INTRODUCTION
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic began in Wuhan, China, in December 2019, and has posed a serious threat to public health around the world (Zhu et al., 2020). SARS-CoV-2 or COVID-19 is a Betacoronavirus belonging to the Coronaviridae family that causes cough, tiredness, myalgia, and fever (Chen et al., 2020; Wu et al., 2020; Wu and McGoogan, 2020). The World Health Organization (WHO) has classified SARS-CoV-2 as a public health emergency at the international level. As of December 3, 2021, a total of 263,563,622 COVID-19 cases had been documented worldwide, with 5,232,562 fatalities (WHO, 2021a). COVID-19 had a massive impact on society, the environment, and the global economy, posing a grave threat to human survival.
Several investigations and approaches have been undertaken to prevent COVID-19 from spreading further and developing safe and effective medications and vaccines. AstraZeneca, Johnson & Johnson, Gamaleya, Novavax, Sinopharm, Sinovac, Moderna, and Pfizer have all produced vaccines to prevent the COVID-19 pandemic (Balkrishna et al., 2021). While current vaccines are assisting in keeping the pandemic under control in some countries, it is also vital to remain prepared for SARS-CoV-2 mutations, as RNA viruses like the coronavirus tend to change and mutate over time (Bollinger and Ray, 2021; WHO, 2021c). Due to mutations in the SARS-CoV-2, the world has seen numerous peaks of the COVID-19 pandemic with higher infectivity and transmissibility.
SARS-CoV-2 variations such as Alpha, Beta, Delta, Eta, Theta, and others have already been identified. In terms of transmissibility and infectivity, the SARS-CoV-2 Delta variant or B.1.617.2, has had the largest impact. In December 2020, it was initially reported in India, and it rapidly spread to 98 countries around the world (CDC, 2021a). On November 13, 2021, Thomas Peacock, a postdoctoral fellow at Imperial College London in the United Kingdom, revealed the sequencing of a novel variant (B.1.1.529) of SARS-CoV-2 acquired from an immune-compromised patient in South Africa (FDA, 2021a). WHO categorized variant B.1.1.529 or Omicron as a variant of concern (VOC) on November 26, 2021 (WHO, 2021c). The B.1.1.529 variant is now most prevalent in COVID-19 patients in South Africa; unfortunately, it has also been documented in more than 20 other countries including Botswana, Belgium, Hong Kong, Israel, etc. (Callaway and Ledford, 2021; Choudhary et al., 2021). SARS-CoV-2 variants are rapidly evolving; this review gives an insight into SARS-CoV-2 variants, with a focus on the highly mutated form Omicron. The importance of bioactive lipids in the battle against COVID-19 is also emphasized.
CATEGORIZATION OF SARS-COV-2 VARIANTS AND ASSOCIATED MUTATIONS
A mutation is defined by the Center for Disease Control and Prevention (CDC) as a change in a virus’s genetic code that occurs naturally over time when an animal or person becomes infected (CDC, 2021a). Viruses like COVID-19 are emerging consistently as alterations in the genetic code (genetic mutations) that arise upon genome replication. These alterations are categorized as lineage and variants. Furthermore, a lineage is a group of viral variations that have a common ancestor and are genetically related, wherein a variation is differentiated from other SARS-CoV-2 variants by one or many mutations (CDC, 2021d). The different variants from SARS-CoV-2 to Omicron, along with their first identification detail are shown in Figure 1.
The WHO and CDC have classified coronavirus variants into the following categories:
Variants of interest (VOI)
The genetic variations in SARS-CoV-2 are considered as VOI when they are observed to alter its characteristics such as immunological escape, the severity of disease, therapeutic or diagnostic escape, enhanced transmissibility, and decreased neutralization by antibodies developed in response to past infection or immunization. The second component is when the SARS-CoV-2 variation is found to produce considerable community transmission or multiple clusters in different nations, with increasing case numbers over time, or other evident epidemiological repercussions, indicating an expanding concern to global public health. VOI may necessitate appropriate public health actions like increasing sequential monitoring, enhancing lab characterization, or epidemiological assessments to identify how conveniently the virus is spreading, the magnitude of disease, the efficiency of therapeutics, and whether commercially available or accredited immunizations offer protection (CDC, 2021d; WHO, 2021c). Lambda and Mu are considered as VOI by WHO (2021c).
