred. Total nine fungal isolates were recovered. Thus cultural, morphological, and microscopic examination was done. Identified isolates were A. fumigatus, H. capsulatum, C. cladosporioides, C. pseudocladosporioides, T. rubrum, P. chrysogenum, A. alternate, N. dimidiatum and A. terreus. Isolates were named alphabetically like A, B, C, D, E, F, G, H, and I as shown in Table 1. Cultural and morphological characteristics have been mentioned in Table 1, while suspected isolates with growth morphology and microscopic examination have been mentioned in Table 2.
 | Table 1. Cultural and morphological characteristics. [Click here to view] |
Broth fermentation and extraction
Broth fermentation was carried for all nine isolates named as A, B, C, D, E, F, G, H, and I. Extraction was done in a beaker. Extracted solvent layer in individual and combination form was directly kept for evaporation. Evaporation took around 3–4 days. At the bottom of all beakers, a sticky substance was obtained, which was removed by a sharp spatula and was collected in vials and refrigerated at 4°C. Before collection, vials were weighed. In Table 3, metabolite concentration has been mentioned.
Metabolite extraction from dry mycelial biomass
Mycelia were collected from a broth fermentation process. It was dried in a hot air oven and powdered by a mortar and pestle. Further extraction of mycelial powder was carried out by using three different solvents like chloroform, ethyl acetate, and ethanol. Extraction was done twice; the filtrate was collected and evaporated. The sticky substance at the bottom of the plate was collected with a spatula and transferred to weighed vials and refrigerated at 4°C. The weight of the extracted metabolite has been mentioned in Table 4.
Antibacterial activity of metabolites extracted by broth fermentation and dry mycelia
Antibacterial activity for both types of metabolites collected from broth fermentation and dry mycelia was done by well diffusion method against E. coli, E. faecalis, E. faecium, K. pneumonia, B. subtilis, and S. aureus. The activity was performed in a range of 10–150 and 2–10 μg/ml. A range of 10–150 μg/ml showed complete inhibition of all the cultures for some extracts which was tested further. So the next range was reduced to 2–10 μg/ml. In terms of few metabolites from broth extraction, inhibition was observed against all selected bacteria, with the only difference in metabolite concentration. Inhibition zone was measured in millimeter. Inhibitory activity by broth fermented extract is shown in Table 5 and Figure 1 and for dry mycelia extracted metabolites it is shown in Table 6 and Figure 2.
