INTRODUCTION
Lung cancer is a type of cancer with the highest incidence after prostate cancer in men and after breast cancer in women (Dela Cruz et al., 2011). However, lung cancer still has the highest mortality among cancer patients worldwide (Bade and Dela Cruz, 2020). The risk factors for lung cancer are industry emissions, air pollution, cigarette smoking, chronic infectious lung diseases, and living lifestyle (Barta et al., 2019). In Indonesia, lung cancer ranks first in men and third in women (The Global Cancer Observatory, 2019) with a mortality rate of 12.6% from 207,210 deaths caused by cancer in 2018 (WHO, 2020).
For more than 40 years, small organic molecules (<3,000 Daltons) derived from microbes and plants have been used as chemotherapy agents for cancer (Kinghorn et al., 2009). The Streptomyces bacteria are the main source of medicinal compounds derived from microbes for nearly 80% of drugs, especially antibiotics (Weber et al., 2015) and anticancer agents (Dhaneesha et al., 2017; Nguyen et al., 2020). In our previous research, we discovered a marine bacterium, Streptomyces sp. GMY01, which has a very high potential active principle to be developed as a therapeutic agent (Herdini et al., 2015). This bacterial extract was also found to contain anticancer activities against T47D and MCF7 breast cancer cell lines (Farida et al., 2007; Werdyani et al., 2017).
In this research, we studied the cytotoxicity of Streptomyces sp. GMY01 extract on human lung cancer cell line A549 and its potential mechanism of action on anticancer drugs during apoptosis and autophagy. In the apoptosis mechanism, poly(ADP-ribose) polymerases (PARPs) are proteins (enzymes) that play a role in the catalytic process of transferring (adenosine diphosphate) ADP- ribose to the target protein. PARP is known to have many important roles in cellular mechanisms, including modulation of chromatin structure, transcription, replication, recombination, and (deoxyribonucleic acid) DNA repair. Meanwhile, in autophagy, the (light chain 3) LC3-II protein indicates the presence of a compound that acts as a trigger for autophagy mechanism. Autophagy plays a critical role in cellular development and differentiation, the response to stress conditions, and tumor suppression (Yang and Klionsky, 2009). To determine the anticancer activities of Streptomyces compounds, we also carried out molecular docking studies to active compounds detected in GMY01 extract. Molecular docking was carried out for the two important protein targets as antiapoptotic, B-cell lymphoma 2 (BCL-2) and B-cell lymphoma-extra large (BCL-XL) (Sathishkumar et al., 2012) and for the two autophagy proteins, namely, mammalian target of rapamycin (mTOR), which consist of two complexes: mTORC1 and mTORC2 (Yun and Lee, 2018). Thus, molecular docking of antiapoptotic and autophagy proteins is one of the methods for finding new anticancer drug candidates.
MATERIALS AND METHODS
Biological materials
Marine actinobacteria Streptomyces sp. GMY01 were isolated from a marine sediment sample collected from Krakal Beach (8°8′'44″"S 110°35′'59″"E), Gunungkidul, Yogyakarta, Indonesia (Farida et al., 2007). GMY01 was deposited in the Indonesian Culture Collection (WDCM 769), Indonesian Institute of Sciences as InaCC A147, and NITE Biological Research Center (NBRC, WDCM 825), Japan, with registration number NBRC 110111. The whole-genome shotgun project of Streptomyces sp. GMY01 has been deposited at DNA Data Bank of Japan/European Nucleotide Archive/GenBank with GenBank accession number: JABBNA000000000. Human lung cancer cell line A549 was obtained from the Department of Cancer Cell Biology, Faculty of Pharmaceutical Sciences, University of Toyama, Japan.
Fermentation, extraction, and fractionation
GMY01 bacterium was maintained in International Streptomyces Project-2 (ISP-2) agar medium (Difco, Sparks, MD). GMY01 was cultured at 28°C and agitated at 180 rpm for 3 days in a 250 ml Erlenmeyer flask containing 100 ml of tryptic soy broth (Difco, Sparks, MD) as the seed medium. For fermentation, the cell culture was transferred into four 1,000 ml flasks containing 500 ml of starch nitrate broth (SNB) medium, incubated for 11 days at 28°C, and agitated at 180 rpm in a shaking incubator (Ghanem et al., 2000). The SNB medium contained 0.5 g of NaCl, 1 g of KNO3, 0.5 g of K2HPO4, 0.5 g of MgSO4·7H2O, 0.01 g of FeSO4·7H2O, and 20 g of soluble starch in 1,000 ml of distilled water. Secondary metabolites were obtained by separating the cell biomass from the liquid using refrigerated centrifuge at 4,137 × g at 4°C for 15 minutes (Farida et al., 2007). The supernatant was extracted twice with an equal volume of ethyl acetate (Merck, Germany) and evaporated using an evaporator machine (Buchi, Switzerland) to obtain the crude extract (CE). The crude ethyl acetate extract was dissolved in methanol (Merck, Germany) and fractionated using an equal volume of n-hexane (Merck, Germany) to separate the polar and nonpolar fractions. The polar fraction was refractionated using flash chromatography (Reveleris™, Buchi, Switzerland) using a 30 μm C-18 column (FlashPure, Buchi, Switzerland) with water-acetonitrile (Merck, Germany) as the mobile phase to obtain fractions. The ethyl acetate–methanol fraction was dissolved in methanol impregnated into celite (Merck, Germany), with a fraction:celite ratio of 1:3 (w/w), and dried. Flash chromatography was carried out on the ethyl acetate–methanol fraction based on the procedure manual for dry loading samples. All the targeted fractions were weighed and evaluated for anticancer assays.