Variant of concern (VOC)
VOC is the one that infects people who have been vaccinated or previously infected. These variations are more likely to enhance transmissibility, cause severe illness (e.g., elevated hospitalizations or mortalities), show resistance against antiviral therapy, circumvent diagnosis or substantially reduce antibodies neutralization produced throughout vaccination or previous infection. Concerning variants may necessitate relevant public health interventions, like reporting to the CDC or intimation to WHO, regional attempts to prevent transmission, rapid diagnosis, or experimentation to assess the efficacy of vaccines and therapies against it. Additional considerations could include developing new diagnostics or altering immunizations or medicines based on the characteristics of the variant (CDC, 2021d; WHO, 2021c). WHO classifies Alpha, Beta, Gamma, Delta (B.1.617.2 and AY lineages), and Omicron (B.1.1.529) as VOC (WHO, 2021c).
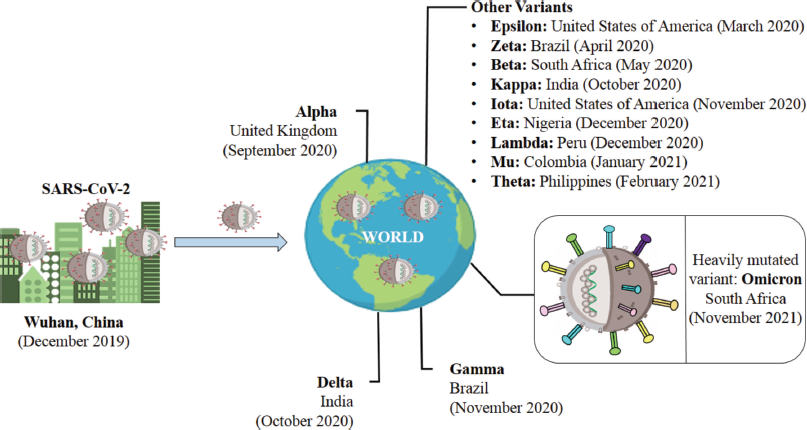 | Figure 1. An overview of variations from SARS-CoV-2 (Wuhan) to Omicron (South Africa). [Click here to view] |
Variant of high consequence (VOHC)
VOHC is one against which existing vaccinations provide no protection. VOHC contains clear proof that precautionary measures or medicinal interventions are much less effective than formerly existing versions. A VOHC would involve information to WHO, reporting to the CDC, disclosure of prevention or containment activities, and considerations to improve therapy and vaccinations (CDC, 2021d; WHO, 2021c). There are no substantial SARS-CoV-2 variations to evaluate under VOHC at this time (CDC, 2021d).
Variants being monitored (VBM)
VBM variations are those for which data implies a possible or apparent influence on allowed or permitted medical treatments, or that have been linked to higher rates of catastrophic disease or transmissions but are no longer detectable, or that exist in relatively small amounts. These variations do not constitute a major or immediate health risk. VOI or VOC may be included in this list after a considerable and consistent decrease in proportions over time or if additional research suggests that the variant will not pose a serious hazard to public health. Alpha (Q lineages and B.1.1.7), Beta (B.1.351 and descendent lineages), Gamma (P.1 and descendent lineages), Epsilon (B.1.427 and B.1.429), Eta (B.1.525), Iota (B.1.526), Kappa (B.1.617.1), Zeta (P.2), Mu (B.1.621.1, B.1.621) are all VBM variations (CDC, 2021d). Table 1 provides a quick review of various mutations found in several SARS-CoV-2 variants.
The majority of the variants were found to be more prevalent in Brazil, South Africa, India, and the United States of America, and these variants were also more transmissible. The B.1.1.529 lineage still has the greatest spike substitutions as compared to previously reported ones, as seen in Table 1, which could affect transmissibility and antibody potency. Stopping Omicron as soon as possible is the only method to prevent future lethal variants.