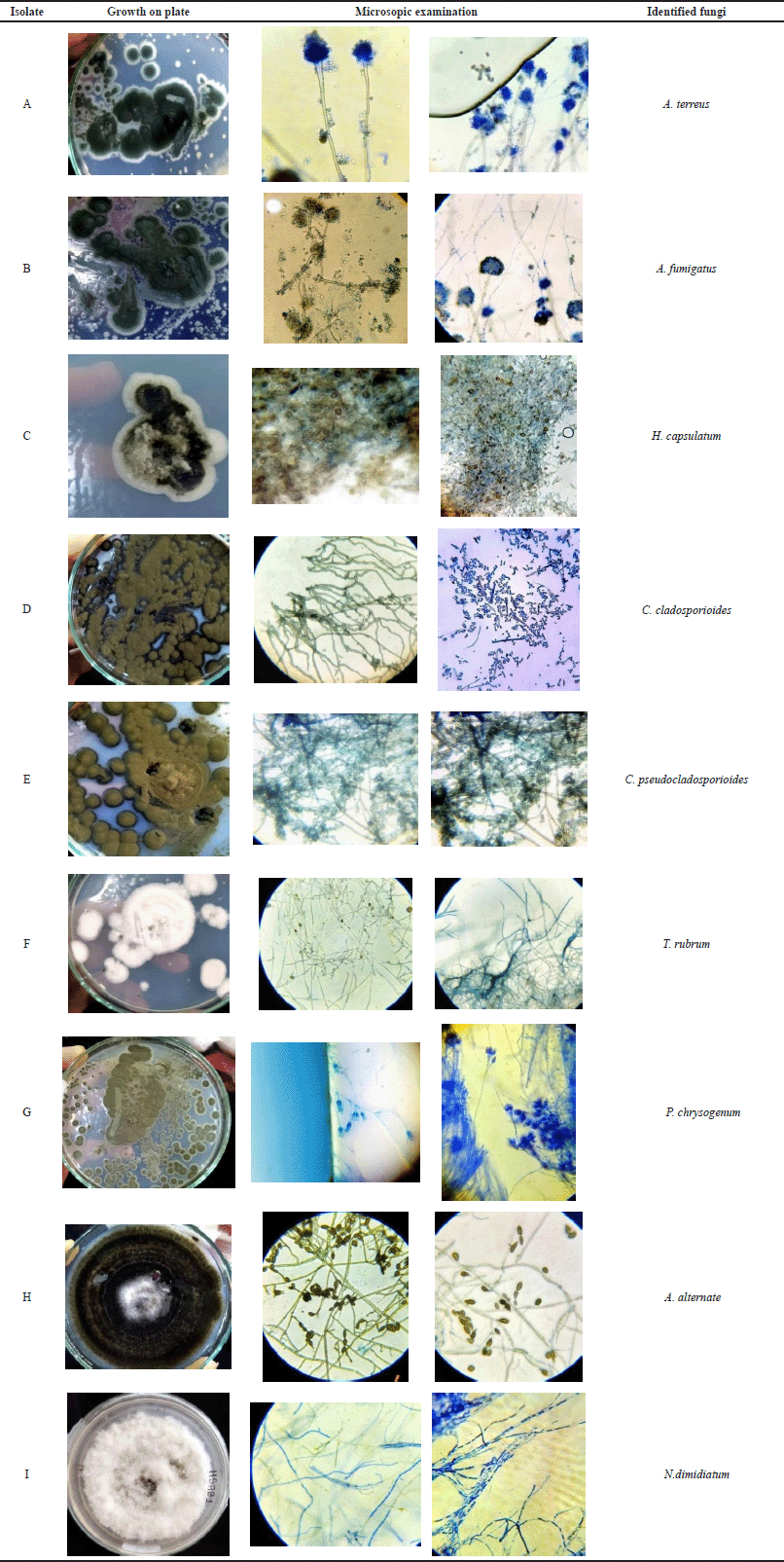 | Table 2. Fungal Isolation and Microscopy. [Click here to view] |
DISCUSSION
Antimicrobial resistance among clinical microorganisms makes it difficult to treat infectious disorders. Since the continuing study of terrestrial sources generated known microorganisms and metabolites in the past, scientists and academics throughout the world have been searching for novel antimicrobials by varying microbial sources. A review of current literature suggests that microorganisms originated from the sea are potential source of bioactive compounds (Rajasekar et al., 2012). Marine fungus is one of the most significant and abundant sources of novel natural compounds for the pharmaceutical and medical sectors as per several studies (Arumugam and Ponnusami, 2017; Aslam et al., 2018). Secondary metabolites are formed and released in response to nutrient deprivation, competition, or any other type of metabolic stress that limits marine fungal growth. Antibiotics, anticancer, and co-stimulatory chemicals are examples of secondary metabolites that can be generated by a variety of metabolic processes (Shabana et al., 2021).
As part of this study, a few of the sea-derived fungi were screened, isolated, and metabolite extraction was done to study their antimicrobial metabolite capacity. As the marine environment is a much-diversified habitat, the isolation of fungi from such an environment can be a challenging task. Here, the major focus in the initial studies was nutrition for the isolates. As many studies have majorly focused on synthetic media for the desired growth of isolates, here we have also used a synthetic medium that is PDA because a nutritionally rich medium can help in the isolation of maximum isolates from a sample (Kossuga et al., 2012). Fungal isolates found to be A. fumigatus, H. capsulatum, C. cladosporioides, C. pseudocladosporioides, T. rubrum, P. chrysogenum, A. alternate, N. dimidiatum and A. terreus. Many studies have isolated fungi from sea samples and all fungi are found to be common. Some studies also says that due to the mixing of water from different sources, some terrestrial and soil-based fungi can enter the marine habitat (Preedanon et al., 2016; Shabana et al., 2021). As the study was focused on metabolite extraction and antimicrobial properties, both were done, and antimicrobial activity was detected in nearly half of the isolates. It includes fungi named A. fumigatus, C. cladosporioides, C. pseudocladosporioides, and P. chrysogenum. These isolates have shown antibacterial activity in previous studies also with differences in concentration of metabolites (Christophersen et al., 1998; Wang et al., 2015).