Cytotoxicity assay
The water-soluble tetrazolium salt (WST) assay was employed to evaluate anticancer activity in vitro (Tominaga et al., 1999) with minor modifications. Selected lung cancer cell line A549 was grown in Roswell Park Memorial Institute 1640 (RPMI 1640) medium. Cells were seeded into a sterile flat bottom 96-well microplate at a density of 1.3 × 104 cells/well and allowed to adhere overnight at a total volume of 100 μl in a humidified incubator (5% CO2, 37°C). One hundred microliters of the sample solution (in 0.1% dimethyl sulfoxide, DMSO) was added to the cells ranging from 0, 12.5, 25, and 50 to 100 μg/ml and then incubated for 24 hours before carrying out the WST-1 assay. Negative control was made using 0.1% DMSO (1 μl DMSO in 1,000 ml RMPI), while positive control was made using a cisplatin standard of 1.8 μl in 1.8 ml (1 mM). All treatments were carried out in triplicate. After aspiration of the liquid medium from the plate, 99 μl of WST-1 solution (90 RPMI 1,640: 1 WST-1) was then added to each well and incubated at 37°C in a CO2 incubator for 4 × 10 minutes. The reaction product was observed spectrophotometrically at 420 and 660 nm using a microplate reader. The collected data were calculated to obtain the ratio of the control (RPMI media) and the percentage of cell growth inhibition according to the following formula:
where A is the mean absorbance of the control (RPMI medium without drug) and B is the mean absorbance of the treated cells.
IC50 values were obtained using nonlinear regression analysis between the log10 concentrations of the extract versus the percent of cell inhibition using GraphPad Prism 8.4.3 software. All the experiments were repeated twice independently.
Protein extraction and Western blot analysis
The protein extraction procedures and Western blot analysis were carried out to evaluate both apoptosis and autophagy mechanisms (Kamal et al., 2015; Williams, 2013). A549 cells were treated with 1 μl of the 100 μg/ml fraction for 6 hours. The cells were separated from the medium and washed with phosphate buffered saline (PBS). The cell lysates were obtained by lysing the cells in 100 μl ice-cold (whole cell extract) WCE option (reductant/inhibitor) consisting of WCE buffer 1.7 ml and WCE + option: 1 M dithiothreitol (DTT) (×1,000) 1.7 μl, sodium orthovanadate (1 M × 1,000) 1.7 μl, Β-glycerophosphate (1M × 50) 34 μl, phenylmethylsulfonyl fluoride (0.1M × 100) 17 μl, leupeptin (10 mg/ml × 1,000) 1.7 μl, and aprotinin (10 mg/ml × 1,000) 1.7 μl. After centrifugation at 14,000 rpm, 4°C for 10 minutes, the protein in the supernatant was quantified by the XL Bradford method by using a microplate reader at 595 nm. The concentration of the protein sample was set to the same concentration by the addition of WCE buffer. The sample solution (150 μl) was added, homogenized, and heat shocked at 95°C for 5 minutes. The protein (10 μg per lane) was applied to 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis, at 500 volts and 120 mA for 75 minutes. After electrophoresis, the protein was transferred to a polyvinylidene difluoride membrane at 400 V and 100 mA for 100 minutes. The membrane was blocked overnight in PBS with 0.1% Tween 20 (phosphate buffer saline – Tween 20) (PBST) and washed with PBST three times. The primary antibody was added to the membrane and incubated on a hot plate for 3 hours for PARP (Cell Signaling Technology) and 2 hours for α-tubulin (Santa Cruz Biotechnology). The membrane was washed again in PBST for 15 minutes three times. The membrane was incubated for 1 additional hour with the corresponding horseradish peroxidase-labeled secondary antibody. The membrane was washed with PBST 0.1%. The protein blots were visualized with enhanced chemiluminescence (ECL) solution 1 and 2 (1:1 ratio) and incubated for 10–20 minutes. The membrane was exposed to a film negative for 10–20 minutes and washed with solution 1 and NaCl 0.3%. Finally, the membrane was fixed with fixative solution and then washed with aqueous solution. Images were captured by using a scanner.
Liquid chromatography high-resolution mass spectrometry (LC-HRMS)
Metabolomic analysis of the CE was carried out using a ultra-high-performance liquid chromatography coupled to targeted high-performance mass spectrometry (Thermo Scientific Dionex Ultimate 3000 RSLC Nano UHPLC paired with Thermo Scientific Q Exactive) (Thermo Fisher Scientific, Bremen, Germany). Targeted LC-HRMS was based on a predicted compound formula which was obtained from the genome mining analysis of Streptomyces sp. GMY01 whole-genome sequence using antiSMASH version 5 (Blin et al., 2019). HRMS was carried out with mobile phases A (water + 0.1% formic acid) and B (acetonitrile + 0.1% formic acid). The column used was a Hypersil Gold aQ analytical column (50 × 1 mm × 1.9 μm) (Thermo Fisher Scientific, Waltham, MA) with a flow rate of 40 μl/minutes, an injection volume of 5 μl, and a gradient with an analysis time of 30 minutes. The gradient was programmed as follows: 2 minutes, 5% B; 15 minutes, 60% B; 22 minutes, 95% B; 25 minutes, 95% B; 25.1 minutes, 5% B; and 30 minutes, 5% B. Experiments were carried out in parallel reaction monitoring at 35,000 the full width at half maximum resolution, heated electrospray ionization, positive ionization, and data processing with Thermo Scientific XCalibur.