EFFICACY OF EXISTING VACCINES AGAINST OMICRON: A MAJOR CONCERN
In recent weeks, with the identification of the B.1.1.529 strain (Omicron), the infections have increased tremendously. There are a lot of mutations in this variant, including the novel spike mutations and antigenic distinctiveness from preceding strains. The variant’s rapid spread in South Africa indicated that it might be immune-evading (WHO, 2021a). The individuals who received any of the three types of vaccinations available in South Africa (Pfizer-BioNTech, Johnson & Johnson, and Oxford-AstraZeneca) have developed breakthrough infections, posing a public health risk. The variant’s rapid transmission in South Africa suggests that it may be able to bypass immunity and existing vaccines. Around a quarter of South Africans are fully vaccinated, and many have been infected in previous COVID-19 waves (Callaway and Ledford, 2021). According to one source, in Hong Kong, two quarantined passengers were found positive for the novel variant even after immunization with the Pfizer vaccine. One of them had come from South Africa (Callaway and Ledford, 2021).
Against all COVID-19 variants, including the super spreader Delta variant, vaccines continue to be crucial in lowering severe illness and mortality. According to WHO, a total of 7,864,123,038 vaccination doses were administered globally up to December 2, 2021 (WHO, 2021a). The role of current immunizations against the Omicron variant is still obscure. To understand the efficacy of current vaccines, researchers are closely tracking the effects of current vaccines on Omicron mutations. They intend to evaluate the virus’s capacity to avoid infection-fighting antibodies and other immunological reactions. Because of the threat posed by Omicron, several rich nations, like the United Kingdom, have accelerated and expanded the distribution of COVID vaccination booster doses. However, it is unknown how successful these dosages will be toward this variant. The novel development of Omicron underscores the significance of immunization and booster shots (CDC, 2021c).
BIOACTIVE LIPIDS AGAINST SARS-COV-2: A RAY OF HOPE
To stop the SARS-CoV-2 pandemic, tremendous attempts are being undertaken to establish both prophylactic and treatment strategies. The scarcity of antiviral drugs, fewer vaccines, and emerging variants necessitates the development of new therapeutic approaches to tackle SARS-CoV-2. Polyunsaturated fatty acids and their derivatives (also known as bioactive lipids) could play an important role in this context. The bioactive lipids include alpha-linolenic acid (ALA), arachidonic acid (AA), dihomo- γ-linolenic acid, docosahexaenoic acid (DHA), eicosapentaenoic acid (EPA), linolenic acid, γ-linolenic acid, leukotrienes, lipoxins, maresins, prostaglandins, protectins, resolvins, and thromboxanes (Das, 2021). Das (2020) suggested that AA and other unsaturated fatty acids, as well as their derivatives, may act as endogenous antiviral agents, and their insufficiency could make individuals vulnerable to SARS-CoV-2. In addition, fatty acids also aid in the inactivation of enveloped viruses (Mousavizadeh and Ghasemi, 2021).
The omega-3 polyunsaturated fatty acids (ALA, EPA, and DHA) were found to have immunomodulatory properties (Chang et al., 2020). Their anti-viral potential is mediated through the inhibition of replication (Shakoor et al., 2021). The underlying mechanism by which unsaturated fatty acids (AA, EPA, and DHA) inactivate the virus is still unknown; nonetheless, the fatty acids trigger bilayer lipid envelope breakdown with enhanced efficacy (Aryan et al., 2021).Das (2021) revealed that AA could be a crucial player in the prevention and management of SARS-CoV-2. He demonstrated multiple mechanisms, such as the production of AA and other bioactive lipids in the lungs inactivate viral pathogens; derivatives of AA (prostaglandin E2, leukotrienes, lipoxin A4), EPA and DHA (maresins, protectins, and resolvins) stimulate the release of pro-inflammatory and anti-inflammatory macrophages. Moreover, AA derivatives and other bioactive lipids also inhibit the production of tumor necrosis factor-α and interleukin-6. These bioactive lipids could be used in the management of new variants of SARS-CoV-2.
