INTRODUCTION
The human genome, a complicated entity made up of 23 chromosomes, is made up of approximately three billion deoxyribose nucleic acid (DNA) base pairs, with an estimated number of 20,000–25,000 genes that code for various proteins. Interestingly, about 99.9% of the human genome is similar in all humans, and the remaining 0.1% difference accounts for each person's uniqueness (MedlinePlus, 2021). Few DNA variants can be detrimental and can result in life-threatening diseases, while others can help in the development of desirable characteristics (Koepsell, 2019). When the variant occurs in more than 1% of the population, it is referred to as genetic polymorphism (MedlinePlus, 2021). The single nucleotide polymorphisms (SNPs) in specific genes impact the expression of various illnesses as well as safety and efficacy of drug therapy (Koofee and Mubarak, 2019). According to reports, at least 10% of the population possesses gene variations that lead to complications (Fridovich-Keil, 2020). Despite significant advances, contemporary pharmacotherapy still confronts several obstacles, including catastrophic or even fatal adverse drug responses and non-responses to the empirical therapy. Interindividual variability in drug treatment has been attributed to genetic polymorphism. Such variabilities in the outcomes of the drug therapy among the different populations are referred as “pharmacoethnicity” and occur due to drug-gene interactions. Various enzymes and transporters involved in drug metabolism and transport exhibit polymorphisms that affect an individual's response to a certain medication. The discovery and analysis of clinically relevant polymorphisms assist in the development of personalized therapy. (Fridovich-Keil, 2020). The benefits of testing for the genetic polymorphisms include improved health care outcomes through the use of gene therapy, the identification of new therapeutic targets, the development of new medications, defining new dosage regimens, and so on (Koepsell, 2019). Recent genetic polymorphism research has focused on leveraging these genetic variations through big data analytics in order to develop tools for identifying critical population diversity patterns connected to treatment response. (Bachtiar, 2019).
The organic cation transporter 1 (OCT1) are the members of the SLC22A family of solute carrier transporters (SLCs). The most essential SLCs implicated in drug elimination are OCTs (Koepsell, 2019). OCT1 is one of the most abundantly expressed poly specific transporter protein in the liver that plays an important role in the hepatic uptake of cationic and neutral small molecules, and also in their renal excretion (Tzvetkov et al., 2009).
OCT1 gene has been reported with about 34 polymorphisms in 10 different ethnic groups reported with altered drug transportation capacity that resulted in interindividual variability in pharmacokinetics (PK) and or pharmacodynamic (PD) outcomes of drugs (Koepsell, 2019). Extensive research on the populations of Japanese, Chinese, and Korean ethnicity were done, and genetic variations have mostly been found in such Asian populations apart from the European population (Mato et al., 2018). The effects of OCT1 polymorphisms on the PK and PD of Metformin in comparison to other medications have been thoroughly studied (Tzvetkov et al., 2009). Indian researchers have examined rs683369 (480 G > C, L160F), rs2282143 (P341L, 1022C > T), rs628031 (M408V, 1222A > G), rs622342 (1386C > A), rs1867351 (156T > C, S52S), and four other intronic variants (Sur, 2014). The allele frequency of the rs628031 polymorphism was reported as 80.3% in the Tamilian population of south India (Umamaheswaran et al., 2011) and the rs628031 polymorphism has been found to affect the responsiveness of substrate medicines such as Metformin, Levodopa, Imatinib, Lamotrigine, and others (Koepsell, 2019).
Only few literature findings are available that suggest heterogeneity in the frequency and impact of the rs628031 SNP among ethnicities, (Chen et al., 2010; Mato et al., 2018; Ningrum et al., 2017; Reséndiz-Abarca et al., 2019; Shokri et al., 2016; Tarasova et al., 2012; Tzvetkov et al., 2009; Umamaheswaran et al., 2011). Among the Indians, only a couple of studies, one from north India and another from south India, have reported the prevalence statistics for rs628031 polymorphisms. (Sur, 2014, Umamaheswaran et al., 2011).
The primary objective of this study is to determine the prevalence of rs628031 polymorphism in healthy south Indian males. The scarcity of research and the difference of effect in the genetic variability of the rs628031 polymorphism in various groups provide compelling reasons to perform the study. The period elapsed since the last study in the south Indian population warrants Umamaheswaran et al. (2011) study included only south Indian Tamilians, whereas this study included participants from all the major states of south India including Tamil Nadu in order to get more generalized results for the south Indian population. As a matter of fact, it is important to investigate the incidence and degree of this SNP's effect in the south Indian population in order to create a medication regimen with improved therapeutic efficacy and minimum side effects for the drugs that are OCT1 substrates.