In silico molecular docking studies
Molecular docking was used to predict the binding affinity of several compounds with proteins that play a role in the apoptosis mechanism. The active compounds of Streptomyces sp. GMY01 were detected using targeted LC-HRMS based on a genome mining study. This study used apoptosis proteins in cancer cells BCL-2 (2w3l) and BCL-XL (2yxj) and autophagy proteins in cancer cells mTORC1 (6BT0) and mTORC2 (6zwm) which were obtained from RCSB database. The 2D structure of the compounds was retrieved from the ChemSpider webserver and converted to an optimized 3D structure using Marvinsketch tools (https://www.chemaxon.com). Both proteins and compounds were prepared using AutoDock tools. The proteins were purified from water, ions, and other small molecules. Hydrogen atoms were added to the polar groups of the proteins to minimize errors caused by ionization and tautomeric states of amino acid residues. Molecular docking was carried out on AutoDock Vina (Trott Oleg and Olson, 2010), while visualization of binding interactions was carried out using the DisCoVery studio visualizer (DS visualizer) tool (Studio, 2015). The computational simulation was carried out on a Windows 10 operating system, with an AMD A8 7410 (Quad-core; 2,2 GHz) processor and 4 GB of RAM. The molecular docking study was observed from the affinity (kcal/mol) value with a root mean square deviation score less than 2 A, which expressed a visualization of the binding interaction between the compounds and the active site of the proteins.
RESULTS AND DISCUSSION
Anticancer activity of Streptomyces sp. GMY01
The fermentation process of Streptomyces sp. GMY01 in 6 l of SNB medium for 11 days obtained a 4.78 g of crude ethyl acetate extract. The results of the refractionation using n-hexane, methanol, and ethyl acetate solvents selected 177.8 mg of methanol-ethyl acetate as an active extract. The refractionation using flash chromatography obtained 17 fractions, five of which were active as anticancer agents. The cytotoxicity assay of Streptomyces sp. GMY01 extract on the lung cancer cell line A549 showed that F5, F6, F7, F8, and F14 fractions and CE had cytotoxic activities at concentrations of 12.5–100 μg/ml (Table 1). At concentrations of 50 and 100 μg/ml, F5 showed the highest inhibitory activity, which was lower than that of 1 mM cisplatin as a positive control. By regression analysis, the CE and two fractions (F5 and F6) showed IC50 value lower than 100 μg/ml. The lowest IC50 value (18.41 μg/ml) was revealed by F5. From these results, the obtained fractions from Streptomyces sp. GMY01 extract had potency as anticancer agents, especially on human lung cancer cells.
In a previous bioassay study, GMY01 exhibited anticancer activity against human breast cancer T47D cells (IC50 value of 19 μg/ml) and MCF7 cells (IC50 value of 7 μg/ml) (Farida et al., 2007). In a previous genome mining study, a whole-genome sequence of Streptomyces sp. GMY01 had 10 non-ribosomal polyketide synthase (NRPS) gene clusters (Herdini et al., 2015). This indicates that Streptomyces sp. GMY01 is a marine organism with biotechnological potential as a drug source.
Molecular mechanism of anticancer
Western blot analysis was carried out to evaluate the fractional effect on protein expression in a human lung cancer cell line (A549). An α-tubulin was used as the control protein in this study. Three selected fractions (F5, F6, and F7) were used for the protein expression assay using Western blot analysis. The results showed that none of the samples induced cleavage of the PARP protein (Fig. 1). On the other hand, all the samples exhibited slight inhibition against LC3-I and an increase in the LC3-II protein level, similar to cisplatin as a positive control. Fractions 6 and 7 showed the highest level of LC3-II protein expression, followed by fraction 5. However, we investigated the apoptosis and autophagy mechanisms in A549 cancer cells.