 | Table 3. Weight of extracted metabolite from Broth fermentation. [Click here to view] |
Metabolite extraction can be done in different ways. Here it has been carried out by the broth fermentation method, along with dry mycelium extraction, which was also done to check the metabolite content of the biomass. Extraction was done by using solvents like chloroform, ethyl acetate, and ethanol with varied polarities. In most of the fungal metabolite studies, extraction was done only in relation to broth fermentation (Rajasekar et al., 2012; Song et al., 2019), a few studies had focus on mycelium extraction (Synytsya et al., 2017; Wong Chin et al., 2021). Both the methods are different concerning protocols as well as results. As we can see in the study, broth fermentation has given a good yield, but when we compare it with dry mycelium extraction, even this has given an almost equal yield. Even though dry mycelium extraction was done using fungal biomass from broth fermentation, an equal yield in less biomass was obtained. So it can be said that for further studies we can just rely on the dry mycelial process. So, for any metabolite related studies, a huge bank of fungal mycelial biomass can be made and used for metabolite studies as the requirement for biomass is less compared to the synthetic broth requirement. For comparison purposes, solvents with varying degrees of increasing polarity were used in both extraction processes. In the broth fermentation process, chloroform has given the highest metabolite yields compared to ethanol. While in the dry mycelial extraction process, ethanol has given the highest yield compared to chloroform. The reason behind these differences will be understood only after the characterization of metabolites, as it will reveal the exact components present in a metabolite and its nature and chemical properties. This will help in comparing the solvent and extraction techniques.
 | Table 4. Weight of extracted metabolite from dry mycelium biomass. [Click here to view] |
 | Table 5. Inhibitory activity of metabolites extracted by broth fermentation. [Click here to view] |
 | Figure 1. Well diffusion method by broth fermented metabolites. [Click here to view] |
Despite the absence of extensive metabolic analysis, the results could provide insight into the metabolic activity of fungi under a variety of solvents and extraction techniques.
Both types of the extracts were tested against two Gram- negative E. coli, K. pneumonia, and four Gram-positive microorganisms, E. faecalis, E. faecium, Bacillus subtilis, and S. aureus. Both Gram-negative and positive samples were selected to check the broad-spectrum activity of metabolites. The range selected was between 10–150 and 2–10 μg/ml. The range of 10–150 μg/ml was found to be quite higher as it completely inhibited all the microbes, so the range was further reduced. Major activity was seen against S. aureus by some of the extracts. In one study, A. fumigatus was found to inhibit S. aureus at concentrations of 15.63, 1.95, and 3.90 μg/ml (Hussein et al., 2022). In current study, inhibition by A. fumigatus metabolite of all solvents was even at 2 μg/ml, so at further reduced concentration might also show inhibition. Here, affectivity is found to be greater. Similarly, P. chrysogenum metabolite extract showed maximum inhibition of up to 6 μg/ml against all microbes. One study shows inhibition of S. aureus by P. chrysogenum metabolite extract between ranges of 31.25 and 1,000 μg/ml (Visamsetti et al., 2016). Also, one study showed inhibition of Bacillus cereus ATCC 11778 and Streptococcus faecalis ATCC 19433 at minimum inhibitory concentration (MIC) values of 32 and 64 μg/ml (Trinh et al., 2018).
Compare to this study, inhibition against S. faecalis was found to be in the range 4–10 μg/ml and inhibition against B. cereus was found to be in the range of 2–10 μg/ml. Regarding C. cladosporioides and C. pseudocladosporioides inhibitory activities, they were tested against other bacteria like Xanthomonas campestris, (Silber et al., 2014) E. coli, (Li et al., 2017) or any other phytobacterial diseases. Here we found its activity against all the selected test organisms, so it can be further studied against human diseases. As per one study, mycelium extract shows more inhibition than broth fermented extract (Synytsya et al., 2017) but here in our study, it showed equal inhibitory activity against all microbes. Not much difference was seen.