MATERIALS AND METHODS
Subjects
Inclusion criteria: Adult male volunteers between the age group of 18 and 35 years in good health based on medical history, physical examination, electrocardiography (ECG), and routine laboratory tests (complete blood count, blood glucose, liver function tests, renal function tests, and urinalysis) were included in the study. The subjects' south Indian heritage was confirmed through interviews, and those who had lived in south India for at least three generations were included. All participants were certified as fit to participate in the study by a competent physician.
Exclusion criteria: Smokers, alcoholics, or subjects taking any drugs for chronic conditions were excluded from the study.
Out of the 60 volunteers screened, 58 were certified as fit to participate in the study out of which 10 volunteers were the drop-outs and thus 48 volunteers participated in the study.
Ethical Clearance
The study protocol was approved by the institutional review board of the academic institution where the study was held (JSSCP/IRB/09/2019-20). All the study participants were well-informed about the purpose, nature, and outcomes of the study prior to the process of getting informed consent forms.
Sampling methods
A qualified phlebotomist withdrew 2 ml of blood from each participant in K3-EDTA (Ethylenediaminetetraacetic acid)-coated blood collection tubes and labelled them accordingly. The blood samples were centrifuged at 12,000 rpm for 1 minute to separate the plasma from the blood cells. The sedimented white blood cells were used for DNA harvesting.
Isolation of DNA from whole blood
The DNA extraction was carried out by Phenol-Chloroform method that involves digesting eukaryotic cells or tissues with proteinase K in the presence of EDTA and solubilizing membrane and denaturing proteins with a detergent such as sodium dodecyl sulphate. The nucleic acids were then purified by the phase extraction method using organic solvents', viz., phenol, chloroform, isoamyl alcohol, and 100% ethanol. Buffers such as saline sodium citrate buffer (sodium chloride, sodium citrate) and Tris-EDTA buffer (Tris-acetate and EDTA (TAE)) were used in the procedure to maintain the pH.
Primers
The primers used for OCT1—rs628031 polymorphism detection were designed using the nucleotide sequences obtained from GENBANK, the accession number for the human OCT1 gene was found as NC_000006.12. The sequences of the forward and reverse primers designed for this study were F5?- TCATCACCATTGACCGCGTG -3? and R5?- ACACTTTCCCCACACTTCGAT -3?, respectively (Sakhala Enterprises, Bengaluru).
DNA amplification
The isolated DNA samples were amplified using polymerase chain reaction (PCR) and restriction fragment length polymorphism technique was used to identify the genetic polymorphism. DNA was amplified using the reaction mixture consisting of 0.1 µl of Taq DNA polymerase, 15 µl of genomic DNA, 1 µl of each dNTP (GENEI LABS, Bengaluru), 0.5 µl of each primer (Sakhala Enterprises, Bengaluru), and 5 µl of PCR buffer with MgCl2 and the volume made to 50µl using sterile water. PCR for OCT1—rs628031 gene was performed by initial denaturation of the sample at 95°C for 5 minutes, followed by 30 cycles of 95°C for 30 seconds denaturation, 54°C for 35 seconds annealing, and 72°C for 60 seconds extension, with a final extension step of 72°C for 5 minutes.
Restriction enzyme
The restriction enzyme was designed using www.restrictionmapper.org. The DNA sequence under question was pasted in the restriction mapper, and a list of all the enzymes that cleaved the sequence was obtained for both wild and variant types. The restriction enzyme that is not common in both lists was selected. The obtained DNA samples were fragmented using restriction enzyme Msc1 (GENEI LABS, Bengaluru) in case of wild type allele.
Genotyping and agarose gel electrophoresis
To detect the presence of polymorphism, 25 µl mixture with 10 µl PCR amplified product, 1µl restriction enzyme (Msc1), 5 µl assay buffer, and 9 µl distilled water was prepared and placed at 37°C bath for about 2 hours to facilitate digestion. After 2 hours, it was placed in a 65°C bath for about 5 minutes to destroy the enzyme and stop digestion. The sample was then allowed to cool. After digestion with a restriction enzyme, the resulting DNA fragments were separated by agarose gel electrophoresis. 1% agarose gel was cast and 2 µl of loading dye was added to 10 µl of PCR product. The PCR product was loaded in the odd-numbered wells, while the PCR products digested by the restriction enzyme were loaded in the adjacent even-numbered wells. The gel was run in a 1× TAE buffer at 50 V by observing the bromophenol blue for the movement of the DNA fragments. The banding patterns were observed for DNA fragmentation under Ultraviolet light (using a photo documentation system) to determine the presence of rs628031 polymorphism.