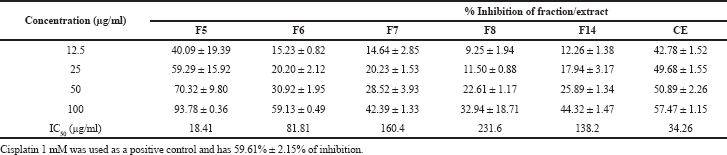 | Table 1. Inhibitory activity and IC50 value of Streptomyces sp. GMY01 fractions against lung cancer cell A549. [Click here to view] |
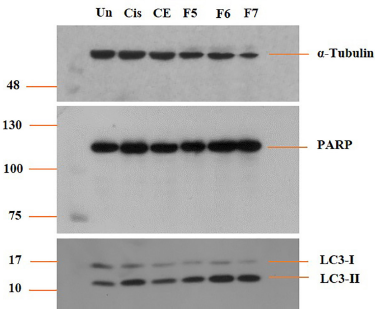 | Figure 1. Effect of Streptomyces sp. GMY01 fraction extract on LC3-I and LC3-II and PARP protein expression. A549 cells were treated with the fraction extract at 1 μl (100 μg/ml) concentration in ml media for 6 hours. LC3-I, LC3-II, and PARP protein levels were examined by Western blot analysis. α-Tubulin was used as an equal loading control. [Click here to view] |
PARP inhibitors have been widely investigated in clinical trials to determine the benefits of this approach in cancer therapy. PARP is often associated with programmed cell death mechanisms characterized by decreased nicotinamide adenine dinucleotide (NAD) ± adenosine triphosphate (ATP), loss of mitochondrial membrane potential, and the presence of apoptosis-inducing factors (Morales et al., 2014). Figure 1 shows that all the cells treated with the fractions, CE, or cisplatin as the positive controls did not display significant differences in the PARP expression level compared with the untreated cells. It can be said that the mechanism of action of the compounds produced by Streptomyces sp. GMY01 is not related to the apoptosis mechanism. In this study, the apoptosis mechanism cannot be seen clearly because there were no smaller PARP proteins as a product of PARP cleavage. Another study mentioned that 4b cinnamido-linked podophyllotoxins were confirmed as anticancer agents in lung A549, with the apoptosis mechanism indicated by cleavage of 116 kDa PARP to cleaved PARP 89 kDa (Kamal et al., 2015).
An anticancer mechanism apart from apoptosis is autophagy, which is a normal process in cells to maintain homeostasis in times of limited nutritional conditions. In autophagy, cells break down unnecessary cell components into metabolites which are useful for the availability of cell nutrients. Microtubule-associated protein 1 light chain 3 (MAP1 LC3) is a very common indicator as a process of autophagy in diseased cells. One of the important markers in the autophagy process is the upregulation and localization of LC3 protein. The carboxyl terminal region of the LC3 is cleaved, showing glycine residue forming LC3I. LC3II is a modified form of LC3I through the addition of phosphatidylethanolamine. The LC3II becomes bound and accumulated in the membrane of the autophagosome (Williams, 2013). In this study, cell treatment was carried out for 6 hours of incubation. The autophagy mechanism was clearly seen in the cells treated with F6 and F7 and slightly seen in F5. Thus, the apoptosis mechanism might not yet be clearly seen because the 6-hour incubation was not sufficient to trigger apoptosis. Autophagy avoids apoptotic mechanism; however, under long-term stress conditions, it can lead to apoptosis (Williams, 2013).
In silico molecular docking
To predict the compounds produced by GMY01 bacteria, we carried out targeted LC-HRMS analysis based on a genome mining study of Streptomyces sp. GMY01 (Supplementary Table S1). Targeted HRMS detected eight compounds in CE (Supplementary Table S2). These compounds are known as antitumor and herbicidal (herboxidiene) (Miller-Wideman et al., 1992; Pokhrel et al., 2015), antibiotic (albaflavenone and grincamycin) (Lai et al., 2018; Zhao et al., 2008), antitumor antibiotic (saframycin A) (Li et al., 2008), aromatic carotenoid (isorenieratene) (Krügel et al., 1999), peptide metabolite (vazabitide A) (Yashiro et al., 2019), and volatile compounds (geosmin and hopene) (Jiang et al., 2006; Seipke and Loria, 2009).
The molecular docking affinity of these detected compounds on the target protein of antiapoptotic proteins BCL-2 and BCL-XL and autophagy proteins mTORC1 and mTORC2 is shown in Tables 2 and 3. Three compounds, namely, grincamycin, isorenieratene, and hopene, showed a higher affinity than other compounds. Among all these compounds, grincamycin (C49H62O18) had the highest affinity with BCL-2 and BCL-XL (−11.8 and −10.4) and with mTORC1 and mTORC2 (−12.1 and −11.5). A three-dimensional illustration of the binding between grincamycin and the target proteins is shown in Figure 2 and Figure 3.
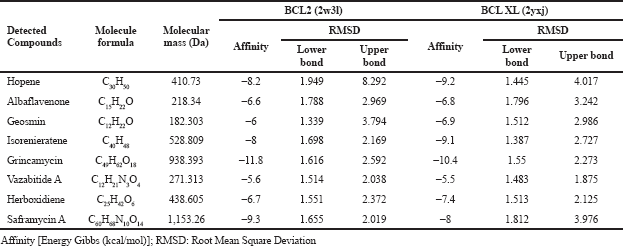 | Table 2. Docking scores of detected compounds in Streptomyces sp. GMY01 extract toward potential binding domain of apoptosis protein. [Click here to view] |
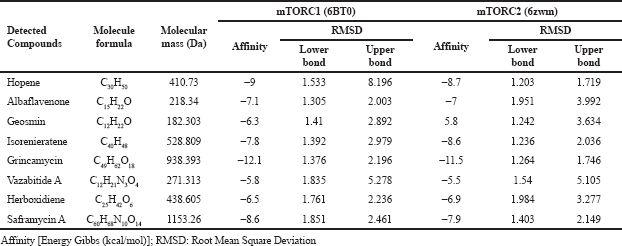 | Table 3. Docking scores of detected compounds in Streptomyces sp. GMY01 extract toward potential binding domain of autophagy protein. [Click here to view] |
BCL-2 and BCL-XL are known as antiapoptotic proteins that bind with proapoptotic proteins to induce apoptosis mechanism. This protein family plays a role as key regulators of programmed cell death or apoptosis (Goreshnik et al., 2011). Several cancers are affected by overexpression of these antiapoptotic proteins (Sathishkumar et al., 2012). High expression of antiapoptotic agents such as BCL-2 and BCL-XL which are commonly found in human cancer causes neoplastic cell expansion and interferes with the therapeutic action of chemotherapy drugs (Huang, 2000). In another study, molecular docking analysis between ginsenosides from Panax ginseng against BCL-2 and BCL-XL resulted in binding energy of −5.62 to −9.77 kcal/mol (Sathishkumar et al., 2012). The result of that study was lower than the binding affinity affinity produced by grincamycin in this study (−10.4 to −11.8 kcal/mol).
