INTRODUCTION
Natural products are unique sources of medicinal ingredients with various chemical structures. The unique nature of the natural product has many advantages for the medical world. Profits in the medical world are supported by the fact that almost 50% of the new drugs produced are derived from natural products and their derivative compounds (Newman and Cragg, 2012). Sources of natural products can come from organisms on terrestrial or marine resources. The marine wealth which is currently a source of very interesting natural products for researchers is sponges. Sponges are multicellular organisms and are the lowest level of marine invertebrates. Most of them live in the sea (80%), while the rest live in freshwater. Sponges can be found in all marine areas from the equator to the poles, in shallow and deep seas (Hooper and Van Soest, 2002). They are marine biota as a source of bioactive compounds. These porous animals host a wide variety of microorganisms. Such association with microbes is one of the factors why sponges can produce secondary metabolites which are bioactive compounds (Taylor et al., 2007).
The development of new drugs in recent decades has been growing very fast. Sponges and their associated microorganisms which are sources of bioactive compounds continue to be explored by researchers. To obtain bioactive compounds to be used for disease treatment, either from sponges or from sponge-associated microorganisms, takes a long time and costs much. Although efforts to make the process faster have been made by using alternative synthetic compounds by isolation, these efforts have still not generated maximum results (Mishra et al., 2008). The discovery of new bioactive compounds derived from sponges and sponge-associated microbes is a major challenge for researchers. This discovery currently focuses on single compounds with certain biological activities that are tested at the molecular level (Verpoorte et al., 2005). This makes researchers and pharmaceutical companies compete to conduct research based on a holistic approach to traditional medicine. Holistically, the preparation of raw materials either directly from marine sponges or from sponge-associated microbes containing many multicomponents is simulated as a single unit of bioactive compounds that have activity on several receptor targets in living organisms. To be able to observe the response of living organisms to these bioactive compounds, it is necessary to conduct a test by observing physiological responses and molecular responses that are used to obtain better complex data, known as the biological system approach (Ulrich-Merzenich et al., 2007; Verpoorte et al., 2005). One of the biological system approaches with the newest “omics” method is metabolomics which is considered the most informative in the biological system approach because it can reflect the genotype (Sumner et al., 2003).
The application of metabolomics in the process of new drug discovery is inseparable from the use of the chemical profile of natural products. Chemical profiles combined with bioactivity data produce complex data to lead to active components of natural products that are useful in the medical field. To obtain a reliable chemical profile that can represent the active components and chemical characteristics, a combination of chromatographic and spectroscopic techniques is required. This combination of instruments can increase selectivity, separation capability, and measurement precision, as well as reduce personal instrument interference (Gong et al., 2001). At any given time, the process of distinguishing the chemical profile of natural products is usually subjective and nonquantitative, so small differences between species may be overlooked (Xu et al., 2006). Possible solutions to this weakness in metabolomics can be accounted for by multivariate analysis combined with chemical profiling, known as chemometrics. Chemometrics in metabolomics plays an important role in providing detailed characteristics of the chemical profile combined with biological activities (Yuliana et al., 2011).
This review aims to present a brief overview of the applications of metabolomics over the past decade to solve problems in the discovery of new drugs from marine sponges and sponge-associated microbes. This review describes, in general terms, the species of sponges and their associated microbes, the method of separation in determining chemical profiles, the chemometric analysis used, and the purpose of using metabolomics in this study. The advantage of this review is to determine the appropriate chemometric method used for research purposes related to determining the chemical profile of marine sponges and their associated microbes, so in the future, it can accelerate the process of using metabolomics in related studies.
METHODS
A systematic search was done to find all publications related to the topic from July 2011 to July 2021 (a decade) on PubMed and Google Scholar. The keywords used to search the articles were “sponge, marine, microorganism, metabolomics” or “sponge, sea, microbe, chemometric.” The data included in this review were original articles, research articles, and main articles on the study of metabolomics in sponges and their associated microbes, as shown in Table 1. Articles were excluded from the main data if they were review articles, conference articles, theses, and dissertations and if there were no data available for retrieval. The variables assessed in this review included species/genera of sponges, species/genera of sponge-associated microorganisms (if any), methods used for chemical profiling, chemometric methods for analysis, and the intended use of metabolomics in the study.
SPONGES AND SPONGE-ASSOCIATED MICROORGANISMS
The search found 62 main articles published from January 2011 to July 2021 (Table 1). We identified 47 genera of sponges that were studied using metabolomics. The sponges most often studied in metabolomic-related research were Geodia, Xestospongia, Agelas, Aplysina, Callyspongia, Haliclona, Plakortis, Sarcotragus, and Spheciospongia. There were 25 out of the 62 main articles using 24 genera of sponge-associated microbes. The associated microorganisms mostly came from actinobacteria (12 genera), bacteria (6 genera), proteobacteria (3 genera), cyanobacteria (1 genus), firmicutes (1 genus), and fungi (1 genus). Figure 1 shows the number of studies related to sponges and their associated microbes by applying metabolomics. In general, there were an increased number of publications in that decade from year to year. The highest number of publications was found in 2020 with 12 articles, followed by 2019 and 2017, each with 10 articles. The increasing number of publications is related to the trend of metabolomic-based research, especially with the objects of sponges and their associated microorganisms. Exploration of sponges and their associated microbes has also become the focus of many scientists, especially in the exploration of microbial associations of marine sponges. This will certainly reduce the exploitation of marine sponges which very slowly grow, thus lowering the possibility of marine sponges extinction if excessive exploration is carried out (Carroll et al., 2019; Samirana et al., 2021b). The bioactive contents derived from marine sponges have been widely used in the medical world and will continue to be developed in the future. Several marine sponges and sponge-associated microorganisms have been shown to have antibacterial, antitumor, antiviral, and anticancer (cytotoxic) activities (Guo et al., 2019; Samirana et al., 2021a; Wang, 2006). Several studies have reported that sponge-associated microorganisms have a major role in sponges in producing secondary metabolites that have biological activities. These sponge-associated microbes are known to be the body tissue of sponges which account for about 40%–50% of the body tissues of marine sponges (Proksch et al., 2002; Samirana et al., 2021b; Thakur and Müller, 2004).
Research related to marine sponge-associated microbes has been carried out. Marine sponge-associated microbes produce bioactive compounds that are almost similar to sponges as the hosts. Marine sponges and their associated microorganisms produce secondary metabolites which are significantly influenced by the environment in which marine sponges grow. The environment can be a state of carbon and nitrogen sources, salinity of seawater, and light sources obtained by sponges and their associated microorganisms (Samirana et al., 2021b). Separating marine sponge-associated microorganisms with their host will certainly affect their metabolism in producing secondary metabolites. Therefore, to allow marine sponge-associated microbes to be able to produce secondary metabolites similar to those in their habitat, the growth medium of these sponge-associated microbes needs to be carefully considered. By making a growth medium that is as similar as possible to the original habitat of the sponge-associated microorganisms, it is expected that these microorganisms can still produce secondary metabolites and the possibility of mutations in these microbes can decrease (Debbab et al., 2011; Huang et al., 2011; Lee et al., 2001; Kjer et al., 2010).
 | Table 1. Summary of data from applications of metabolomics to sponges and sponge-associated microorganisms. [Click here to view] |
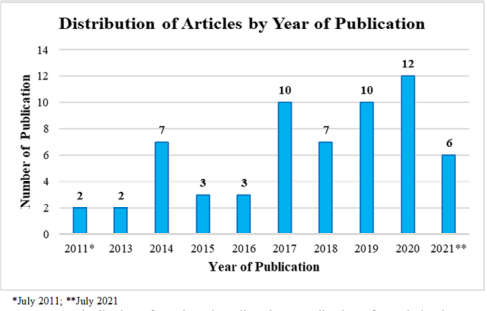 | Figure 1. Distribution of conducted studies about application of metabolomics on sponges and their associated microorganisms. *July 2011; **July 2021. [Click here to view] |
ANALYTICAL METHODS FOR METABOLOMICS
Metabolomics is a new alternative method that facilitates the multitarget analysis of endogenous cellular metabolites. Metabolomics, as a new “omics” field, is a combination of genomics, transcriptomics, and proteomics. The main objective of metabolomics is to carry out qualitative and quantitative analysis of all the metabolites (metabolome) contained in an organism at a given time and to a certain effect. Such an approach represents a paradigm shift for understanding the pathophysiological processes in organisms. In addition, this method can detect metabolite profiles with different phenotypes (Colquhoun, 2007; Isgut et al., 2018; Ulrich-Merzenich et al., 2007). Metabolomics provides a (semi)quantitative measurement of multiparametric metabolic responses in living systems simultaneously to monitor changes in hundreds of low-molecular-weight metabolites (i.e., small organic molecules with MW < 1,500 Da). In marine research, the application of metabolomics aims to identify the biomarkers with certain phenotypes of organisms in the sea. However, the diversity and complexity of types of chemical structures make metabolomic analysis a challenge in the future. Significant strides have been made in the analysis of complex metabolomic data (Favre et al., 2017; Kaplan et al., 2004; Rangel-Huerta and Gil, 2016).