Statistical analysis
The average baseline characteristics of the study subjects were calculated using descriptive statistics such as the mean and standard deviation. Hardy–Weinberg equilibrium was used to compare the genotype data with the expected frequencies. Chi-square test was used to compare the differences between observed and expected frequencies. A p-value of less than 0.05 was considered to be statistically significant.
RESULTS
Patient characteristics
A total of 60 volunteers were screened for fitness. Based on the physician’s advice, two volunteers were excluded from the study due to lack of fitness. The average age of the subjects was 18.5 + 0.91 years. All general parameters and biochemical investigations revealed that all such parameters were within the normal limits.
Genotyping
In 48 samples, DNA was isolated, and genetic polymorphism was studied. The genotype frequencies of rs628031 GG homozygous variant, AG heterozygous variant, and AA homozygous wild are 66.66%, 2.08%, and 31.25% (n = 32, 1 and 15), respectively, as shown in figure 1–6. p, q CHWE: Check was used to calculate Hardy–Weinberg equilibrium. To compare the expected and observed values, the Chi-square test was used. The observed and expected values differed significantly, and the p-value was greater than 0.05, indicating rs628031 deviation from Hardy–Weinberg equilibrium.
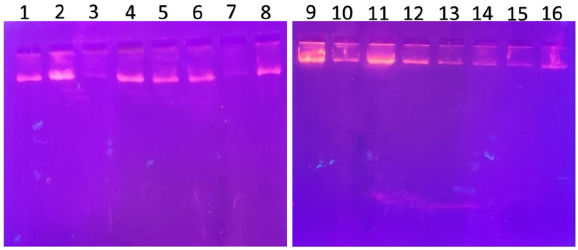 | Figure 1. PCR amplified-RFLP treated DNA samples of OCT1 rs628031 gene results by agarose gel electrophoresis (1%): The lanes 1, 3, 5, 7, 9, 11, 13, 15 are loaded with PCR amplified DNA and the lanes 2, 4, 6, 8, 10, 12, 14, 16 contain restriction enzyme-treated DNA of the volunteers. From the results, a single clear band was observed, this indicates, lanes 2, 4, 6, 8, 10, 12, 14, 16, all samples found to be GG homozygous variants. No fragmentation was detected after treatment with restriction enzyme specific for OCT1 polymorphism. [Click here to view] |
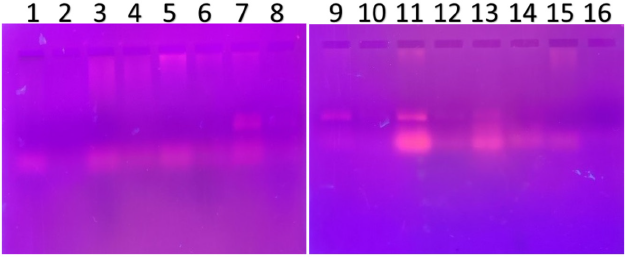 | Figure 2. PCR amplified-RFLP treated DNA samples of OCT1 gene results by agarose gel electrophoresis (1%): The lanes 1, 3, 5, 7, 9, 11, 13, 15 are loaded with PCR amplified DNA and the lanes 2, 4, 6, 8, 10, 12, 14, 16 contain restriction enzyme-treated DNA of the volunteers. From the results, clear evidence of fragments was observed, this indicates, lanes 2, 4, 6, 8, 10, 12, 14, 16, all samples were found to be AA homozygous wild. Fragmentation was detected after treatment with a restriction enzyme specific for OCT1 polymorphism [Click here to view] |
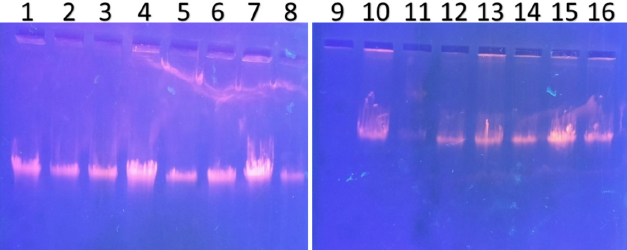 | Figure 3. PCR amplified-RFLP treated DNA samples of OCT1 gene results by agarose gel electrophoresis (1%): The lanes 1,3,5,7,9,11,13,15 are loaded with PCR amplified DNA and the lanes 2, 4, 6, 8, 10, 12, 14, 16 contain restriction enzyme-treated DNA of the volunteers. From the results, a single clear band was observed, this indicates, lanes 2, 4, 6, 8, 10, 12, 14, 16, all samples found to be GG homozygous variants. No fragmentation was detected after treatment with restriction enzyme specific for OCT1 polymorphism. [Click here to view] |