mTOR is a protein associated with cell proliferation, stress, and cancer progression. This protein controlled the autophagy mechanism. mTOR protein consists of mTORC1 and mTORC2 as a complex. Activated mTORC1 leads to the inhibition of autophagy (Yun and Lee, 2018). Inhibition of mTORC1 protein is associated with enhanced autophagy. Another study showed that silencing mTORC2-induced apoptosis (Chawsheen and Dash, 2021). The mTORC1 and mTORC2 inhibitor compounds are used as models in the design and development of lung cancer treatments (Chaube et al., 2021). Two compounds that have been shown to be mTORC inhibitors include tetrahydroquinoline derivatives (Chaube et al., 2021) in lung cancer and MG-2477 in A549 cell (Viola et al., 2012).
In this study, grincamycin compounds had a higher affinity for autophagy proteins than for apoptosis-related proteins. This is in line with the results of the analysis of protein expression using Western blot where the LC3-I and LC3-II apoptosis-related proteins were higher in expression than the PARP-related apoptosis-related proteins. This was explained in a previous study which states that when mTORC1 is inhibited, the protein complex associated with autophagy activation recruits LC3 protein which will be converted into active cytosolic isoform LC3-I and transformed into LC3-II. LC3 II protein is located in the autophagosome membrane and allows the binding of degraded substrates. Autophagosomes will fuse with lysosomes and form autolysosomes which play a role in the autophagy process (Yun and Lee, 2018).
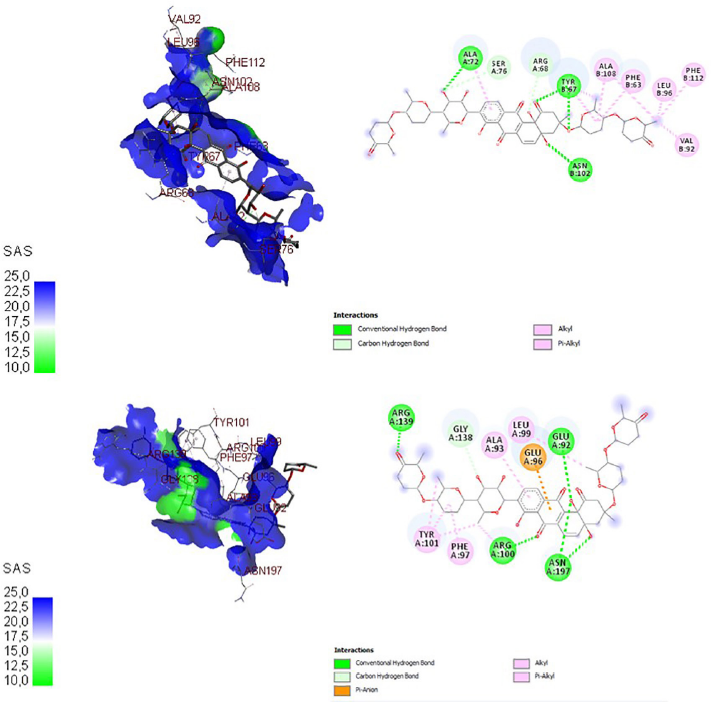 | Figure 2. Three-dimensional and two-dimensional docking visualization of grincamycin on BCL-2 (top) and BCL-XL (bottom) proteins. [Click here to view] |
The docking visualization of grincamycin on antiapoptosis proteins BCL-2 and BCL-XL is shown in Figure 2 and on autophagy proteins mTORC1 and mTORC2 is shown in Figure 3. In three- and two-dimensional docking visualization, grincamycin binds with alanine (72), serine (76), arginine (68), tyrosine (67), asparagine (102), alanine (108), phenylalanine (63), leucine (96), phenylalanine (112), valine (92) amino acids of BCL-2 protein and binds with arginine (139), glycine (138), alanine (93), glutamic acid (96), leucine (99), glutamic acid (92), tyrosine (101), phenylalanine (97), arginine (100), and asparagine (197) amino acids of BCL-XL. This compound also binds isoleucine (90), lysine (9,151), leucine (170), histidine (9,100), leucine (9,104), lysine (169), methionine (125), glutamic acid (126), threonine (88), serine (89), lysine (91) of mTORC1, arginine (1,665), isoleucine (1,629), histidine (1,454), tryptophan (1,456), proline (1,426), arginine (2,117), arginine (2,322), glycine (1,425), lysine (2,218), tyrosine (1,393), phenylalanine (2,314), and leucine (1,423) of mTORC2. This finding showed that grincamycin had high affinity antiapoptotic proteins and autophagy proteins. The presence of grincamycin binds with BCL-2, BCL-XL, mTORC1, and mTORC2 areas expected to inhibit these proteins, leading to apoptosis and autophagy induction.