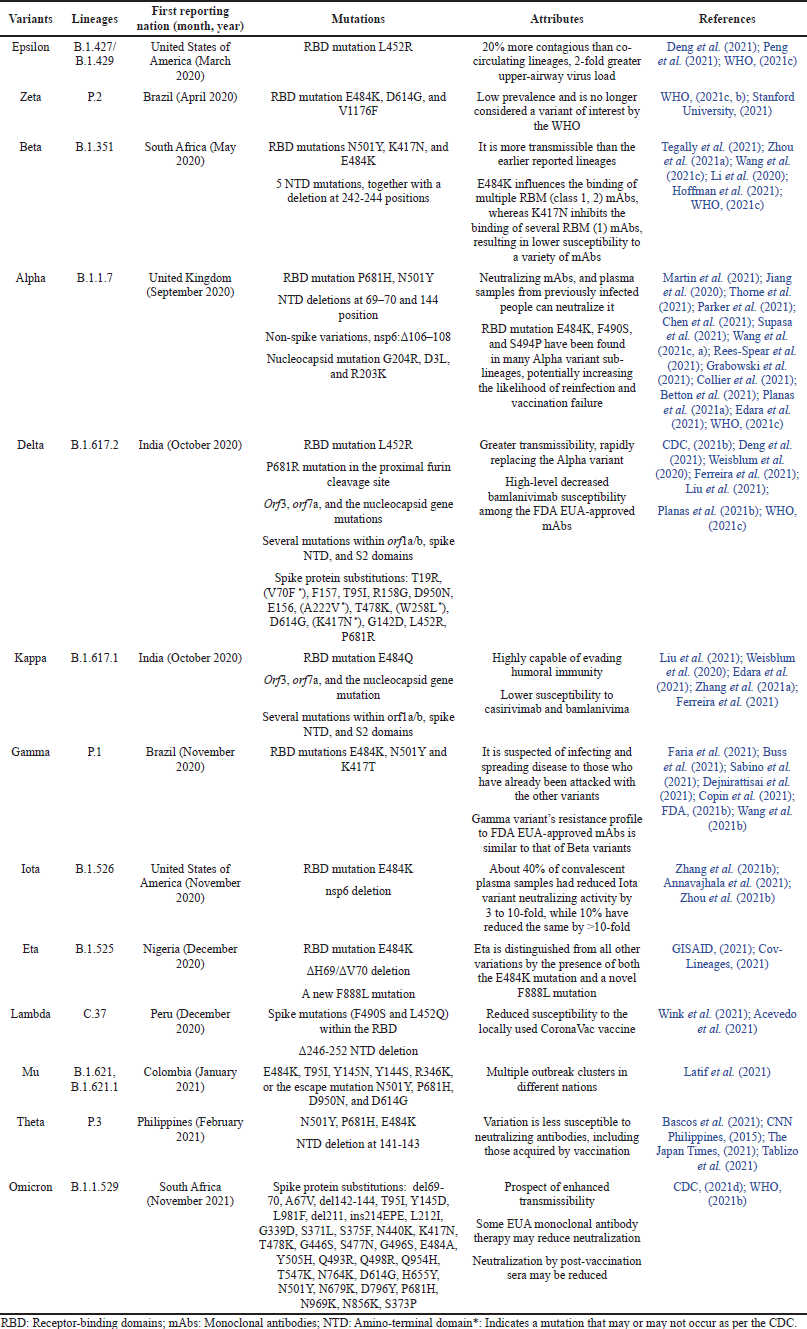 | Table 1. An overview of various variants of SARS-CoV-2. [Click here to view] |
CONCLUSION AND RECOMMENDATIONS
Undoubtedly, it would take days to weeks to determine the severity of the Omicron. Although it is too early to determine Omicron’s fatality potential based on preliminary findings, its rapid transmission cannot be ignored, as evidenced by the increase in cases in South Africa. Despite vaccination, the rate of transmission has increased, raising concerns among researchers and nations. Many developed nations are detecting this variant through genome sequencing; however, accurate data about Omicron appears to be a significant issue in the developing countries due to poor infrastructure. Omicron was discovered to be the heavily mutant variant. Until the facts regarding the Omicron are exposed, masking, social distancing, and vaccination are strongly suggested. To acquire a better understanding of Omicron’s transmissibility, researchers are monitoring how it spreads in the rest of South Africa and around the world. To halt Omicron from spreading further, governments and researchers must play a critical role. The interplay between bioactive lipids and SARS-CoV-2 has opened up a new front in the fight against the pandemic.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FUNDING
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Acevedo ML, Alonso-Palomares L, Bustamante A, Gaggero A, Paredes F, Cortés CP, Valiente-Echeverría F, Soto-Rifo R. Infectivity and immune escape of the new SARS-CoV-2 variant of interest Lambda. Medrxiv, 2021, vol. 2021. CrossRef
Annavajhala MK, Mohri H, Zucker JE, Sheng Z, Wang P, Gomez-Simmonds A, Ho DD, Uhlemann AC. A novel SARS-CoV-2 variant of concern, B. 1.526, identified in New York. MedRxiv, 2021, vol. 2021. CrossRef
Aryan H, Saxena A, Tiwari A. Correlation between bioactive lipids and novel coronavirus: constructive role of biolipids in curbing infectivity by enveloped viruses, centralizing on EPA and DHA. Syst Microbiol Biomanuf, 2021; 1:186–92. CrossRef
Balkrishna A, Arya V, Rohela A, Kumar A, Verma R, Kumar D, Nepovimova E, Kuca K, Thakur N, Thakur N, Kumar P. Nanotechnology interventions in the management of COVID-19: prevention, diagnosis and virus-like particle vaccines. Vaccines, 2021; 9(10):1129. CrossRef
Bascos NAD, Mirano- Bascos D, Saloma CP. Structural analysis of spike protein mutations in an emergent SARS-CoV-2 variant from the Philippines. BioRxiv, 2021, vol. 2021. CrossRef