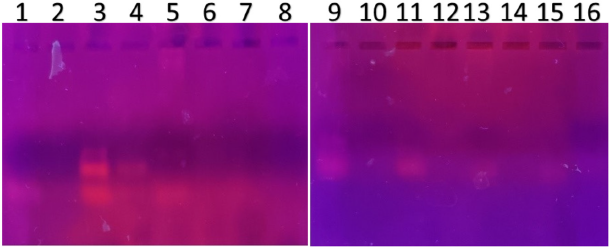 | Figure 4. PCR amplified-RFLP treated DNA samples of OCT1 gene results by agarose gel electrophoresis (1%): The lanes 1, 3, 5, 7, 9, 11, 13, 15 are loaded with PCR amplified DNA and the lanes 2, 4, 6, 8, 10, 12, 14, 16 contain restriction enzyme-treated DNA of the volunteers. Lane 4 was found to be an AG heterozygous variant; evidence of fragmented DNA strand was detected after restriction enzyme treatment specific for OCT1 polymorphism. The remaining 2, 6, 8, 10, 12, 14, 16 lanes’ samples were found to be AA homozygous wild with clear evidence of fragmented DNA strand after restriction enzyme digestion. [Click here to view] |
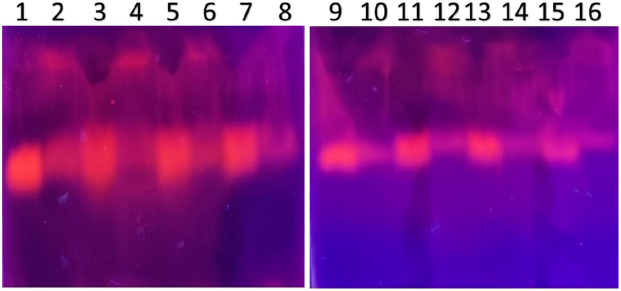 | Figure 5. PCR amplified-RFLP treated DNA samples of OCT1 gene results by agarose gel electrophoresis (1%): The lanes 1, 3, 5, 7, 9, 11, 13, 15 are loaded with PCR amplified DNA and the lanes 2, 4, 6, 8, 10, 12, 14, 16 contain restriction enzyme-treated DNA of the volunteers. From the results, a single clear band was observed, this indicates, lanes 2, 4, 6, 8, 10, 12, 14, 16, all samples found to be GG homozygous variants. No fragmentation was detected after treatment with restriction enzyme specific for OCT1 polymorphism. [Click here to view] |
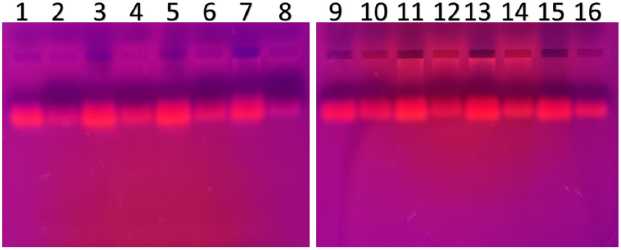 | Figure 6. PCR amplified-RFLP treated DNA samples of OCT1 gene results by agarose gel electrophoresis (1%): The lanes 1, 3, 5, 7, 9, 11, 13, 15 are loaded with PCR amplified DNA and the lanes 2, 4, 6, 8, 10, 12, 14, 16 contain restriction enzyme-treated DNA of the volunteers. From the results, a single clear band was observed, this indicates, lanes 2, 4, 6, 8, 10, 12, 14, 16, all samples found to be GG homozygous variants. No fragmentation was detected after treatment with restriction enzyme specific for OCT1 polymorphism. [Click here to view] |
DISCUSSION
OCT1 (rs628031) genetic polymorphism influences the PK and PD of its substrate drugs. Multiple studies have shown significant interethnic differences in OCT1 genetic variants. In this study, the minor allele frequency of rs628031 polymorphism was found to be 0.67 in the south Indian population. This frequency of rs628031 polymorphism is higher when compared to that predicted by the Hardy–Weinberg equilibrium that indicates a deviation. The Hardy–Weinberg equilibrium is a concept that states that in the absence of disrupting influences, genetic variation in a population will remain constant from generation to generation. Though statistical variance from Hardy–Weinberg predictions typically implies a violation of the theorem's assumptions, the reverse is not always true. The possible causes of this deviation include the study’s small sample size or the possibility of natural selection of the minor allele over the generations based on the five principles of Hardy–Weinberg equilibrium. The frequency of the minor allele of rs628031 was determined to be 60.47% in the Javanese population, which is similar to the findings of this study (Ningrum et al., 2017). In a study among the diabetic population, in the Han Chinese community, found a frequency of about 72% (Zhou et al., 2015). In an Iranian population, the G allele frequency of rs628031 was 41% in the metformin-responding group and 51% in the non-responding group (Shokri et al., 2016). The minor allele frequency for rs628031 in a Latvian population was found to be 0.42 in the control group and 0.275 in the case group (Tarasova et al., 2012). Findings from a 1,000 