In previous studies, the grincamycin compounds from an actinomycete from the sea, namely Streptomyces lusitanus SCSIO LR32, were found. Grincamycin compounds show antitumor activity against several human cancer cells, namely MDA-MB-435, MDA-MB-231, NCI-H460, HCT-116, HepG2, and MCF10A normal human breast epithelial cells with IC50 values ranging from 0.4 to 6.9 μM (Lai et al., 2018). In another study, grincamycins B–F also were isolated from S. lusitanus SCSIO LR32 and havehad cytotoxicity on human cancer cell lines: liver carcinoma HepG2, pancreatic carcinoma SW-1990, cervixal carcinoma HeLa, and breast carcinoma NCI-H460 and MCF-7, and also the mouse melanoma cell line B16, with IC50 being in the range of 1.1–31 μM (Huang et al., 2012). Previous findings also showed that Streptomyces dengpaensis XZHG99 produces a high level of compounds: grincamycins L, M, and N (Patent CN108949610A) available online at https://patents.google.com/patent/CN108949610A/en?q=grincamycin&oq=grincamycin (accessed on 16 January 2021). In another medical field, it is known that the application of grincamycin B compounds functions as an antiglioma drug. This compound has a selective activity on glioblastoma cells. In the cell cycle, grincamycin is able to induce apoptosis in tumor cells and tumor stem cells, block cells at stages G0–G1, and inhibit glioma stem cell regeneration. Thus, grincamycin has been claimed to be a potential anticancer drug with a new mechanism of action for targeted cancer stem cell oncotherapy (Patent CN103550231B) available online at https://patents.google.com/patent/CN103550231B/en?q=grincamycin&oq=grincamycin (accessed on 16 January 2021). However, in these studies, grincamycin has never been tested as an anticancer compound in lung cancer cell lines, especially A549. Thus, this finding becomes important for the possible use of grincamycin as a candidate for lung cancer anticancer drugs in humans.
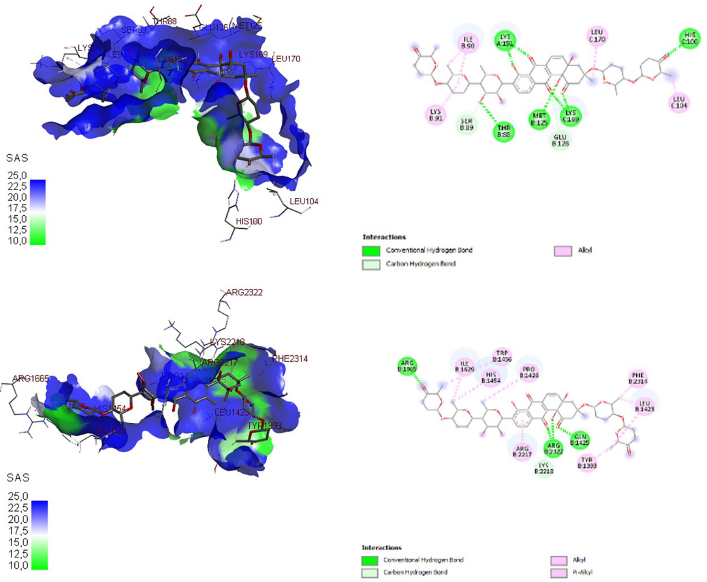 | Figure 3. Three-dimensional and two-dimensional docking visualization of gricamycin on mTORC1 (top) and mTORC2 (bottom) proteins. [Click here to view] |
CONCLUSION
Marine-derived Streptomyces sp. GMY01 exhibited potential anticancer activity against lung cancer cell line A549-induced autophagy mechanism. In silico molecular docking also showed that detected compounds on GMY01 extract have an affinity with autophagy proteins mTORC1 and mTORC2 higher than with antiapoptotic protein BCL-2 and BCL-XL of cancer cells. This finding revealed that detected compound grincamycin from Streptomyces sp. GMY01 has the highest affinity on autophagy and antiapoptotic protein and has a promising anticancer agent against human lung cancer.
SUPPLEMENTARY MATERIALS
The following are available online: Table S1: Detected compounds on targeted LC-HRMS of the ethyl acetate extract produced by Streptomyces GMY01. Table S2: Genome mining analysis of the Streptomyces sp. GMY01 genome using antiSMASH 5.0.
AUTHORS’ CONTRIBUTIONS
Concept and design: ED, KN, M, and JW; data acquisition: ED, KN, SH, and RF; data analysis/interpretation: ED, KN, SH, RTD, and RF; drafting manuscript: ED and KN; critical revision of the manuscript: KN, SH, M, AD and JW; statistical analysis: ED; funding: JW and KN; admin, technical, and material support: ED, KN, RTD, and JW; supervision: KN, M, AD, and JW; final approval: KN, M, AD, and JW.