Betton M, Livrozet M, Planas D, Fayol A, Monel B, Védie B, Bruel T, Tartour E, Robillard N, Manuguerra JC, Blanchard A. Sera neutralizing activities against SARS-CoV-2 and multiple variants six month after hospitalization for COVID-19. Clin Infect Dis, 2021; 2021:e1337–44.
Bollinger R, Ray S. COVID variants: what you should know, 2021. Available via https://www.hopkinsmedicine.org/health/conditions-and-diseases/coronavirus/a-new-strain-of-coronavirus-what-you-should-know (Accessed 4 December 2021). CrossRef
Buss LF, Prete CA, Abrahim CM, Mendrone A, Salomon T, de Almeida-Neto C, França RF, Belotti MC, Carvalho MP, Costa AG, Crispim MA. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science, 2021; 371(6526):288–92.
Callaway E, Ledford H. How bad is Omicron? What scientists know so far. Nature, 2021; 600(7888):197–9. Available via https://www.nature.com/articles/d41586-021-03614-z (Accessed 07 December 2021).
CDC. CDC data tracker, 2021a. Available via https://covid.cdc.gov/covid-data-tracker/#variant-proportions (Accessed 26 November 2021).
CDC. CDC delta variant: what we know about the science, 2021b. Available via https://www.cdc.gov/coronavirus/2019-ncov/variants/delta-variant.html (Accessed 07 December 2021).
CDC. CDC omicron variant: what you need to know, 2021c. Available via https://www.cdc.gov/coronavirus/2019-ncov/variants/omicron-variant.html (Accessed 07 December 2021).
CDC. CDC SARS-CoV-2 variant classifications and definitions, 2021d. Available via https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fcases-updates%2Fvariant-surveillance%2Fvariant-info.html#anchor_1632158885160 (Accessed 07 December 2021). CrossRef
Chang JPC, Pariante CM, Su KP. Omega-3 fatty acids in the psychological and physiological resilience against COVID-19. Prostaglandins Leukot Essent Fatty Acids, 2020; 161:102177. CrossRef
Choudhary OP, Dhawan M, Priyanka. Omicron variant (B.1.1.529) of SARS-CoV-2: threat assessment and plan of action-correspondence. Int J Surg, 2021; 97:106187; doi:10.1016/j.ijsu.2021.1061 CrossRef
Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Yu T. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet, 2020; 395(10223):507–13. CrossRef
Chen X, Chen Z, Azman AS, Sun R, Lu W, Zheng N, Zhou J, Wu Q, Deng X, Zhao Z, Chen X. Neutralizing antibodies against SARS-CoV-2 variants induced by natural infection or vaccination: a systematic review and individual data meta-analysis. Clin Infect Dis, 2021; 2021:ciab646.