A metabolome is a component of an organism that can be seen from the end product of gene expression, so it can be used as a tool to monitor the gene function of the organism. The genetic profile of an organism can be analyzed using the polymerase chain reaction (PCR) method which will produce a genetic pattern of an organism that is usually specific for each individual. A metabolomic analysis of the genetic profile produced by PCR can be done using a multivariate analysis, namely, a cluster analysis. A cluster analysis helps group organisms that share the same genetic profile. Identification using this PCR technique has limited information because it only identifies mRNA and protein based on sequence similarity and database matching. Therefore, the application of metabolomics was further developed using other instruments to provide an integrated understanding of information about an organism (Sumner et al., 2003; Ulrich-Merzenich et al., 2007). The current metabolomic methodology uses instruments based on chromatography and spectroscopy. The use of these methods in metabolomics is an ideal means of chemical screening and subsequent detailed comparison of the secondary metabolism of organisms. Different spectroscopic techniques [nuclear magnetic resonance (NMR), mass spectroscopy (MS) and chromatographic techniques [high-performance liquid chromatography, GC, gas chromatography-mass spectrometry (GC-MS), liquid chromatography-mass spectrometry (LC-MS), Thin Layer Chromatography (TLC), etc.] are widely used in the metabolomic analysis of natural products for quality assurance and discovery of new compounds (Shyur and Yang, 2008). Spectroscopic techniques such as NMR and MS can provide early-stage structural information for further identification of compounds. The observed fragmentation pattern of the MS spectrum can be a guide for the evaluation of molecular networks related to the identification of compound relationships produced by several individual types through spectral correlation (Kim et al., 2011; Sidebottom et al., 2013; Valentino et al., 2020). The chromatographic techniques are usually used when the compound has a structure that has previously not been identified, so it is unavoidable to use a chromatographic technique to characterize the structure of the compound. Chromatographic techniques are usually used at the beginning of the separation and are guided later by the MS and NMR techniques for structural determination (Geng et al., 2014; Grkovic et al., 2014). Figure 2 presents a workflow for metabolomic-guided sponges and their associated microorganisms.
The research on the application of metabolomics to sponges and their associated microbes during the last decade shows that most of the analytical methods still use chromatographic techniques in determining chemical profiles combined with MS. Of the 62 main articles obtained, there were 54 articles using the LC technique combined with MS, 19 articles using the PCR technique in determining the genetic profile, nine articles using the NMR technique, and six articles using the gas chromatography technique combined with MS.
Chromatographic techniques are often used to separate complex compounds before detection. LC-MS and GC-MS are the most commonly used compound analysis techniques that can cover a wide range of metabolites. LC is most suitable for the separation of complex compounds from scratch, although it has a limited polarity window. Therefore, many early metabolomics studies used the LC technique combined with MS for chemical profiling. The GC technique requires that the separated metabolites are volatile and thermostable, but many secondary metabolites do not have thermostable and volatile properties, making detection difficult. Therefore, GC is least used in metabolomics research as an analytical method because not many metabolites are separated by this technique. The LC and GC techniques are usually combined with mass spectroscopy to detect the metabolites obtained. The combination of MS with chromatography (LC and GC) provides a clearer picture of the identification of compounds, especially for new compounds. The use of high-resolution MS will provide information on the molecular formula of a compound that helps identify compounds, although low-weight molecules will have many compounds with the same molecular weight and the molecular weights of many new compounds are not known (Badjakov et al., 2008; Han et al., 2009). The use of chromatographic techniques combined with MS (LC-MS and GC-MS) in metabolomic applications on sponges and their associated microorganisms in general aims at determining the chemical profile of the sample. The determination of chemical profiles aims at conducting bioassay-guided isolation. In conducting bioassay-guided isolation, the use of a dereplication model of the active compound profile becomes very important. The metabolomic approach here plays a very important role in facilitating the bioassay-guided isolation process by utilizing the multivariate data analysis so it can shorten the isolation route of an active compound, especially in the identification and dereplication steps. The most time-consuming step in metabolomic work is the identification of metabolites in the fractionated mixture or extract. Therefore, the availability of the spectral library of a compound is very important in accelerating such identification. Several studies on metabolomics in sponges and their associated microbes reported that the use of LC-MS and GC-MS resulted in complex chemical profiles, thus requiring a reliable spectral library to assist the identification process; several studies used molecular networking methods combined with a cluster analysis in the identification of compounds (Alkhalifah, 2021; Erngren et al., 2021; Fagundes et al., 2021; Ho et al., 2021; Yuliana et al., 2011). Therefore, it is highly recommended to apply metabolomics on sponges and their associated microbes whose active compounds are not known using chromatographic techniques combined with MS by conditioning the type of compounds to be separated and conducting chemical profiles to facilitate a multivariate data analysis. In addition, there are still few metabolomic studies on sponges and their associated microbes.
 | Figure 2. Workflows of metabolomics-guided on sponges and their associated microorganisms. [Click here to view] |
The next technique that is often used in metabolomic applications on sponges and their associated microbes is the PCR technique. This technique is a technique for determining the genetic profile of an organism by checking its mRNA sequence and protein composition. This technique has the advantage of determining species of organisms that have morphological similarities, especially for groups of microorganisms. Microorganisms currently in the discovery of modern drugs play an important role because they are a source of antibiotic and antiproliferative agents in the clinical world, in the form of either direct compounds isolated from microbes or their semisynthetic derivative products, including sponge-associated microorganisms that have been widely proven to have biological activities in the clinical field (Debbab et al., 2011; Huang et al., 2011; Newman and Cragg, 2016). There will certainly be errors in isolating and culturing the desired microorganism based on morphological similarities. Therefore, a technique is needed to be able to distinguish species genetically, namely, the PCR technique. This method can distinguish individuals who are morphologically similar but genetically different; genetic differences will cause individual microorganisms to produce different secondary metabolites. Advances in the genome sequencing technology in the PCR techniques make it easier to distinguish microorganisms that have active and nonactive metabolites and discover microorganisms with new genetic makeup. This analytical method can reveal several microorganisms that have the potential to produce more secondary metabolites than usual albeit with almost the same morphological similarities (Baral et al., 2018; Sekurova et al., 2019). Over the past decade, marine organisms, especially sponge species, have become an important source of highly diverse and unusual, and often highly complex, natural products, for which the number of new chemical structures being reported from sponges and their associated microbes is increasing steadily. Sponges are known to be the host of many microorganisms, be it fungi, bacteria, actinomycetes, and others, so many new chemical structures will be found from these associated microorganisms (Achlatis et al., 2019; Raimundo et al., 2018). Therefore, the PCR technique plays an important role in the process of determining the specific species of sponge-associated microorganisms. The results of the genome sequencing from the PCR technique will usually be combined with a multivariate data analysis in the form of a cluster analysis. This analysis cluster is usually in the form of a phylogenetic tree of microorganisms which can later be explained genetically; the genetically analyzed organisms have similarities to the microorganisms already listed in the gene bank data. This metabolomic application certainly greatly facilitates the determination of the species of marine sponge-associated microorganisms. Genomic characterization in several studies on marine sponge-associated microbes is very helpful in sorting out microorganisms that will produce active secondary metabolites based on the existing studies (Matroodi et al., 2020; Nouioui et al., 2017). Therefore, this PCR technique is chosen in metabolomic applications usually on species-specific characterization of organisms, especially for sponges and their associated microbes.
NMR is an analytical technique that is considered ideal for metabolic work. This is due to its excellent reproducibility and its database that can be used openly by the public which aims to add to the data obtained. NMR spectroscopy provides the most detailed information on the chemical compounds present, and by applying the 2D NMR technique, identification and explanation of the chemical structure in a sample mixture can be performed repeatedly. The advantages of using the NMR technique are simple and fast preanalytical sample preparation, short measurement times, and the ability to describe the chemical structure of a compound from a complex mixture. The main advantage of using NMR is that each proton will give the same signal intensity, so the quantification process is simple and only requires an internal standard, whereas other methods require a calibration curve for every compound (Colquhoun, 2007; Verpoorte et al., 2007). Like the LC chromatography technique, the NMR technique also has a weakness in terms of the limited polarity of the solvent. The solution to it is the use of solid-phase NMR, but the resulting signals are wide and overlap. In metabolomic applications on sponges and their associated microbes, the NMR technique provides initial information on the chemical structure of compounds in the form of NMR spectral profiles for compound identification. The step that can be taken is to compare the 1H NMR chemical shift information with the available literature or database. The next step after the structure has been known is to identify the discriminator without requiring a time-consuming and expensive isolation process; thus, metabolomic applications can function as dereplication, which is then continued with the help of 2D NMR techniques. If the compound has a core structure that has never been identified, then chromatographic isolation is unavoidable for structural characterization, followed by a multivariate data analysis as a tool to track the desired compound target (Bayona et al., 2018; Grkovic et al., 2014; Olatunji et al., 2021; Verpoorte et al., 2007). The use of NMR techniques in metabolomics is the ideal solution, but there are still very few metabolomics studies that fully use this method. There are several possibilities why this method is still not commonly used. First, this technique costs quite much because it uses specific and expensive materials for NMR. Second, there are still many compound structures that have not been previously identified, so researchers prefer to use a chromatographic technique combined with MS as an alternative for early identification.