genomes project in the Chinese - Japanese community, the minor allele frequency of rs628031 was determined to be 0.15. (Chen et al., 2010). Minor allele frequencies of 0.43 and 0.80 were found in two earlier investigations done in India (Sur, 2014; Umamaheswaran et al., 2011). To summarize, studies in Chinese, Mexican, and Japanese populations, as well as a previous study in south India, revealed a greater frequency (73%–83%) of variant allele rs628031 than Chen et al., 2010; Reséndiz-Abarca et al., 2019; Umamaheswaran et al.; 2011, Zhou et al. 2015) whereas, the prevalence of this polymorphism was lower (39%–40%) in European nations (Becker et al., 2011; Tarasova et al., 2012) as well as among (43%–51%) Iranian and North Indian groups (Shokri et al., 2016; Sur 2014). Based on the study's findings and the existing literature, the rs628031 polymorphism has a diverse distribution. To further understand the prevalence and clinical significance of rs628031, genotyping studies should be undertaken in smaller, more focused areas, as a generalized model cannot be developed by extrapolating existing data.
Limitations
This study was only able to examine 48 samples, as compared to the 60 samples that were scheduled to be analyzed at the beginning of the project. A larger sample size would allow for more generalizable findings on the prevalence of the rs628031 polymorphism in the south Indian population.
CONCLUSION
The study results show that the minor allele frequency of rs628031 polymorphism was 0.67 which is comparable to that of the Javanese population. At the same time, it differs from the prevalence found in other populations. This demonstrates the importance of conducting polymorphism studies in a specific population.
Further studies are needed to determine the effect of the rs628031 polymorphism on the PK and PD of drugs widely transported by the OCT1, such as Metformin. These types of research can help in the development of precision medicine and designing customized dosing regimen in individuals known to have the rs628031 polymorphism. Given the high prevalence of the rs628031 polymorphism among the study subjects, individuals who take OCT1 substrate drugs may be urged to undertake a genotyping test in the future, if its effects on PK and PD of relevant drugs are studied in a wider population.
ACKNOWLEDGEMENT
The authors acknowledge the JSS AHER for the research grant. The authors also acknowledge Dr. Ashish D Wadhwani, Professor and Head, Dr. Raman Rajesh Kumar, Lecturer, Department of Pharmaceutical Biotechnology. It is our privilege to express our bunch of embedded feelings of affection and thanks to Mr. Rajesh J, the postgraduate student of the same department for the technical support towards conducting this study.
CONFLICT OF INTERESTS
The authors have no conflict of interest to declare.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
ETHICAL APPROVALS
The study protocol was approved by the institutional review board of the academic institution where the study was held (JSSCP/IRB/09/2019-20).
FUNDING
This study was funded by JSS Academy of Higher Education & Research grants (REG/DIR(R)/URG/54/2011-12/5293).
REFERENCES
Al-Koofee DAF, Mubarak SMH. The Recent Topics in genetic polymorphisms. In: Öz GC, Çal??kan M, Erol O (eds.). IntechOpen, Najaf, Iraq, IQ, 2019; 1-15.