CNN Philippines: DOH confirms new COVID-19 mutations in Central Visayas, 2015. Available via https://www.cnnphilippines.com/news/2021/2/18/two-covid-mutations-of-concern-cebu-doh-central-visayas.html (Accessed 05 December 2021). CrossRef
Collier DA, De Marco A, Ferreira IA, Meng B, Datir RP, Walls AC, Kemp SA, Bassi J, Pinto D, Silacci-Fregni C, Bianchi S. Sensitivity of SARS-CoV-2 B. 1.1. 7 to mRNA vaccine-elicited antibodies. Nature, 2021; 593(7857):136–41. CrossRef
Copin R, Baum A, Wloga E, Pascal KE, Giordano S, Fulton BO, Zhou A, Negron N, Lanza K, Chan N, Coppola A. The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell, 2021; 184(15):3949–61.
Cov-lineages.org “B.1.525”, 2021. Available via https://cov-lineages.org/global_report_B.1.525.html (Accessed 05 December 2021). CrossRef
Das UN. Can bioactive lipids inactivate coronavirus (COVID-19)? Arch Med Res, 2020; 51(3):282–6. CrossRef
Das UN. Bioactive lipids in COVID-19-further evidence. Arch Med Res, 2021; 52(1):107–20. CrossRef
Dejnirattisai W, Zhou D, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HM, Tuekprakhon A, Nutalai R, Wang B. Antibody evasion by the P. 1 strain of SARS-CoV-2. Cell, 2021; 184(11):2939–54. CrossRef
Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, Sotomayor-González A, Glasner DR, Reyes KR, Gliwa AS, Reddy NP. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. MedRxiv, 2021, CrossRef
Edara VV, Hudson WH, Xie X, Ahmed R, Suthar MS. Neutralizing antibodies against SARS-CoV-2 variants after infection and vaccination. JAMA, 2021; 325(18):1896–8. CrossRef
Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DD, Mishra S, Crispim MA, Sales FC, Hawryluk I, McCrone JT, Hulswit RJ. Genomics and epidemiology of the P. 1 SARS-CoV-2 lineage in Manaus, Brazil. Science, 2021; 372(6544):815–21. CrossRef
Ferreira IA, Kemp SA, Datir R, Saito A, Meng B, Rakshit P, Takaori-Kondo A, Kosugi Y, Uriu K, Kimura I, Shirakawa K Abdullahi A, Agarwal A, Ozono S, Tokunaga K, Sato K, Gupta RK; CITIID-NIHR BioResource COVID-19 Collaboration, Indian SARS-CoV-2 Genomics Consortium; Genotype to Phenotype Japan (G2P-Japan) Consortium. SARS-CoV-2 B. 1.617 mutations L452R and E484Q are not synergistic for antibody evasion. J Infect Dis, 2021; 224(6):989–94.
Food and Drug Administration (FDA). Coronavirus (COVID-19) update: monoclonal antibodies for treatment of COVID-19, 2021a. Available via https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibodies-treatment-covid-19 (Accessed 26 November 2021).
Food and Drug Administration (FDA). Fact sheet for health care provider’s emergency use authorization (EUA) of bamlanivimab and etesevimab, 2021b. Available via https://www.fda.gov/media/ 145802/download (Accessed 26 November 2021).
GISAID. COVID 19 variants, 2021. Available via https://www.gisaid.org/hcov19-variants/ (Accessed 05 December 2021). CrossRef
Grabowski F, Preibisch G, Gizi?ski S, Kocha?czyk M, Lipniacki T. SARS-CoV-2 variant of concern 202012/01 has about twofold replicative advantage and acquires concerning mutations. Viruses, 2021; 13(3):392. CrossRef
Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, Krüger N, Graichen L, Hofmann-Winkler H, Kempf A, Winkler MS. SARS-CoV-2 variants B. 1.351 and P. 1 escape from neutralizing antibodies. Cell, 2021; 184(9):2384–93. CrossRef
Jiang HW, Zhang HN, Meng QF, Xie J, Li Y, Chen H, Zheng YX, Wang XN, Qi H, Zhang J, Wang PH. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell Mol Immunol, 2020; 17(9):998–1000.