Through the application of metabolomic applications to sponges and their associated microorganisms, many single compounds have been discovered. These compounds can come from the sponge itself or can come from its associated microorganisms. The single compounds that have been isolated and identified from marine sponges using metabolomic applications are as follows: halisulfate 1; halisulfate (3-5); suvanine (Ali et al., 2013); balibaloside; 6″-O-acetylbalibaloside; 6″′-O-acetylbalibaloside; 6″,6″′-O-diacetylbalibaloside (Audoin et al., 2013); barettin; 8,9-dihydrobarettin; bromobenzisoxazolone-barettin; N-acyl-taurine geodiataurine (Olsen et al., 2015); dienone (2-(3,5-dibromo-1-hydroxy-4-oxocyclohexa-2,5-dien-1-yl)acetamide); 3,4-dihydroxyquinoline-2-carboxylic acid; aerophobin-1; aerophobin-2; aplysinamisin-1; 11-OH-aerothionin; aerothionin; homoaerothionin; 11-deoxyfistularin-3 (Reverter et al., 2016); crambescin; crambescidin (Ternon et al., 2016); demethylfurospongin-4; furofficin; furospongin-1; spongialactam A; spongialactam B (Bauvais et al., 2017); haliclamine, cyclostellettamine, viscosaline; viscosamine (Einarsdottir et al., 2017); monobromoagelaspongin; (−)-equinobetaine B (Sauleau et al., 2017); tsitsikammamine A; 16,17-dehydrotsitsikammamine A (Li et al., 2018); hyrtiodoline A (Shady et al., 2018); smenamide F; smenamide G (Caso et al., 2019); 5-bromo trisindoline; 6-bromo trisindoline (El-Hawary et al., 2019); (−)-discorhabdin L; (+)-discorhabdin A; (+)-discorhabdin Q; (−)-2-bromo-discorhabdin D; (−)-1-acetyl-discorhabdin L; (+)-1-octacosatrienoyl-discorhabdin L (Li et al., 2019); smenamides A; smenamides B; smenothiazoles A; smenothiazoles B (Teta et al., 2019); stylissamide A; stylissoside A (Abdelhameed et al., 2020); xestenone; spongialactam A; spongialactam B (Chaudhari and Kumar, 2020); (−)-cyclo(L-trans-Hyp-L-Ile); cyclo(L-trans-Hyp-L-Phe); 1-O-hexadecyl-sn-glycero-3-phosphocholine; 1-O-octadecanoyl-sn-glycero-3-phosphocholine; 3β-hydroxycholest-5-ene-7,24-dione; (22E)-3β-hydroxycholesta-5,22-diene-7,24-dione; loliolide; 5-epi-loliolide (Kouchaksaraee et al., 2020); tridiscorhabdin; didiscorhabdin (Li et al., 2020); sarasinoside A1; sarasinoside B1 (Mohanty et al., 2020b); hachijodine E; nakadomarin A; amphimic acid A; manzamine H; amphilactam A (Shady et al., 2020); pateamine A; peloruside A; mycalamide A (Storey et al., 2020). The single compounds that have been isolated and identified from associated microorganisms from marine sponges using metabolomic applications are as follows: actinosporins A; actinosporins B (Abdelmohsen et al., 2014); 4,10-dihydroxy-10-methyl-dodec-2-en-1,4-olide; 4,11-dihydroxy-10-methyl-dodec-2-en-1,4-olide; 4-hydroxy-10-methyl-11-oxo-dodec-2-en-1,4-olide (Viegelmann et al., 2014); petrocidin A; 2,3-dihydroxybenzoic acid; 2,3-dihydroxybenzamide; maltol (Cheng et al., 2017); fridamycins H; fridamycins I; actinosporin C; actinosporin D; actinosporin G (Tawfike et al., 2019b); phenazine-1,6-dicarboxylate; phencomycin; tubermycin; N-(2-hydroxyphenyl)-acetamide; p-anisamide (Hifnawy et al., 2020); N-palmitoyl-l-leucine; N-palmitoyl-l-phenylalanine, N-palmitenoyl-l-phenylalanine; N-oleyl-l-phenylalanine (Vitale et al., 2020); ethyl plakortide Z; ethyl didehydro-seco-plakortide Z; manadoperoxide H; acanthosterol sulfate F; acanthosterol sulfate G (Alkhalifah, 2021).
CHEMOMETRIC METHODS FOR METABOLOMICS
Chemometrics is a branch of science that relates the measurement of chemical systems or processes to certain conditions through the application of mathematical or statistical methods. Chemometrics is basically classified into two main categories according to the intended uses. A qualitative use is known as a pattern recognition method (without supervision) while a quantitative use aims at multivariate calibration (Brereton, 2003; Gemperline, 2006). In its application in metabolomics, chemometrics has a very important role in data processing. The data can be in the form of a chemical profile consisting of retention time and peak area on the chromatogram, absorbance, or transmittance in spectroscopy. These chemical data are displayed in variables (multivariate data) which are plotted in the same number of dimensions as the existing variables. Chemometrics can be used to design or select optimal procedures and tests, as well as to extract as much chemical information as possible from the data. Multivariate data measurement has a lot to do with chemometrics, where multivariate data are the results from measuring many variables in the same sample. The steps in a chemometric analysis include experimental design, data preprocessing, classification, and calibration (Hanrahan and Gomez, 2010; Rohman, 2014). Modeling with chemometrics requires instruments and software to interpret patterns in the data. Several methods in chemometrics that are commonly used include principal component analysis (PCA); chemometrics regression [partial least square (PLS), PCR, MLR, and 3-way PLS) and prediction analysis; soft independent modeling by class analogy and partial least square-discriminant analysis (PLS-DA) classification; design of experimental, analysis of variance, and response surface methodology; multivariate curve resolution and clustering (K-means) (Berrueta et al., 2007). The use of chemometrics in the form of a multivariate data analysis in metabolomic applications on sponges and their associated microbes is for sequencing the PCR results and analyzing the chemical profiles obtained from the separation by chromatographic and spectroscopic techniques. A multivariate data analysis is used to represent the statistical weights of all significant variables and distributed among individuals according to their biochemical contents.
Research on the application of metabolomics to sponges and their associated microbes over the past decade has demonstrated the use of a chemometric analysis that is inseparable. Of the 62 main articles obtained, similarity analysis (SA) was found to be the most frequently used chemometric technique, with 40 articles using the analysis. The next most commonly used technique was PCA, with 24 articles using it. In addition, hierarchical cluster analysis (HCA) was used in 22 articles. Next, 10 articles used orthogonal projections to latent structures-discriminant analysis (OPLS-DA), and three articles used orthogonal projections to latent structures (OPLS). There were eight articles that used PLS-DA, and one article used partial least square (PLS). Finally, there was one article that used linear discriminant analysis (LDA).
SA is the most widely used technique in the metabolomic applications on sponges and their associated microbes. This chemometric technique is usually used in unsupervised metabolomic applications that aim to explore compounds to detect similarities in chemical structures through the resulting chemical profiles. The first step of this analysis is to determine the similarity between objects using the SA technique, which is based on the correlation coefficient r. This is implemented using fingerprints on references from standard compounds or extracts, but this data library is still not widely available. An alternative step that can be taken is to determine the average or median fingerprint from the collected data and perform SA based on all available fingerprint profiles, relative retention time, and the peak area of the peak to be characterized (Chen et al., 2010; Li et al., 2010; Zhu et al., 2010). This may appear to be a subjective approach as the fingerprint profile depends on the composition and size of the data set, which can affect the results. Another weakness is the high contribution of the similarity value to the main peak, so it covers the similarity value from the smaller peak. Despite these drawbacks, SA is a fast and easy-to-use technique that is useful for the initial analysis of datasets. Therefore, this technique is preferred by most researchers who perform metabolomic applications on sponges and their associated microbes (Gan and Ye, 2006; Parejo et al., 2004). Along with the development of technology and the availability of many databases over the last decade, the SA technique was developed into a molecular networking database that makes it easier to perform a similarity analysis of available chemical profiles. The chemical profile is usually spectrally derived from MS. These chemical profile data are then processed by the database library, and the resulting grouping of similar chemical profiles makes it easier to carry out a subsequent analysis. Molecular networking has been used by several recent studies to analyze the metabolomic application on sponges and their associated microbes. The chemical shift profile of the NMR spectroscopy allows it to be analyzed with this molecular networking technique, provided that the available database is sufficient (Fagundes et al., 2021; Ho et al., 2021; Kouchaksaraee et al., 2020; Li et al., 2020).
PCA is a form of data interpretation technique in a chemometric or metabolomic analysis. The purpose of PCA is to reduce the large dimensions of the data space (observed variables) to smaller dimensions of the data space (independent variables), to describe the data more simply (Pratiwi and Harjoko, 2013). PCA is an interpretation of data that is carried out with data reduction, in which the number of variables in a matrix is reduced to produce new variables while maintaining the information held by the data. The resulting new variable is in the form of a score or main component. This technique can reduce the influence of noise and take advantage of subtle differences from spectrum data (Che Man et al., 2011). There are two ways to determine the number of principal components (PCs) to be used for the analysis: first by looking at a minimum of 80% of the total proportion that can be explained and second by observing the scree plot, namely, by looking at the elbow fracture of the scree plot (Johnson and Wichern, 2007). PCA interpretation can be obtained from a loading analysis. Loading is the correlation between the original variable and the new variable. Loading provides an indication about which original variables are very important or have an effect on the formation of new variables. The higher the loading value, the more influential the old variable on the formation of new variables (Sharma, 1996). There are four important pieces of information obtained from the biplot display, including the closeness between the observed objects, the diversity of variables, the correlation between variables, and the value of variables on an object (Che Man et al., 2011; Sartono et al., 2003). In metabolomic applications on sponges and their associated microbes, the large number of chemical profile data from LC-MS makes it difficult to process them; the PCA method is convenient to summarize these data without losing the uniqueness of each datum. PCA is used to represent the statistical weights of all significant variables and their distributions based on their biochemical contents. A PCA analysis including unsupervised metabolomics is applied to reduce the dimensions of the data while preserving most of the variation in the dataset. One of the advantages of the PCA technique in metabolomics is its ability to determine the relationship between the chemical profile and the given biological activity. This is certainly very helpful in the search for new active compounds from sponges and their associated microbes. Several studies have shown that the PCA technique can determine the relationship between the bioactivity of sponges and their associated microbes with their chemical profile (Einarsdottir et al., 2017; Ivanisevic et al., 2011; Reverter et al., 2016).
The next chemometric analysis technique that is often used in metabolomic applications on sponges and their associated microorganisms is HCA. This chemometric technique is in the form of a hierarchical grouping based on the creation of branched structures, which are called a dendrogram. A dendrogram displays data qualitatively and allows showing cluster visualization and correlations between samples. HCA in its application uses two main methods in comparing samples. The first is the agglomeration technique, in which each observation starts in its own individual cluster and joins the others as they move up the hierarchy. The second is the division technique, which starts with all the samples in one cluster and then splits down the hierarchy. To determine when clusters should be split or merged, a measure of (dis)similarity between samples is required for a relationship criterion that determines the association between clusters. The main objective of HCA is to visualize data in a cluster that is in a two-dimensional polarity (Beebe et al., 1998; Brereton, 2003). The HCA technique in the metabolomic application on sponges and their associated microbes is usually coupled with the PCR technique for the purpose of a genome sequence analysis in determining the genotype of an organism. The HCA technique in metabolomics coupled with the PCR technique will be described in a phylogenetic tree, where this phylogenetic tree visualizes the relationships between individuals or specific genotypes of a sponge or its associated microorganisms (Matroodi et al., 2020; Nouioui et al., 2017). Therefore, the HCA technique in the application of metabolomics on sponges and their associated microbes aims at individual clusters as well as species-specific determination.