Bachtiar M, Ooi BNS, Wang J, Jin Y, Tan TW, Chong SS, Lee CGL. Towards precision medicine: interrogating the human genome to identify drug pathways associated with potentially functional, population-differentiated polymorphisms. Pharmacogenomics J, 2019; 19(6):516–27. CrossRef
Becker ML, Visser LE, van Schaik RHN, Hofman A, Uitterlinden AG, Stricker BHC. OCT1 polymorphism is associated with response and survival time in anti-Parkinsonian drug users. Neurogenetics, 2011; 12(1):79–82. CrossRef
Chen L, Takizawa M, Chen E, Schlessinger A, Seigenthaler JR, Choi JH, Sali A, Kubo M, Nakamura S, Iwamoto Y, Iwasaki N, Giacomini KM. Genetic polymorphisms in organic cation transporter 1 (OCT1) in Chinese and Japanese populations exhibit altered function. J Pharmacol Exp Ther, 2010; 335(1):42–50. CrossRef
Fridovich-Keil JL, Fridovich I, Robinson A. “Human genetic disease”. Encyclopedia Britannica, 2020. Available via https://www.britannica.com/science/human-genetic-disease (Accessed 25 May 2021).
Koepsell H. Organic Cation Transporters in Health and Disease. Pharmacol Rev, 2019; 72(1):253–319. CrossRef
Mato EPM, Guewo-Fokeng M, Essop MF, Owira PMO. Genetic polymorphisms of organic cation transporter 1 (OCT1) and responses to metformin therapy in individuals with type 2 diabetes: a systematic review. Medicine, 2018; 97(27):e11349. CrossRef
MedlinePlus. 2021. Available via https://medlineplus.gov/genetics/understanding/basics/gene/#:~:text=In%20humans%2C%20genes%20vary%20in,between%2020%2C000%20and%2025%2C000%20genes (Accessed 23 May 2021).
Ningrum VDA, Ikawati Z, Sadewa AH, Ikhsan MR. Allele frequencies of two main metformin transporter genes: SLC22A1 rs628031 A>G and SLC47A1 rs2289669 G>A among the Javanese population in Indonesia. CPPM, 2017; 15(2):121–8. CrossRef
Reséndiz-Abarca CA, Flores-Alfaro E, Suárez-Sánchez F, Cruz M, Valladares-Salgado A, Alarcón-Romero LDC, Vázquez-Moreno MA, Wacher-Rodarte NA, Gómez-Zamudio JH. Altered Glycemic control associated with polymorphisms in the SLC22A1 (OCT1) gene in a Mexican population with type 2 diabetes Mellitus treated with Metformin: a cohort study. J Clin Pharmacol, 2019; 59(10):1384–90. CrossRef
Shokri F, Ghaedi H, Ghafouri Fard S, Movafagh A, Abediankenari S, Mahrooz A, Kashi Z, Omrani MD. Impact of ATM and SLC22A1 polymorphisms on therapeutic response to Metformin in Iranian diabetic patients. Int J Mol Cell Med, 2016; 5:1–7.
Sur D. A tale of genetic variation in the human Slc22a1 gene encoding OCT1 among type 2 diabetes mellitus population groups of West Bengal, India. IMPACT: IJRANSS, 2014; 2(5):97–106.
Tarasova L, Kalnina I, Geldnere K, Bumbure A, Ritenberga R, Nikitina-Zake L, Fridmanis D, Vaivade I, Pirags V, Klovins J. Association of genetic variation in the organic cation transporters OCT1, OCT2 and multidrug and toxin extrusion 1 transporter protein genes with the gastrointestinal side effects and lower BMI in metformin-treated type 2 diabetes patients. Pharmacogenet Genomics, 2012; 22(9):659–66. CrossRef
Tzvetkov MV, Vomfelde SV, Balen D, Meineke I, Schmidt T, Sehrt D, Saboli? I, Koepsell H, Brockmöller J. The effects of genetic polymorphisms in the organic cation transporters OCT1, OCT2, and OCT3 on the renal clearance of metformin. Clin Pharmacol Ther, 2009; 86(3):299–306. CrossRef
Umamaheswaran G, Praveen RG, Arunkumar AS, Das AK, Shewade DG, Adithan C. Genetic analysis of OCT1 gene polymorphisms in an Indian population. Indian J Hum Genet, 2011; 17(3):164–8. CrossRef
Zhou Y, Ye W, Wang Y, Jiang Z, Meng X, Xiao Q, Zhao Q, Yan J. Genetic variants of OCT1 influence glycemic response to metformin in Han Chinese patients with type-2 diabetes mellitus in Shanghai. Int J Clin Exp Pathol, 2015; 8(8):9533–42.