Latif AA, Mullen LJ, Alkuzweny M, Tsueng G, Cano M, Haag E, Zhou J, Zeller M, Hufbauer E, Matteson N, Wu C, Andersen KG, Andrew ISu, Karthik G, Laura DH, and the Center for Viral Systems Biology. Mu variant report, 2021 Available via https://outbreak.info/situation-reports/Mu (Accessed 05 December 2021). CrossRef
Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, Zhao C, Zhang Q, Liu H, Nie L, Qin H. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell, 2020; 182(5):1284–94. CrossRef
Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A, Nutalai R, Zhou D, Mentzer AJ, Zhao Y, Duyvesteyn HM. Reduced neutralization of SARS-CoV-2 B. 1.617 by vaccine and convalescent serum. Cell. 2021; 184(16):4220–36. CrossRef
Martin DP, Weaver S, Tegally H, San EJ, Shank SD, Wilkinson E, Giandhari J, Naidoo S, Pillay Y, Singh L, Lessells RJ. The emergence and ongoing convergent evolution of the N501Y lineages coincides with a major global shift in the SARS-CoV-2 selective landscape. MedRxiv, 2021, vol. 2021. CrossRef
Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID19: their roles in pathogenesis. J Microbiol Immunol Infect, 2021; 54(2):159–63. CrossRef
Parker MD, Lindsey BB, Shah DR, Hsu S, Keeley AJ, Partridge DG, Leary S, Cope A, Johnson K, Ali N, Raghei R. Altered sub-genomic RNA expression in SARS-CoV-2 B. 1.1. 7 infections. BioRxiv, 2021, vol. 2021. CrossRef
Peng J, Liu J, Mann SA, Mitchell AM, Laurie MT, Sunshine S, Pilarowski G, Ayscue P, Kistler A, Vanaerschot M, Li LM, McGeever A, Chow ED, Marquez C, Nakamura R, Rubio L, Chamie G, Jones D, Jacobo J, Rojas S, Rojas S, Tulier-Laiwa V, Black D, Martinez J, Naso J, Schwab J, Petersen M, Havlir D, DeRisi J; IDseq Team. Estimation of secondary household attack rates for emergent spike L452R SARS- CoV-2 variants detected by genomic surveillance at a community- based testing site in San Francisco. Clin Infect Dis, 2021; 2021:ciab283. CrossRef
Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, Porrot F, Planchais C, Buchrieser J, Rajah MM, Bishop E, Albert M. Sensitivity of infectious SARS-CoV-2 B. 1.1. 7 and B. 1.351 variants to neutralizing antibodies. Nat Med, 2021a; 27(5):917–24. CrossRef
Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M. Reduced sensitivity of SARS-CoV-2 variant delta to antibody neutralization. Nature, 2021b; 596(7871):276–80. CrossRef
Rees-Spear C, Muir L, Griffith SA, Heaney J, Aldon Y, Snitselaar JL, Thomas P, Graham C, Seow J, Lee N, Rosa A. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep, 2021; 34(12):108890.
Sabino EC, Buss LF, Carvalho MP, Prete CA, Crispim MA, Fraiji NA, Pereira RH, Parag KV, da Silva Peixoto P, Kraemer MU, Oikawa MK. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet, 2021; 397(10273):452–5. CrossRef
Shakoor H, Feehan J, Al Dhaheri AS, Ali HI, Platat C, Ismail LC, Apostolopoulos V, Stojanovska L. Immune-boosting role of vitamins D, C, E, zinc, selenium and omega-3 fatty acids: could they help against COVID-19? Maturitas, 2021; 143:1–9. CrossRef
Stanford University. Coronavirus antiviral and resistance database, 2021. Available via https://covdb.stanford.edu/page/mutation-viewer/#sec_zeta (Accessed 07 December 2021).