ACKNOWLEDGMENTS
The authors thank Professor Hiroaki Sakurai and Dr. Yue Zhue at the Cancer Cell Biology Laboratory, Postgraduate School of Medicine and Pharmacy, Toyama University, Japan, for technical assistance on the anticancer assay.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
FUNDING
This research was funded by the Indonesian Ministry of Research, Technology and Higher Education, grant number 2784/UN1.P.III/DIT-LIT/LT/2019 awarded to JK. A part of this work was supported by Tailor Made Courses 2019 awarded to KN, SH, and ED.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Bade BC, Dela Cruz CS. Lung cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med, 2020; 41(1):1–24; doi:10.1016/j.ccm.2019.10.001 CrossRef
Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health, 2019; 85(1):8; doi:10.5334/aogh.2419 CrossRef
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. AntiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res, 2019; 47(W1):W81–7; doi:10.1093/nar/gkz310 CrossRef
Chaube UJ, Rawal R, Jha AB, Variya B, Bhatt HG. Design and development of Tetrahydro-Quinoline derivatives as dual mTOR-C1/C2 inhibitors for the treatment of lung cancer. Bioorg Chem, 2021; 106:104501; doi:10.1016/j.bioorg.2020.104501 CrossRef
Chawsheen MA, Dash PR. mTOR modulates resistance to gemcitabine in lung cancer in an MTORC2 dependent mechanism. Cell Signal, 2021; 81:109934; doi:10.1016/j.cellsig.2021.109934 CrossRef
Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med, 2011; 32(4):605–44; doi:10.1016/j.ccm.2011.09.001 CrossRef
Dhaneesha M, Benjamin Naman C, Krishnan KP, Sinha RK, Jayesh P, Joseph V, Bright Singh IS, Gerwick WH, Sajeevan TP. Streptomyces artemisiae MCCB 248 isolated from Arctic fjord sediments has unique PKS and NRPS biosynthetic genes and produces potential new anticancer natural products. 3 Biotech, 2017; 7(1):32; doi:10.1007/s13205-017-0610-3 CrossRef
Farida Y, Widada J, Meiyanto E. Combination methods for screening marine actinomycetes producing potential compounds as anticancer. Indones J Biotechnol, 2007; 12:988–97; doi:10.22146/ijbiotech.7772
Ghanem NB, Sabry SA, El-Sherif ZM, Abu El-Ela GA. Isolation and enumeration of marine actinomycetes from seawater and sediments in Alexandria. J Gen Appl Microbiol, 2000; 46:105–11; https://doi.org/10.2323/jgam.46.105 CrossRef
Goreshnik I, Brock AM, Maly DJ. Biochemical and pharmacological profiling of the pro-survival protein Bcl-xL. Bioorg Med Chem Lett, 2011; 21(17):4951–5; doi:10.1016/j.bmcl.2011.06.134 CrossRef
Herdini C, Hartanto S, Mubarika S, Hariwiyanto B, Wijayanti N, Hosoyama A, Yamazoe A, Nojiri H, Widada J. Diversity of nonribosomal peptide synthetase genes in the anticancer producing actinomycetes isolated from marine aediment in Indonesia. Indones J Biotechnol, 2015; 20:34; doi:10.22146/ijbiotech.15266 CrossRef
Huang H, Yang T, Ren X, Liu J, Song Y, Sun A, Ma J, Wang B, Zhang Y, Huang C, Zhang C, Ju J. Cytotoxic angucycline class glycosides from the deep sea actinomycete Streptomyces lusitanus SCSIO LR32. J Nat Prod, 2012; 75(2):202–8; https://doi.org/10.1021/np2008335 CrossRef
Huang Z. Bcl-2 family proteins as targets for anticancer drug design. Oncogene, 2000; 19(56):6627–31; doi:10.1038/sj.onc.1204087 CrossRef
Jiang J, He X, Cane DE. Geosmin biosynthesis. Streptomyces coelicolor germacradienol/germacrene D synthase converts farnesyl diphosphate to geosmin. J Am Chem Soc, 2006; 128:8128–9; doi:10.1021/ja062669x CrossRef
Kamal A, Nayak VL, Bagul C, Vishnuvardhan MVPS, Mallareddy A. Investigation of the mechanism and apoptotic pathway induced by 4b cinnamido linked podophyllotoxins against human lung cancer cells A549. Apoptosis, 2015; 20(11):1518–29; doi:10.1007/s10495-015-1173-6 CrossRef
Kinghorn AD, Chin YW, Swanson SM. Discovery of natural product anticancer agents from biodiverse organisms. Curr Opin Drug Discov Devel, 2009; 12(2):189–96.