PLS or partial least square is usually used in estimating the dependent variable (response) of a large number of predictor independent variables that have a linear or nonlinear systematic structure with or without missing data and have high collinearity (Gemperline, 2006). PLS is used in multivariate calibration because of the quality of the resulting calibration model and its easy implementation. In the PLS chemometric technique, the selected variable is a variable that has a good correlation with the response, so the variable will provide a more effective prediction (Adams, 2004). This PLS method is a linear combination of predictive variables selected from variables that have a high correlation with the response variable and explains the variation in the predictive variable (Miller and Miller, 2010). Regression in PLS is carried out using a least-squares algorithm that connects two matrices, namely, the spectra data on the X matrix and the reference value on the Y matrix. PLS forms a new variable called a latent variable or component, where each component is a linear combination of independent variables. The main purpose of PLS is to form components that can capture information from variables to predict response variables (Garthwaite, 1994). An alternative technique of PLS that discriminates more against variables that are very influential in a system is the PLS-DA. Both the PLS and PLS-DA techniques provide advantages in the form of the formation of a PLS regression component that can describe the correlation between the X and Y variables. In metabolic applications on sponges and their associated microbes, both PLS and PLS-DA (especially PLS-DA) are used to extract information from chemical profiles from both MS and NMR spectra which are complex with overlapping peaks, impurities, and noise from the instrument used (Ali et al., 2013; Bojko et al., 2019; Ternon et al., 2017).
The analytical technique in chemometrics that can also be used in metabolomics is OPLS. This technique is still very new in chemometrics which is used for the projection of supervised multivariate data which is used to relate a set of predictor variables (X) with one or more responses (Y). Basically, OPLS has similarities with PLS, but OPLS has the ability to extract maximum information that reflects variations in the dataset, while assuming the presence of a small subset of hidden variables in the X data to predict the response variable. This subset is widely known as the latent variable (LV) because it is not measurable. The concept of hidden structures in this dataset is derived from a well-known chemometric technique, namely, PCA. The OPLS technique uses orthogonal signal correction to maximize the covariance described in the first LV, while the remaining LV captures the variance in the orthogonal predictors, which are not statistically correlated with the response variable. The OPLS technique, unlike PLS, which handles random noise quite well, allows structured noise filtering in the dataset by separately modeling the variation of the correlated and uncorrelated X predictor with Y response. In conclusion, the OPLS technique reduces the complexity of the model by decreasing the number of LV and allows for the identification, analysis, and investigation of orthogonal primary sources (Brereton, 2003; Tapp and Kemsley, 2009). A complementary technique of OPLS is OPLS-DA, which aims at filtering out the differential variables responsible for the differentiation between groups after scaling. The loading-plot and S-plot generated from the OPLS-DA model are used to visualize the relative importance of the differential variables and obtain a list of peak indices (Yang et al., 2017; Wu et al., 2018). In the application of metabolomics on sponges and their associated microbes, the use of the OPLS and OPLS-DA techniques (especially OPLS-DA) is very useful and promising. The use of these two techniques can determine and predict fractions or extracts derived from active and inactive sponges or their associated microbes based on the tested bioactivity. Thus, bioactivity tests will be easier and simpler when the number of extract samples or fractions is quite large. The application of metabolomics to sponges and their associated microbes using the OPLS-DA chemometric technique has been done in many studies to be able to predict active or inactive samples (Ali et al., 2013; Bayona et al., 2018; Erngren et al., 2021; Tawfike et al., 2019a).
OBJECTIVES OF METABOLOMICS RESEARCH
Initially, metabolomic applications were used to analyze higher plants in the land zone, but over time this application can also be applied to groups of organisms in the marine zone. Metabolomics plays a crucial role in the study of marine organisms, especially for studies of sponges and their associated microorganisms. In general, the roles of metabolomic applications on marine organisms, especially sponges and their associated microbes, include explaining some specific metabolites of a species for the development of metabolic profiles, identifying and explaining natural products of sponges and their associated microbes and biomarkers that have biological effects, and providing a mechanical understanding of the effects of the environment in which sponges and their associated microbes grow, such as carbon and nitrogen sources, salinity, and required micronutrients (Samirana et al., 2021b; Yuliana et al., 2013).
The purpose of research related to metabolomics on natural products includes all organisms that exist on land and in the sea. There are many purposes of research with metabolomic application on natural products that will be grouped into seven groups, including research with metabolomic application on sponges and their associated microorganisms. The groupings are as follows: (a) metabolomics for identification and dereplication, (b) metabolomics for quality control, (c) metabolomics to link chemical profile and bioactivity pattern, (d) metabolomics for identification of active compounds and quantitative prediction of bioactivity, (e) metabolomics for proof of efficacy and mode of action identification, (f) metabolomics for bioavailability and fate of natural compounds assessment, and (g) metabolomics for identification of safety and toxicity (Yuliana et al., 2011). Of the 62 main articles obtained, most of them (58 articles) applied metabolomics on sponges and their associated microbes with the purpose of the identification and dereplication. In addition, 11 articles had the purpose of quality control, and 10 articles applied metabolomics to link chemical and bioactivity profiles. Unfortunately, no metabolomic research aiming at quantitative prediction of bioactivity, bioavailability, safety, and toxicity in sponges and their associated microbes during the last decade was found.
For the purpose of identification and dereplication, the metabolomic application is carried out with a data reduction approach and it is currently widely applied in research on new drug discovery from natural products using bioassay-guided isolation. In this approach, rapid dereplication of known and identified active components is essential. The use of various analytical techniques combined with appropriate multivariate analytical data can be used to shorten the isolation route based on bioassays with data reduction approaches, especially in the identification and dereplication steps. Several studies on sponges and their associated microbes, which have a large number of chemical profiles or genome profiles, use the multivariate data analysis technique, particularly HCA more than PCA, because the number of the main components in the first technique is 50% more than that of the second technique. The PCA technique is more appropriate for similar large datasets. The use of this reduced clustering technique can accelerate the dereplication step and avoid overanalyzing for the selection of isolation techniques in the discovery of new natural products, especially from sponges and their associated microbes (Cheng et al., 2015; Ellis et al., 2017; Mehbub et al., 2016; Romoli et al., 2014; Yuliana et al., 2011). The most time-consuming step in metabolomic research is the identification of metabolites when a mixture fraction has not been available. In relation to the use of MS and NMR spectra, in several studies of metabolomic applications on sponges and their associated microbes, the spectra data are then analyzed with the help of MS or NMR spectra databases available in the spectral data library using a SA. Currently, a SA analysis has been developed into a molecular networking analysis which is very helpful in analyzing chemical structures obtained through MS or NMR spectra data (Bayona et al., 2020; Kouchaksaraee et al., 2020; Olatunji et al., 2021; Storey et al., 2020).
The most popular application of metabolomics today is natural product quality control. The metabolite profile of an organism can have differences due to variations between species or varieties, environmental changes during the growing or harvesting period, postharvest treatment, extraction processes, and sample preparation methods. These factors have a very significant effect on the chemical profile and bioactivity of the samples, especially samples derived from sponges and their associated microbes (Wang et al., 2005). The application of the NMR and MS techniques combined with LC and GC followed with a multivariate analysis to detect variations due to changes in the environment where sponges and their associated microbes grow has been widely reported. The multivariate data analysis techniques that are often used in metabolomic applications for quality control of sponges and their associated microbes are PCA, PLS-DA, SA, and OPLS-DA. Almost all research on the application of metabolomics for quality control on sponges and their associated microbes is related to the environment in which they grow which later affects the chemical profile of both the MS and NMR spectra. This way, to conduct metabolomic research on the influence of the environment where sponges and their associated microbes grow, using the NMR or MS techniques combined with LC or GC, and PCA, PLS-DA, SA, and OPLS-DA, is recommended (Bojko et al., 2019; Bayona et al., 2018, 2020; Fiore et al., 2017; Mehbub et al., 2016; Reverter et al., 2018; Ternon et al., 2017, 2016).
The implementation of metabolomics in linking chemical profiles and bioactivity, especially in sponges and their associated microbes, has been reported in several studies. With the existence of a data reduction approach that focuses on active compounds, in which no single compound or group of compounds is found to be responsible for its bioactivity, there is a high possibility of synergistic activity and the presence of prodrugs in the chemical content of a sample. Therefore, a holistic approach would be useful in solving this mystery in terms of the therapeutic efficacy of complex samples (Ulrich-Merzenich et al., 2007; Verpoorte et al., 2005). Recent research developments reveal that extracts from natural products have different biological activities even though they have the same chemical profile pattern. In solving this problem, researchers use a holistic approach and find that there is a change in the pattern of the genome, transcriptome, and proteome, resulting in changes in the metabolome, resulting in changes in the chemical structure of the active compound. Although it is only a slight change, it has a major effect on its bioactivity. Changes in the genome, transcriptome, and proteome can be detected by the PCR technique. This has been proven in several studies on sponges and their associated microbes, in which the genome sequences in several individuals were examined using the PCR technique, followed by a multivariate analysis using HCA and linked to other multivariate analysis such as SA and OPLS-DA, yielding information that the genome plays a very important role in the production of natural products from sponges and their associated microbes, so it affects the level of bioactivity. Therefore, linking the pattern of chemical profiles with bioactivity in metabolomics is important, and it will be better if the genomic profile of the organism under research is also examined in order to synchronize the relationship between the chemical profile and the resulting bioactivity (Betancur et al., 2017; Costantini et al., 2017; Einarsdottir et al., 2017; Ellis et al., 2017; Velasco-Alzate et al., 2019).