As antibacterial activity of both extracts was performed here, if we try to compare antibacterial metabolites from both methods, metabolites extracted from broth fermentation have shown the highest inhibitory activity against microbes as compared to metabolites from the mycelial extraction method.
 | Table 6. Inhibitory activity of metabolites extracted by dry mycelia. [Click here to view] |
 | Figure 2. Well diffusion method by mycelial extracted metabolites. [Click here to view] |
By analyzing the results, it was found that diversified fungi that are present in the marine environment can produce metabolites. Further study on MIC needs to be done for specific concentration. As a result, future research should take these findings into account and investigate them using more diverse methods and criteria. Further studies can be performed by exploring more marine samples and different isolates.
CONCLUSION
The study revealed the antibacterial potential of fungal metabolites against both Gram-positive and Gram-negative bacteria. The marine fungal metabolites could lead to the development of natural and novel drugs and help to combat drug resistance menace. Further, metabolomics needs to be carried out to characterize the bioactive compounds.
ACKNOWLEDGMENT
Authors are grateful to Department of Microbiology, Sandip University Nashik for providing all necessary support to carry out this research.
LIST OF ABBREVIATIONS
ATCC, American Type Culture Collection; MIC, Minimum Inhibitory Concentration; MTCC, Microbial Type Culture Collection and Gene Bank; PDA, Potato Dextrose Agar; rpm, rotation per minute.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FUNDING
There is no funding to report.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Arumugam A, Ponnusami V. Production of biodiesel by enzymatic transesterification of waste sardine oil and evaluation of its engine performance. Heliyon, 2017; 3(12):1–18; doi:10.1016/j.heliyon.2017.e00486 CrossRef
Arumugam GK, Srinivasan SK, Joshi G, Gopal D, Ramalingam K. Production and characterization of bioactive metabolites from piezotolerant deep sea fungus Nigrospora sp. in submerged fermentation. J Appl Microbiol, 2015; 118(1):99–111; doi:10.1111/jam.12693 CrossRef
Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, Nisar MA, Alvi RF, Aslam MA, Qamar MU, Salamat MK. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist, 2018; 11:1645–58; doi:10.2147/IDR.S173867 CrossRef
Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal, 2016; 6(2):71–9; doi:10.1016/j.jpha.2015.11.005 CrossRef
Christophersen C, Crescente O, Frisvad JC, Gram L, Nielsen J, Nielsen PH, Rahbæk L. Antibacterial activity of marine-derived fungi. Mycopathologia, 1998; 143:135–8. CrossRef
Demain AL. Pharmaceutically active secondary metabolites of microorganisms. Appl Microbiol Biotechnol, 1999; 52(4):455–63; doi:10.1007/s002530051546 CrossRef
Hussein ME, Mohamed OG, El-Fishawy AM, El-Askary HI, El-Senousy AS, El-Beih AA, Nossier ES, Naglah AM, Almehizia AA, Tripathi A, Hamed AA. Identification of antibacterial metabolites from endophytic fungus Aspergillus fumigatus, isolated from Albizia lucidior leaves (Fabaceae), utilizing metabolomic and molecular docking techniques. Molecules, 2022; 8:1–21. CrossRef
Kasanah N, Hamann MT. Development of antibiotics and the future of marine microorganisms to stem the tide of antibiotic resistance. Curr Opin Investig Drugs, 2004; 5(8):827–37.
Kossuga MH, Romminger S, Xavier C, Milanetto MC, Valle MZ, Pimenta EF, Morais RP, Carvalho ED, Mizuno CM, Coradello LF, Barroso VD, Vacondio B, Javaroti DCD, Seleghim MHR, Cavalcanti BC, Pessoa C, Pessoa MO, Lima BA, Gonçalves R, Bonugli-Santos RC, Sette LD, Berlinck RGS. Evaluating methods for the isolation of marine-derived fungal strains and production of bioactive secondary metabolites. Rev Bras Farmacogn, 2012; 22(2):257–67; doi:10.1590/S0102-695X2011005000222 CrossRef
Li HL, Li XM, Mandi A, Antus S, Li X, Zhang P, Liu Y, Kurtan T, Wang BG. Characterization of cladosporols from the marine algal-derived endophytic fungus Cladosporium cladosporioides EN-399 and configurational revision of the previously reported cladosporol derivatives. J Org Chem, 2017; 82(19):9946–54; doi:10.1021/acs.joc.7b01277 CrossRef
Luo Y, Wei X, Yang S, Gao YH, Luo ZH. Fungal diversity in deep-sea sediments from the Magellan seamounts as revealed by a metabarcoding approach targeting the ITS2 regions. Mycology, 2020; 11(3):214–29; doi:10.1080/21501203.2020.1799878 CrossRef
Manimegalai K, Devi NA, Padmavathy S. Marine fungi as a source of secondary metabolites of antibiotics. Int J Biotechnol Bioeng Res, 2013; 4(3):2231–8. Available via http://www.ripublication.com/
Preedanon S, Phongpaichit S, Sakayaroj J, Rukachaisirikul V, Khamthong N, Trisuwan K, Plathong S. Antimicrobial activities of fungi derived from the gorgonian sea fan Annella sp. & their metabolites. Indian J Geo-Mar Sci, 2016; 45(11):1491–8.