Supasa P, Zhou D, Dejnirattisai W, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HM, Nutalai R, Tuekprakhon A, Wang B. R educed neutralization of SARS- CoV-2 B.1.617 by vaccine a neutralization of SARS-CoV-2 B. 1.1. 7 variant by convalescent and vaccine sera. Cell, 2021; 184(8):2201–11. CrossRef
Tablizo FA, Kim KM, Lapid CM, Castro MJ, Yangzon MS, Maralit BA, Ayes ME, Cutiongco-de la Paz EM, De Guzman AR, Yap JM, Llames JH. Genome sequencing and analysis of an emergent SARS-CoV-2 variant characterized by multiple spike protein mutations detected from the Central Visayas Region of the Philippines. MedRxiv, 2021, vol. 2021. CrossRef
Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature, 2021; 592(7854):438–43. CrossRef
The Japan Times. Japanese authorities discover a new coronavirus variant in traveler from Philippines, 2021. Available via https://www.japantimes.co.jp/news/2021/03/13/national/new-variant-philippines/ (Accessed 05 December 2021).
Thorne LG, Bouhaddou M, Reuschl AK, Zuliani-Alvarez L, Polacco B, Pelin A, Batra J, Whelan MV, Ummadi M, Rojc A, Turner J. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B. 1.1. 7 UK variant. BioRxiv, 2021. CrossRef
Wang GL, Wang ZY, Duan LJ, Meng QC, Jiang MD, Cao J, Yao L, Zhu KL, Cao WC, Ma MJ. Susceptibility of circulating SARS-CoV-2 variants to neutralization. NEJM, 2021a; 384(24):2354–6. CrossRef
Wang P, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, Liu L, Kwong PD, Huang Y, Shapiro L, Ho DD. Increased resistance of SARS-CoV-2 variant P. 1 to antibody neutralization. Cell Host Microbe, 2021b; 29(5):747–51. CrossRef
Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, Graham BS. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7. Nature, 2021c; 593(7857):130–5. CrossRef
Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann HH, Michailidis E, Gaebler C. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife, 2020; 9:e61312. CrossRef
Wink PL, Volpato FC, Monteiro FL, Willig JB, Zavascki AP, Barth AL, Martins AF. First identification of SARS-CoV-2 Lambda (C. 37) variant in southern Brazil. Infect Control Hosp Epidemiol, 2021; 1–2. CrossRef
World Health Organization (WHO). Coronavirus (COVID-19) dashboard. WHO, Geneva, Switzerland, 2021a. Available via https://covid19.who.int/ (Accessed 4 December 2021).
World Health Organization (WHO). Classification of omicron (B.1.1.529): SARS-CoV-2 variant of Concern. WHO, Geneva, Switzerland, 2021b. Available via https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
World Health Organization (WHO). Tracking SARS-CoV-2 variants. WHO, Geneva, Switzerland, 2021c. Available via https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/FDA
Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe, 2020; 27(3):325–8. CrossRef
Wu Z, McGoogan J M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72,314 cases from the Chinese Center for Disease Control and Prevention. JAMA, 2020; 323:1239–42. CrossRef
Zhang JZ, Yeh HW, Walls AC, Wicky BI, Sprouse K, VanBlargan LA, Treger R, Quijano-Rubio A, Pham MN, Kraft JC, Haydon IC. Detection of antibodies neutralizing historical and emerging SARS-CoV-2 strains using a thermodynamically coupled de novo biosensor system. BioRxiv, 2021a, vol. 2021. CrossRef
Zhang L, Cui Z, Li Q, Yu Y, Wu J, Nie J, Ding R, Wang H, Zhang Y, Liu S, Chen Z. Comparison of 10 emerging SARS-CoV-2 variants: infectivity, animal tropism, and antibody neutralization. Research Square, 2021b, vol. 2021. CrossRef
Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HM, Tuekprakhon A, Nutalai R, Wang B. Evidence of escape of SARS-CoV-2 variant B. 1.351 from natural and vaccine-induced sera. Cell, 2021a; 184(9):2348–61. CrossRef
Zhou H, Dcosta BM, Samanovic MI, Mulligan MJ, Landau NR, Tada T. B. 1.526 SARS-CoV-2 variants identified in New York City are neutralized by vaccine-elicited and therapeutic monoclonal antibodies. BioRxiv, 2021b, vol. 2021. CrossRef
Zhu H, Wei L, Niu P. The novel coronavirus outbreak in Wuhan, China. Glob Health Res Policy, 2020; 5:1–3. CrossRef