Other purposes of metabolomic research on sponges and their associated microbes, namely, identification of active compounds and quantitative prediction of bioactivity, identification of proof of efficacy and mode of action, assessment of bioavailability and fate of natural compounds, and identification of safety and toxicity, have not been found in the last decade. This is because the development of research on sponges and their associated microbes has not yet reached the in vivo testing stage and safety testing in humans has not been carried out. Therefore, some of the intended metabolic applications have not been found in the research conducted in the last decade. On the other hand, there have been many metabolomic studies on herbal plants in the terrestrial zone because they have been widely used for health purposes, so both in vivo and safety testing are absolutely necessary (Li et al., 2008).
CONCLUSION
The data presented in the review of metabolomic applications on sponges and sponge-associated microorganisms show that in the last decade there has been quite a lot of research on metabolomics related to sponges and sponge-associated microbes. The sponges most often studied in metabolomic research are Geodia, Xestospongia, and Agelas, while the sponge-associated microbe most often studied is actinobacteria. The LC-MS technique is an analytical technique widely used in metabolomic research on sponges and sponge-associated microorganisms. The most frequently used multivariate data analysis method is SA. Mostly, metabolomic research has an objective of identification and dereplication. In the future, metabolomic research on sponges and sponge-associated microorganisms will increase along with the number of new compounds that have potential bioactivities. Therefore, it is possible that metabolomic application can be done for bioavailability determination, quantitative bioactivity determination, and safety and toxicity testing.
ACKNOWLEDGMENTS
The author would like to acknowledge the funding support from PDUPT-DIKTI Indonesia 1622/UN1/DITLIT/DIT- LIT/PT/2021.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
ABBREVIATIONS
LC-MS: Liquid chromatography-mass spectroscopy
NMR: Nuclear magnetic resonance
PCR: Polymerase chain reaction
GC-MS Gas chromatography-mass spectroscopy
SA: Similarity analysis
PCA: Principal component analysis
HCA: Hierarchical cluster analysis
PLS: Partial least square
PLS-DA: Partial least square-discriminant analysis
OPLS: Orthogonal projections to latent structures
OPLS-DA: Orthogonal projections to latent structures-discriminant analysis
LDA: Linear discriminant analysis.
REFERENCES
Abdelhameed RFA, Habib ES, Eltahawy NA, Hassanean HA, Ibrahim AK, Mohammed AF, Fayez S, Hayallah AM, Yamada K, BeheryFA, Al-Sanea MM, Alzarea SI, Bringmann G, Ahmed SA, Abdelmohsen UR. New cytotoxic natural products from the red sea sponge stylissa carteri. Mar Drugs, 2020; 18:1–13; http//doi.org/10.3390/md18050241 CrossRef
Abdelmohsen UR, Cheng C, Viegelmann C, Zhang T, Grkovic T, Ahmed S, Quinn RJ, Hentschel U, Edrada-Ebel R. Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins a and B from a marine sponge associated-Actinokineospora sp. EG49. Mar Drugs, 2014; 12:1220–44; http//doi.org/10.3390/md12031220 CrossRef
Achlatis M, Pernice M, Green K, De Goeij JM, Guagliardo P, Kilburn MR, Hoegh-Guldberg O, Dove S. Single-cell visualization indicates direct role of sponge host in uptake of dissolved organic matter. Proc R Soc B Biol Sci, 2019; 286:1–9; http//doi.org/10.1098/rspb.2019.2153 CrossRef
Adams MJ. Chemometrics in analytical spectroscopy. 2nd edition, The Royal Society of Chemistry, Cambridge, UK, 2004.
Ali K, Iqbal M, Yuliana ND, Lee YJ, Park S, Han S, Lee J-W, Lee H-S, Verpoorte R, Choi YH. Identification of bioactive metabolites against adenosine A1 receptor using NMR-based metabolomics. Metabolomics, 2013; 9:778–85; http//doi.org/10.1007/s11306-013-0498-9 CrossRef
Alkhalifah DHM. Sponge-associated sp. RM66 metabolome induction with N-acetylglucosamine: antibacterial, antifungal and anti-trypanosomal activities. Saudi J Biol Sci, 2021; 28:4691–8; http//doi.org/10.1016/j.sjbs.2021.04.082 CrossRef
Audoin C, Bonhomme D, Ivanisevic J, De La Cruz M, Cautain B, Monteiro MC, Reyes F, Rios L, Perez T, Thomas OP. Balibalosides, an original family of glucosylated sesterterpenes produced by the Mediterranean sponge Oscarella balibaloi. Mar Drugs, 2013; 11:1477–89; http//doi.org/10.3390/md11051477 CrossRef
Badjakov I, Nikolova M, Gevrenova R, Kondakova V, Todorovska E, Atanassov A. Bioactive compounds in small fruits and their influence on human health. Biotechnol Biotechnol Equip, 2008; 22:581–7; http//doi.org/10.1080/13102818.2008.10817517 CrossRef
Baral B, Akhgari A, Metsä-Ketelä M. Activation of microbial secondary metabolic pathways: avenues and challenges. Synth Syst Biotechnol, 2018; 3:163–78; http//doi.org/10.1016/j.synbio.2018.09.001 CrossRef
Bauvais C, Bonneau N, Blond A, Pérez T, Bourguet-Kondracki ML, Zirah S. Furanoterpene diversity and variability in the marine sponge Spongia officinalis, from untargeted LC-MS/MS metabolomic profiling to furanolactam derivatives. Metabolites, 2017; 7:1–20; http//doi.org/10.3390/metabo7020027 CrossRef
Bayona LM, Van Leeuwen G, Erol Ö, Swierts T, Swierts T, Van Der Ent E, de Voogd NJ, Choi YH. Influence of geographical location on the metabolic production of giant barrel sponges (Xestospongia spp.) revealed by metabolomics tools. ACS Omega, 2020; 5:12398–408; http//doi.org/10.1021/acsomega.0c01151 CrossRef
Bayona LM, Videnova M, Choi YH. Increasing metabolic diversity in marine sponges extracts by controlling extraction parameters. Mar Drugs, 2018; 16:1–12; http//doi.org/10.3390/md16100393. CrossRef
Beebe KR, Pell RJ, Seasholtz MB. Chemometrics: a practical guide. Wiley, New York, 1998.
Berrueta LA, Alonso-Salces RM, Héberger K. Supervised pattern recognition in food analysis. J Chromatogr A, 2007; 1158:196–214; http//doi.org/10.1016/j.chroma.2007.05.024 CrossRef
Betancur LA, Naranjo-Gaybor SJ, Vinchira-Villarraga DM, Moreno-Sarmiento NC, Maldonado LA, Suarez-Moreno ZR, Acosta-González A, Padilla-Gonzalez GF, Puyana M, Castellanos L, Ramos FA. Marine Actinobacteria as a source of compounds for phytopathogen control: an integrative metabolic-profiling/bioactivity and taxonomical approach. PLoS One, 2017; 12:1–25; http//doi.org/10.1371/journal.pone.0170148 CrossRef
Bojko B, Onat B, Boyaci E, Psillakis E, Dailianis T, Pawliszyn J. Application of in situ solid-phase microextraction on mediterranean sponges for untargeted exometabolome screening and environmental monitoring. Front Mar Sci, 2019; 6:1–13; http//doi.org/10.3389/fmars.2019.00632 CrossRef
Bose U, Hewavitharana AK, Ng YK, Shaw PN, Fuerst JA, Hodson MP. LC-MS-based metabolomics study of marine bacterial secondary metabolite and antibiotic production in salinispora arenicola. Mar Drugs, 2015; 13:249–66; http//doi.org/10.3390/md13010249 CrossRef
Brereton RG. Chemometrics?: data analysis for laboratory and chemical plant. John Wiley and Sons Ltd., Chichester, UK, 2003.