Rajasekar T, Balaji S, Kumaran S, Deivasigamani B, Pugzhavendhan SR. Isolation and characterization of marine fungal metabolites against clinical pathogens. Asian Pac J Trop Dis, 2012; 2(SUPPL.1):6–11; doi:10.1016/S2222-1808(12)60187-X CrossRef
Roy S, Banerjee D. Bioactive metabolites of Actinomycetes–screening from genomic and metabolomic approach. J Postgenomics Drug Biomarker Dev, 2017; 7(03):e154; doi:10.4172/2153-0769.1000e154 CrossRef
Schmidt FR. The challenge of multidrug resistance: actual strategies in the development of novel antibacterials. Appl Microbiol Biotechnol, 2004; 63(4):335–43; doi:10.1007/s00253-003-1344-1 CrossRef
Shabana S, Lakshmi KR, Satya AK. An updated review of secondary metabolites from marine fungi. Mini Rev Med Chem, 2021; 21:602–42; doi:10.2174/1389557520666200925142514 CrossRef
Shamly V, Kali A, Srirangaraj S, Umadevi S. Comparison of microscopic morphology of fungi using lactophenol cotton blue, iodine glycerol and Congo red formaldehyde staining. J Clin Diagn Res, 2014; 8(7):7–8; doi:10.7860/JCDR/2014/8521.4535 CrossRef
Silber J, Ohlendorf B, Labes A, Wenzel-Storjohann A, Näther C, Imhoff JF. Malettinin E, an antibacterial and antifungal tropolone produced by a marine Cladosporium strain. Front Mar Sci, 2014; 1:1–6; doi:10.3389/fmars.2014.00035 CrossRef
Song R, Wang J, Sun L, Zhang Y, Ren Z, Zhao B, Lu H. The study of metabolites from fermentation culture of Alternaria oxytropis. BMC Microbiol, 2019; 19(1):4–11; doi:10.1186/s12866-019-1408-8 CrossRef
Synytsya A, Monkai J, Bleha R, Macurkova A, Ruml T, Ahn J, Chukeatirote E. Antimicrobial activity of crude extracts prepared from fungal mycelia. Asian Pac J Trop Biomed, 2017; 7(3):257–61; doi:10.1016/j.apjtb.2016.12.011 CrossRef
Trinh PT, Chanh NT, Ngoc NT, Tien PQ, Ly BM, Van TT. Secondary metabolites from a marine-derived fungus Penicillium chrysogenum 045-357-2. Vietnam J Sci Technol, 2018; 55(1A):65; doi:10.15625/2525-2518/55/1a/12383 CrossRef
VanderMolen KM, Raja HA, El-Elimat T, Oberlies NH. Evaluation of culture media for the production of secondary metabolites in a natural products screening program. AMB Express, 2013; 3:1–7; doi:10.1186/2191-0855-3-71Visamsetti A, Ramachandran SS, Kandasamy D. Penicillium chrysogenum DSOA associated with marine sponge (Tedania anhelans) exhibit antimycobacterial activity. Microbiol Res, 2016; 185:55–60; doi:10.1016/j.micres.2015.11.001 CrossRef
Wang YT, Xue YR, Liu CH. A brief review of bioactive metabolites derived from deep-sea fungi. Mar Drugs, 2015; 13(8):4594–616; doi:10.3390/md13084594 CrossRef
Wong Chin JM, Puchooa D, Bahorun T, Jeewon R. Antimicrobial properties of marine fungi from sponges and brown algae of Mauritius. Mycology, 2021; 12(4):231–44; doi:10.1080/21501203.2021.1895347 CrossRef