Cantrell TP, Freeman CJ, Paul VJ, Agarwal V, Garg N. Mass spectrometry-based integration and expansion of the chemical diversity harbored within a marine sponge. J Am Soc Mass Spectrom, 2019; 30:1373–84; http//doi.org/10.1007/s13361-019-02207-5 CrossRef
Carroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR. Marine natural products. Nat Prod Rep, 2019; 36:122–73; http//doi.org/10.1039/c8np00092a CrossRef
Caso A, Esposito G, Sala G Della, Pawlik JR, Teta R, Mangoni A, Costantino V. Fast detection of two smenamide family members using molecular networking. Mar Drugs, 2019; 17:1–12; http//doi.org/10.3390/md17110618 CrossRef
Chaudhari S, Kumar MS. Marine sponges Sarcotragus foetidus, Xestospongia carbonaria and Spongia obscura constituents ameliorate IL?1 β and IL?6 in lipopolysaccharide?induced RAW 264.7 macrophages and carageenan?induced.pdf. Inflammopharmacology, 2020; 28:1091–119. CrossRef
Che Man YB, Rohman A, Mansor TST. Differentiation of lard from other edible fats and oils by means of Fourier transform infrared spectroscopy and chemometrics. JAOCS, J Am Oil Chem Soc, 2011; 88:187–92; http//doi.org/10.1007/s11746-010-1659-x CrossRef
Chen Q, Zhao J, Guo Z, Wang X. Determination of caffeine content and main catechins contents in green tea (Camellia sinensis L.) using taste sensor technique and multivariate calibration. J Food Compos Anal, 2010; 23:353–8; http//doi.org/10.1016/j.jfca.2009.12.010 CrossRef
Cheng C, Macintyre L, Abdelmohsen UR, Horn H, Polymenakou PN, Edrada-Ebel R, Hentschel U. Biodiversity, anti-trypanosomal activity screening, and metabolomic profiling of actinomycetes isolated from Mediterranean sponges. PLoS One, 2015; 10:1–21; http//doi.org/10.1371/journal.pone.0138528 CrossRef
Cheng C, Othman EM, Stopper H, Edrada-Ebel RA, Hentschel U, Abdelmohsen UR. Isolation of petrocidin a, a new cytotoxic cyclic dipeptide from the marine sponge-derived bacterium Streptomyces sp. SBT348. Mar Drugs, 2017; 15:1–9; http//doi.org/10.3390/md15120383 CrossRef
Colquhoun IJ. Use of NMR for metabolic profiling in plant systems. J Pestic Sci, 2007; 32:200–12; http//doi.org/10.1584/jpestics.R07-03 CrossRef
Costantini S, Guerriero E, Teta R, Capone F, Caso A, Sorice A, Romano G, Ianora A, Ruocco N, Budillon A, Costantino V, Costantini M . Evaluating the effects of an organic extract from the mediterranean sponge Geodia cydonium on human breast cancer cell lines. Int J Mol Sci, 2017; 18:1–16; http//doi.org/10.3390/ijms18102112 CrossRef
Debbab A, Aly AH, Proksch P. Bioactive secondary metabolites from endophytes and associated marine derived fungi. Fungal Divers, 2011; 49:1–12; http//doi.org/10.1007/s13225-011-0114-0 CrossRef
Einarsdottir E, Magnusdottir M, Astarita G, Köck M, Ögmundsdottir HM, Thorsteinsdottir M, Rapp HT, Omarsdottir S, Paglia G. Metabolic profiling as a screening tool for cytotoxic compounds: Identification of 3-alkyl pyridine alkaloids from sponges collected at a shallow water hydrothermal vent site North of Iceland. Mar Drugs, 2017; 15:1–15; http//doi.org/10.3390/md15020052 CrossRef
El-Hawary SS, Sayed AM, Mohammed R, Hassan HM, Rateb ME, Amin E, Mohammed TA, El-Mesery M, Bin Muhsinah A, Alsayari A, Wajant H, Anany MA, Abdelmohsen UR. Bioactive brominated oxindole alkaloids from the red sea sponge Callyspongia siphonella. Mar Drugs, 2019; 17:1–13; http//doi.org/10.3390/md17080465 CrossRef
Ellis GA, Thomas CS, Chanana S, Adnani N, Szachowicz E, Braun DR, Harper MK, Wyche TP, Bugni TS. Brackish habitat dictates cultivable Actinobacterial diversity from marine sponges. PLoS One, 2017; 12:1–19; http//doi.org/10.1371/journal.pone.0176968 CrossRef
Elsayed Y, Refaat J, Abdelmohsen UR, Othman EM, Stopper H, Fouad MA. Metabolomic profiling and biological investigation of the marine sponge-derived bacterium Rhodococcus sp. UA13.pdf. Phytochem Anal, 2018:1–6. CrossRef
Erngren I, Smit E, Pettersson C, Cárdenas P, Hedeland M. The effects of sampling and storage conditions on the metabolite profile of the marine sponge Geodia barretti. Front Chem, 2021; 9:1–19; http//doi.org/10.3389/fchem.2021.662659 CrossRef
Fagundes T da SF, da Silva LRG, Brito M de F, Schmitz LSS, Rigato DB, Jimenez PC, Soares AR, Costa-Lotufo LV, Muricy G, Vasconcelos TRA, Cass QB, Valverde AL. Metabolomic fingerprinting of Brazilian marine sponges: a case study of Plakinidae species from Fernando de Noronha Archipelago. Anal Bioanal Chem, 2021; 413:4301–10; http//doi.org/10.1007/s00216-021-03385-6 CrossRef
Favre L, Ortalo-Magné A, Greff S, Pérez T, Thomas OP, Martin JC, Culioli G. Discrimination of four marine biofilm-forming bacteria by lc-ms metabolomics and influence of culture parameters. J Proteome Res, 2017; 16:1962–75; http//doi.org/10.1021/acs.jproteome.6b01027 CrossRef
Fiore CL, Freeman CJ, Kujawinski EB. Sponge exhalent seawater contains a unique chemical profile of dissolved organic matter. PeerJ, 2017; 2017:1–22; http//doi.org/10.7717/peerj.2870 CrossRef
Gan F, Ye R. New approach on similarity analysis of chromatographic fingerprint of herbal medicine. J Chromatogr A, 2006; 1104:100–5; http//doi.org/10.1016/j.chroma.2005.11.099 CrossRef
Garthwaite PH. An interpretation of partial least squares. J Am Stat Assoc, 1994; 89:122–7; http//doi.org/10.2307/2291207 CrossRef
Gemperline P. Practical guide to chemometrics, vol. 37, 2nd edition, Taylor & Francis Group, New York, 2006; http//doi.org/10.2307/1269627 CrossRef
Geng CA, Chen XL, Zhou NJ, Chen H, Ma YB, Huang XY, Huang XY, Zhang XM, Chen JJ. LC-MS guided isolation of (±)-sweriledugenin a, a pair of enantiomeric lactones, from Swertia leducii. Org Lett, 2014; 16:370–3; http//doi.org/10.1021/ol403198d CrossRef
Gong F, Liang YZ, Xu QS, Chau FT, Ng KM. Evaluation of separation quality in two-dimensional hyphenated chromatography. Anal Chim Acta, 2001; 450:99–114; http//doi.org/10.1016/S0003-2670(01)01368-X CrossRef
Grkovic T, Pouwer RH, Vial ML, Gambini L, Noël A, Hooper JNA, Wood SA, Mellick GD, Quinn RJ. NMR fingerprints of the drug-like natural-product space identify iotrochotazine a: a chemical probe to study Parkinson’s disease. Angew Chemie—Int Ed, 2014; 53:6070–4; doi.org/10.1002/anie.201402239 CrossRef
Guo C, Wang P, Lin X, Salendra L, Kong F, Liao S, Yang B, Zhou X, Wang J, Liu Y. Phloroglucinol heterodimers and bis-indolyl alkaloids from the sponge-derived fungus: Aspergillus sp. SCSIO 41018. Org Chem Front, 2019; 6:3053–9; http//doi.org/10.1039/c9qo00351g CrossRef
Han J, Datla R, Chan S, Borchers CH. Mass spectrometry-based technologies for high-throughput metabolomics. Bioanalysis, 2009; 1:1665–84; http//doi.org/10.4155/bio.09.158 CrossRef
Hanrahan G, Gomez FA. Chemometric methods in capillary electrophoresis. Wiley & Sons, Inc., Hoboken, NJ, 2010. CrossRef
Hifnawy MS, Hassan HM, Mohammed R, Fouda MM, Sayed AM, Hamed AA, F AbouZid S, Rateb ME, Alhadrami HA, Abdelmohsen UR. Induction of antibacterial metabolites by co-cultivation of two red-sea-sponge-associated actinomycetes Micromonospora sp. UR56 and Actinokinespora sp. EG49. Mar Drugs, 2020; 18:1–17; http//doi.org/10.3390/md18050243 CrossRef
Ho XY, Katermeran NP, Deignan LK, Phyo MY, Ong JFM, Goh JX, Ng JY, Tun K, Tan LT. Assessing the diversity and biomedical potential of microbes associated with the neptune’s cup sponge, Cliona patera. Front Microbiol, 2021; 12:1–17; http//doi.org/10.3389/fmicb.2021.631445 CrossRef
Hooper JNA, Van Soest RWM. Systema porifera: a guide to the classification of sponges. Kluwer Academic, Plenum Publisher, New York, NY, vol. 1, 2002; http//doi.org/10.1007/978-1-4615-0747-5_1 CrossRef
Huang J, Lu C, Qian X, Huang Y, Zheng Z, Shen Y. Effect of salinity on the growth, biological activity and secondary metabolites of some marine fungi. Acta Oceanol Sin, 2011; 30:118–23; http//doi.org/10.1007/s13131-011-0126-3 CrossRef
Isgut M, Rao M, Yang C, Subrahmanyam V, Rida PCG, Aneja R. Application of combination high-throughput phenotypic screening and target identification methods for the discovery of natural product-based combination drugs. Med Res Rev, 2018; 38:504–24; http//doi.org/10.1002/med.21444 CrossRef
Ivaniševi? J, Thomas OP, Lejeusne C, Chevaldonné P, Pérez T. Metabolic fingerprinting as an indicator of biodiversity: Towards understanding inter-specific relationships among Homoscleromorpha sponges. Metabolomics, 2011; 7:289–304; http//doi.org/10.1007/s11306-010-0239-2 CrossRef
Ivanisevic J, Thomas OP, Pedel L, Pénez N, Ereskovsky AV, Culioli G, Perez T. Biochemical trade-offs: evidence for ecologically linked secondary metabolism of the sponge Oscarella balibaloi. PLoS One, 2011; 6:1–11; http//doi.org/.1371/journal.pone.0028059 CrossRef
Johnson RA, Wichern DW. Applied multivariate statistical analysis. 6th edition, Pearson Education Limited, Upper Saddle River, NJ, 2007.
Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL. Exploring the temperature-stress metabolome. Plant Physiol, 2004; 136:4159–68; http//doi.org/10.1104/pp.104.052142.1 CrossRef
Keller-Costa T, Jousset A, Van Overbeek L, Van Elsas JD, Costa R. The freshwater sponge Ephydatia fluviatilis harbours diverse Pseudomonas species (Gammaproteobacteria, Pseudomonadales) with broad-spectrum antimicrobial activity. PLoS One, 2014; 9:1–15; http//doi.org/10.1371/journal.pone.0088429 CrossRef
Kim HK, Choi YH, Verpoorte R. NMR-based plant metabolomics: where do we stand, where do we go? Trends Biotechnol, 2011; 29:267–75; http//doi.org/10.1016/j.tibtech.2011.02.001 CrossRef
Kjer J, Debbab A, Aly AH, Proksch P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat Protoc, 2010; 5:479–90; http//doi.org/10.1038/nprot.2009.233 CrossRef
Kouchaksaraee RM, Farimani MM, Li F, Nazemi M, Tasdemir D. Integrating molecular networking and 1H NMR spectroscopy for isolation of bioactive metabolites from the persian gulf sponge axinella sinoxea. Mar Drugs, 2020; 18:1–15; http//doi.org/10.3390/MD18070366 CrossRef
Lee YK, Lee J, Lee HK. Minireview: microbial symbiosis in marine sponges. J Microbiol, 2001; 39:254–64.
Li F, Janussen D, Peifer C, Pérez-Victoria I, Tasdemir D. Targeted isolation of tsitsikammamines from the antarctic deep-sea sponge latrunculia biformis by molecular networking and anticancer activity. Mar Drugs, 2018; 16:1–17; http//doi.org/10.3390/md16080268 CrossRef
Li F, Pandey P, Janussen D, Chittiboyina AG, Ferreira D, Tasdemir D. Tridiscorhabdin and didiscorhabdin, the first discorhabdin oligomers linked with a direct C-N bridge from the sponge Latrunculia biformis collected from the deep sea in Antarctica. J Nat Prod, 2020; 83:706–13; http//doi.org/10.1021/acs.jnatprod.0c00023 CrossRef
Li F, Peifer C, Janussen D, Tasdemir D. New discorhabdin alkaloids from the antarctic deep-sea sponge Latrunculia biformis. Mar Drugs, 2019; 17:1–19; http//doi.org/10.3390/md17080439 CrossRef
Li P, Qi LW, Liu EH, Zhou JL, Wen XD. Analysis of Chinese herbal medicines with holistic approaches and integrated evaluation models. Trends Anal Chem, 2008; 27:66–77; http//doi.org/10.1016/j.trac.2007.11.005 CrossRef
Li Y, Wu T, Zhu J, Wan L, Yu Q, Li X, Cheng Z, Guo C. Combinative method using HPLC fingerprint and quantitative analyses for quality consistency evaluation of an herbal medicinal preparation produced by different manufacturers. J Pharm Biomed Anal, 2010; 52:597–602; http//doi.org/10.1016/j.jpba.2010.01.018 CrossRef
Macintyre L, Zhang T, Viegelmann C, Martinez IJ, Cheng C, Dowdells C, Abdelmohsen UR, Gernert C, Hentschel U, Edrada-Ebel R. Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Mar Drugs, 2014; 12:3416–48; http//doi.org/10.3390/md12063416 CrossRef
Matroodi S, Siitonen V, Baral B, Yamada K, Akhgari A, Metsä-Ketelä M. Genotyping-guided discovery of persiamycin a from sponge-associated halophilic Streptomonospora sp. PA3. Front Microbiol, 2020; 11:1–15; http//doi.org/10.3389/fmicb.2020.01237 CrossRef
McCauley E, Radjas OK, Trianto A, Crews MS, Smith A, Smith GC, Zerebinski P, Sabdono A, Crews P. The UNDIP-UCSC campaign to culture chemically prolific gram-negative bacteria from Indonesian Jaspis sponges. Arkivoc, 2018; 2018:123–31; http//doi.org/10.24820/ark.5550190.p010.505 CrossRef
Mehbub MF, Tanner JE, Barnett SJ, Franco CMM, Zhang W. The role of sponge-bacteria interactions: the sponge Aplysilla rosea challenged by its associated bacterium Streptomyces ACT-52A in a controlled aquarium system. Appl Microbiol Biotechnol, 2016; 100:10609–26; http//doi.org/10.1007/s00253-016-7878-9 CrossRef
Miller JN, Miller JC. Statistics and chemometrics for analytical chemistry. 6th edition, Pearson Education Limited, Harlow, UK, 2010; http//doi.org/10.1198/tech.2004.s248
Mishra KP, Ganju L, Sairam M, Banerjee PK, Sawhney RC. A review of high throughput technology for the screening of natural products. Biomed Pharmacother, 2008; 62:94–8; http//doi.org/10.1016/j.biopha.2007.06.012 CrossRef
Mohanty I, Moore SG, Yi D, Biggs JS, Gaul DA, Garg N, Agarwal V. Precursor-guided mining of marine sponge metabolomes lends insight into biosynthesis of pyrrole−imidazole alkaloids. ACS Chem Biol, 2020a; 15:2185–94. CrossRef
Mohanty I, Podell S, Biggs JS, Garg N, Allen EE, Agarwal V. Multi-omic profiling of melophlus sponges reveals diverse metabolomic and microbiome architectures that are non-overlapping with ecological neighbors. Mar Drugs, 2020b; 18:124; http//doi.org/10.3390/md18020124 CrossRef
Mohanty I, Tapadar S, Moore SG, Biggs JS, Freeman CJ, Gaul DA, Agarwal V. Presence of bromotyrosine alkaloids in marine sponges is independent of metabolomic and microbiome architectures. MSystems, 2021; 6:1–17; http//doi.org/10.1128/msystems.01387-20 CrossRef
Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod, 2016; 79:629–61; http//doi.org/10.1021/acs.jnatprod.5b01055 CrossRef
Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod, 2012; 75:311–35; http//doi.org/10.1021/np200906s CrossRef
Ng YK, Hodson MP, Hewavitharana AK, Bose U, Shaw PN, Fuerst JA. Effects of salinity on antibiotic production in sponge-derived Salinispora actinobacteria. J Appl Microbiol, 2014; 117:109–25; http//doi.org/10.1111/jam.12507 CrossRef
Nouioui I, Rückert C, Willemse J, van Wezel GP, Klenk HP, Busche T, Kalinowski J, Bredholt H, Zotchev SB. Actinoalloteichus fjordicus sp. nov. isolated from marine sponges: phenotypic, chemotaxonomic and genomic characterisation. Antonie van Leeuwenhoek, Int J Gen Mol Microbiol, 2017; 110:1705–17; http//doi.org/10.1007/s10482-017-0920-9 CrossRef
Olatunji OO, Brecker L, Plubrukarn A. Metabolomics approach towards the chemical distribution in the sponge penares cf. nux. Songklanakarin J Sci Technol, 2021; 43:696–702.
Olsen EK, Søderholm KL, Isaksson J, Andersen JH, Hansen E. Metabolomic profiling reveals the N-acyl-taurine geodiataurine in extracts from the marine sponge Geodia macandrewii (Bowerbank). J Nat Prod, 2015; 79:1285–91; http//doi.org/10.1021/acs.jnatprod.5b00966 CrossRef
Ong JFM, Goh HC, Lim SC, Pang LM, Chin JSF, Tan KS, Liang Z-X, Yang L, Glukhov E, Gerwick WH, Tan LT. Integrated genomic and metabolomic approach to the discovery of potential anti-quorum sensing natural products from microbes associated with marine samples from Singapore. Mar Drugs, 2019; 17:2–15; http//doi.org/10.3390/md17010072 CrossRef
Parejo I, Viladomat F, Bastida J, Schmeda-Hirschmann G, Burillo J, Codina C. Bioguided isolation and identification of the nonvolatile antioxidant compounds from fennel (Foeniculum vulgare Mill.) waste. J Agric Food Chem, 2004; 52:1890–7; http//doi.org/10.1021/jf030717g CrossRef
Pratiwi DE, Harjoko A. Implementasi Pengenalan Wajah Menggunakan PCA (principal component analysis). IJEIS, 2013; 3:175–84; http//doi.org/10.1002/jlac.19335020105
Proksch P, Edrada RA, Ebel R. Drugs from the seas—current status and microbiological implications. Appl Microbiol Biotechnol, 2002; 59:125–34; http//doi.org/10.1007/s00253-002-1006-8 CrossRef
Raimundo I, Silva SG, Costa R, Keller-Costa T. Bioactive secondary metabolites from octocoral-associated microbes—new chances for blue growth. Mar Drugs, 2018; 16:1–25; http//doi.org/10.3390/md16120485 CrossRef
Rangel-Huerta OD, Gil A. Nutrimetabolomics: an update on analytical approaches to investigate the role of plant-based foods and their bioactive compounds in non-communicable chronic diseases. Int J Mol Sci, 2016; 17; http//doi.org/10.3390/ijms17122072 CrossRef
Reverter M, Perez T, Ereskovsky AV, Banaigs B. Secondary metabolome variability and inducible chemical defenses in the mediterranean sponge Aplysina cavernicola. J Chem Ecol, 2016; 42:60–70; http//doi.org/10.1007/s10886-015-0664-9 CrossRef
Reverter M, Tribalat MA, Pérez T, Thomas OP. Metabolome variability for two Mediterranean sponge species of the genus Haliclona: specificity, time, and space. Metabolomics, 2018; 14:1–12; http//doi.org/10.1007/s11306-018-1401-5 CrossRef
Rohman A. Statistika dan Kemometrika Dasar dalam Analisis Farmasi. Pustaka Pelajar, Yogyakarta, Indonesia, 2014.
Romoli R, Papaleo MC, de Pascale D, Tutino ML, Michaud L, LoGiudice A, Fani R, Bartolucci G. GC-MS volatolomic approach to study the antimicrobial activity of the antarctic bacterium Pseudoalteromonas sp. TB41. Metabolomics, 2014; 10:42–51; http//doi.org/10.1007/s11306-013-0549-2 CrossRef
Ruiz C, Ivaniševi? J, Chevaldonné P, Ereskovsky A V., Boury-Esnault N, Vacelet J, Thomas OP, Perez T. Integrative taxonomic description of Plakina kanaky, a new polychromatic sponge species from New Caledonia (Porifera: Homoscleromorpha). Mar Ecol, 2014; 36:1129–43; http//doi.org/10.1111/maec.12209 CrossRef
Salvatore MM, Nicoletti R, Salvatore F, Naviglio D, Andolfi A. GC–MS approaches for the screening of metabolites produced by marine-derived Aspergillus. Mar Chem, 2018; 206:19–33; http//doi.org/10.1016/j.marchem.2018.08.003 CrossRef
Samirana PO, Murti YB, Istighfari Jenie R, Prawita Setyowati E. Antibacterial and cytotoxic activities of supernatant and mycelium extracts from fermentation of fungal symbiont Trichoderma reesei TV221. J Appl Pharm Sci, 2021a; 11:90–9; http//doi.org/10.7324/japs.2021.1101207 CrossRef
Samirana PO, Murti YB, Jenie RI, Setyowati EP. Marine sponge-derived fungi: fermentation and cytotoxic activity. J Appl Pharm Sci, 2021b; 11:21–39; http//doi.org/10.7324/japs.2021.110103
Sartono B, Affendi FM, Syafitri UD, Sumertajaya I., Angraeni Y. Analisis Peubah Ganda. IPB Press, Bogor, Indonesia, 2003.
Sauleau P, Moriou C, Al Mourabit A. Metabolomics approach to chemical diversity of the Mediterranean marine sponge Agelas oroides. Nat Prod Res, 2017; 31:1625–32; http//doi.org/10.1080/14786419.2017.1285298 CrossRef
Sekurova ON, Schneider O, Zotchev SB. Novel bioactive natural products from bacteria via bioprospecting, genome mining and metabolic engineering. Microb Biotechnol, 2019; 12:828–44; http//doi.org/10.1111/1751-7915.13398 CrossRef
Shady NH, Fouad MA, Ahmed S, Pimentel-Elardo SM, Nodwell JR, Kamel MS, Abdelmohsen UR. A new antitrypanosomal alkaloid from the Red Sea marine sponge Hyrtios sp. J Antibiot (Tokyo), 2018; 71:1036–9; http//doi.org/10.1038/s41429-018-0092-5 CrossRef
Shady NH, Khattab AR, Ahmed S, Liu M, Quinn RJ, Fouad MA, Kamel MS, Muhsinah AB, Krischke M, Mueller MJ, Abdelmohsen UR. Hepatitis c virus ns3 protease and helicase inhibitors from red sea sponge (Amphimedon) species in green synthesized silver nanoparticles assisted by in silico modeling and metabolic profiling. Int J Nanomedicine, 2020; 15:3377–89; http//doi.org/10.2147/IJN.S233766 CrossRef
Sharma S. Applied multivariate techniques. John Wiley and Sons Inc., Hoboken, NJ, 1996.
Shyur LF, Yang NS. Metabolomics for phytomedicine research and drug development. Curr Opin Chem Biol, 2008; 12:66–71; http//doi.org/10.1016/j.cbpa.2008.01.032 CrossRef
Sidebottom AM, Johnson AR, Karty JA, Trader DJ, Carlson EE. Integrated metabolomics approach facilitates discovery of an unpredicted natural product suite from Streptomyces coelicolor M145. ACS Chem Biol, 2013; 8:2009–16; http//doi.org/10.1021/cb4002798 CrossRef
Storey MA, Andreassend SK, Bracegirdle J, Brown A, Keyzers RA, Ackerley DF, Northcote PT, Owen JG. Metagenomic exploration of the marine sponge mycale hentscheli uncovers multiple polyketide-producing bacterial symbionts. MBio, 2020; 11:1–16; http//doi.org/10.1128/mBio.02997-19 CrossRef
Sumner LW, Mendes P, Dixon RA. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry, 2003; 62:817–36; http//doi.org/10.1016/S0031-9422(02)00708-2 CrossRef
Tapp HS, Kemsley EK. Notes on the practical utility of OPLS. TrAC—Trends Anal Chem, 2009; 28:1322–7; http//doi.org/10.1016/j.trac.2009.08.006 CrossRef
Tawfike A, Attia EZ, Desoukey SY, Hajjar D, Makki AA, Schupp PJ, Edrada-Ebel RA, Abdelmohsen UR. New bioactive metabolites from the elicited marine sponge-derived bacterium Actinokineospora spheciospongiae sp. nov. AMB Express, 2019a; 9:1–9. http//doi.org/10.1186/s13568-018-0730-0 CrossRef
Tawfike AF, Romli M, Clements C, Abbott G, Young L, Schumacher M, Diederich M, Farag M, Edrada-Ebel R. Isolation of anticancer and anti-trypanosome secondary metabolites from the endophytic fungus Aspergillus flocculus via bioactivity guided isolation and MS based metabolomics. J Chromatogr B, 2019b; 1106–7:71–83; http//doi.org/10.1016/j.jchromb.2018.12.032 CrossRef
Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev, 2007; 71:295–347; http//doi.org/10.1128/mmbr.00040-06 CrossRef
Ternon E, Perino E, Manconi R, Pronzato R, Thomas OP. How environmental factors affect the production of guanidine alkaloids by the mediterranean sponge crambe crambe. Mar Drugs, 2017; 15:1–15; http//doi.org/10.3390/md15060181 CrossRef
Ternon E, Zarate L, Chenesseau S, Croué J, Dumollard R, Suzuki MT, Thomas OP. Spherulization as a process for the exudation of chemical cues by the encrusting sponge C. crambe. Sci Rep, 2016; 6:1–11; http//doi.org/10.1038/srep29474 CrossRef
Teta R, Sala G Della, Esposito G, Via CW, Mazzoccoli C, Piccoli C, Bertin MJ, Costantino V, Mangoni A. A joint molecular networking study of a Smenospongia sponge and a cyanobacterial bloom revealed new antiproliferative chlorinated polyketides. Org Chem Front, 2019; 6:1762–74; http//doi.org/10.5860/choice.36-5099 CrossRef
Thakur NL, Müller WEG. Biotechnological potential of marine sponges. Curr Sci, 2004; 86:1506–12.
Ulrich-Merzenich G, Zeitler H, Jobst D, Panek D, Vetter H, Wagner H. Application of the “-Omic-” technologies in phytomedicine. Phytomedicine, 2007; 14:70–82; http//doi.org/10.1016/j.phymed.2006.11.011 CrossRef
Valentino G, Graziani V, D’Abrosca B, Pacifico S, Fiorentino A, Scognamiglio M. NMR-based plant metabolomics in nutraceutical research: an overview. Molecules, 2020; 25:1–22; http//doi.org/10.3390/molecules25061444 CrossRef
Velasco-Alzate KY, Bauermeister A, Tangerina MMP, Lotufo TMC, Ferreira MJP, Jimenez PC, Padilla G, Lopes NP, Costa-Lotufo LV. Marine bacteria from Rocas Atoll as a rich source of pharmacologically active compounds. Mar Drugs, 2019; 17; http//doi.org/10.3390/md17120671 CrossRef
Verpoorte R, Choi YH, Kim HK. NMR-based metabolomics at work in phytochemistry. Phytochem Rev, 2007; 6:3–14; http//doi.org/10.1007/s11101-006-9031-3 CrossRef
Verpoorte R, Choi YH, Kim HK. Ethnopharmacology and systems biology: a perfect holistic match. J Ethnopharmacol, 2005; 100:53–6; http//doi.org/10.1016/j.jep.2005.05.033 CrossRef
Viegelmann C, Margassery LM, Kennedy J, Zhang T, O’Brien C, O’Gara F, Morrissey JP, Dobson AD, Edrada-Ebel R . Metabolomic profiling and genomic study of a marine sponge-associated Streptomyces sp. Mar Drugs, 2014; 12:3323–51; http//doi.org/10.3390/md12063323 CrossRef
Villegas-Plazas M, Wos-Oxley ML, Sanchez JA, Pieper DH, Thomas OP, Junca H. Variations in microbial diversity and metabolite profiles of the tropical marine sponge Xestospongia muta with season and depth. Microb Ecol, 2019; 78:243–56; http//doi.org/10.1007/s00248-018-1285-y CrossRef
Vitale GA, Sciarretta M, Cassiano C, Buonocore C, Festa C, Mazzella V, Núñez Pons L, D’Auria MV, de Pascale D. Molecular network and culture media variation reveal a complex metabolic profile in pantoea cf. Eucrina d2 associated with an acidified marine sponge. Int J Mol Sci, 2020; 21:1–18; http//doi.org/10.3390/ijms21176307. CrossRef
Wang G. Diversity and biotechnological potential of the sponge-associated microbial consortia. J Ind Microbiol Biotechnol, 2006; 33:545–51; http//doi.org/10.1007/s10295-006-0123-2 CrossRef
Wang M, Lamers RJAN, Korthout HAAJ, Van Nesselrooij JHJ, Witkamp RF, Van Der Heijden R, Voshol PJ, Havekes LM, Verpoorte R, van der Greef J. Metabolomics in the context of systems biology: bridging traditional Chinese medicine and molecular pharmacology. Phyther Res, 2005; 19:173–82; http//doi.org/10.1002/ptr.1624 CrossRef
Wu H, Chen Y, Li Z, Liu X. Untargeted metabolomics profiles delineate metabolic alterations in mouse plasma during lung carcinoma development using UPLCQTOF/MS in MSE mode. R Soc Open Sci, 2018; 5:1–13; http//doi.org/10.1098/rsos.181143 CrossRef
Xu CJ, Liang YZ, Chau FT, Heyden Y Vander. Pretreatments of chromatographic fingerprints for quality control of herbal medicines. J Chromatogr A, 2006; 1134:253–9; http//doi.org/10.1016/j.chroma.2006.08.060 CrossRef
Yang Q, Lin SS, Yang JT, Tang LJ, Yu RQ. Detection of inborn errors of metabolism utilizing GC-MS urinary metabolomics coupled with a modified orthogonal partial least squares discriminant analysis. Talanta, 2017; 165:545–52; http//doi.org/10.1016/j.talanta.2017.01.018 CrossRef
Yuliana ND, Jahangir M, Verpoorte R, Choi YH. Metabolomics for the rapid dereplication of bioactive compounds from natural sources. Phytochem Rev, 2013; 12:293–304; http//doi.org/10.1007/s11101-013-9297-1 CrossRef
Yuliana ND, Khatib A, Choi YH, Verpoorte R. Metabolomics for bioactivity assessment of natural products. Phyther Res, 2011; 25:157–69; http//doi.org/10.1002/ptr.3258 CrossRef
Zhu H, Wang Y, Liang H, Chen Q, Zhao P, Tao J. Identification of Portulaca oleracea L. from different sources using GC-MS and FT-IR spectroscopy. Talanta, 2010; 81:129–35; http//doi.org/10.1016/j.talanta.2009.11.047 CrossRef