Krügel H, Krubasik P, Weber K, Saluz HP, Sandmann G. Functional analysis of genes from Streptomyces griseus involved in the synthesis of isorenieratene, a carotenoid with aromatic end groups, revealed a novel type of carotenoid desaturase. Biochim Biophys Acta, 1999; 1439(1):57–64; doi:10.1016/S1388-1981(99)00075-X CrossRef
Lai Z, Yu J, Ling H, Song Y, Yuan J, Ju J, Tao Y, Huang H. Grincamycins I-K, cytotoxic angucycline glycosides derived from marine-derived actinomycete Streptomyces lusitanus SCSIO LR32. Planta Med, 2018; 84(3):201–7; https://doi.org/10.1055/s-0043-119888
Li L, Deng W, Song J, Ding W, Zhao QF, Peng C, Song WW, Tang GL, Liu W. Characterization of the saframycin a gene cluster from Streptomyces lavendulae NRRL 11002 revealing a nonribosomal peptide synthetase system for assembling the unusual tetrapeptidyl skeleton in an iterative manner. J Bacteriol, 2008; 190(1):251–63 https://doi.org/10.1128/JB.00826-07 CrossRef
Miller-Wideman M, Makkar N, Tran M, Isaac B, Biest N, Stonard R. Herboxidiene, a new herbicidal substance from Streptomyces chromofuscus a7847 taxonomy, fermentation, isolation, physico-chemical and biological properties. J Antibiot (Tokyo), 1992; 45(6):914–21. https://doi.org/10.7164/antibiotics.45.914
Morales JC, Li L, Fattah FJ, Dong Y, Bey EA, Patel M, Gao J, Boothman DA. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr, 2014; 24(1):15–28; https://doi.org/10.1615/CritRevEukaryotGeneExpr.2013006875
Nguyen HT, Pokhrel AR, Nguyen CT, Pham VTT, Dhakal D, Lim HN, Jung HJ, Kim TS, Yamaguchi T, Sohng JK. Streptomyces sp. VN1, a producer of diverse metabolites including non-natural furan-type anticancer compound. Sci Rep, 2020; 10(1756):1–14; doi:10.1038/s41598-020- 58623-1. CrossRef
Pokhrel AR, Dhakal D, Jha AK, Sohng JK. Herboxidiene biosynthesis, production, and structural modifications: prospect for hybrids with related polyketide. Appl Microbiol Biotechnol, 2015; 99(20):8351–62; doi:10.1007/s00253-015-6860-2 CrossRef
Sathishkumar N, Sathiyamoorthy S, Ramya M, Yang DU, Lee HN, Yang DC. Molecular docking studies of anti-apoptotic BCL-2, BCL-XL, and MCL-1 proteins with ginsenosides from Panax ginseng. J Enzyme Inhib Med Chem, 2012; 27(5):685–92; doi:10.3109/14756366.2011.608663 CrossRef
Seipke RF, Loria R. Hopanoids are not essential for growth of Streptomyces scabies 87-22. J Bacteriol, 2009; 191(16):5216–23; doi:10.1128/JB.00390-09 CrossRef
Studio D. Dassault systemes BIOVIA, discovery studio modelling environment, release 4.5. Accelrys Softw. Inc, San Diego, CA, 2015.
The Global Cancer Observatory. Source: GLOBOCAN 2018. World Health Organization, Geneva, Switzerland, 2019.
Tominaga H, Ishiyama M, Ohseto F, Sasamoto K, Hamamoto T, Suzuki K, Watanabe M. A water-soluble tetrazolium salt useful for colorimetric cell viability assay. Anal Commun, 1999; 36:47–50; doi:10.1039/a809656b CrossRef
Trott O, Olson AJ. AutoDock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem, 2010; 31(2):455–61; doi.org/10.1002/jcc CrossRef
Viola G, Bortolozzi R, Hamel E, Moro S, Brun P, Castagliuolo I, Ferlin MG, Basso G. MG-2477, a new tubulin inhibitor, induces autophagy through inhibition of the Akt/mTOR pathway and delayed apoptosis in A549 cells. Biochem Pharmacol, 2012; 83:16–26; doi:10.1016/j.bcp.2011.09.017 CrossRef
Weber T, Charusanti P, Musiol-Kroll EM, Jiang X, Tong Y, Kim HU, Lee SY. Metabolic engineering of antibiotic factories: New tools for antibiotic production in actinomycetes. Trends Biotechnol, 2015; 33:15–26; doi:10.1016/j.tibtech.2014.10.009 CrossRef
Werdyani S, Wijayanti N, Fitria A, Rahmawati S. Cytotoxic effects of ethyl acetate fractions from secondary metabolites of Streptomyces sp. GMY01 on human breast cancer MCF7 cell lines. Asian J Pharm Clin Res, 2017; 10:9–11; doi:10.22159/ajpcr.2017v10s3.21351 CrossRef
WHO. Cancer Country profile 2020 Mexico. International Agency for Research on Cancer, Lyon, France, 2020.
Williams CJ. LC3I and LC3II as autophagy markers for the development and improvement of products and techniques used in research. ProQuest Diss Theses, Of Wayne State University, Detroit, MI, 2013.
Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol, 2009; 335:1–32; doi:10.1007/978-3-642-00302-8-1 CrossRef
Yashiro T, Sakata F, Sekimoto T, Shirai T, Hasebe F, Matsuda K, Kurosawa S, Suzuki S, Nagata K, Kasakura K, Nishiyama M, Nishiyama C. Immunosuppressive effect of a non-proteinogenic amino acid from Streptomyces through inhibiting allogeneic T cell proliferation. Biosci Biotechnol Biochem, 2019; 83(6):1111–6; doi:10.1080/09168451.2019.1591262 CrossRef
Yun CW, Lee SH. The roles of autophagy in cancer. Int J Mol Sci, 2018; 19(11):3466; doi:10.3390/ijms19113466 CrossRef
Zhao B, Lin X, Lei L, Lamb DC, Kelly SL, Waterman MR, Cane DE. Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor A3(2). J Biol Chem, 2008; 283(13):8183–9; doi:10.1074/jbc.M710421200 CrossRef
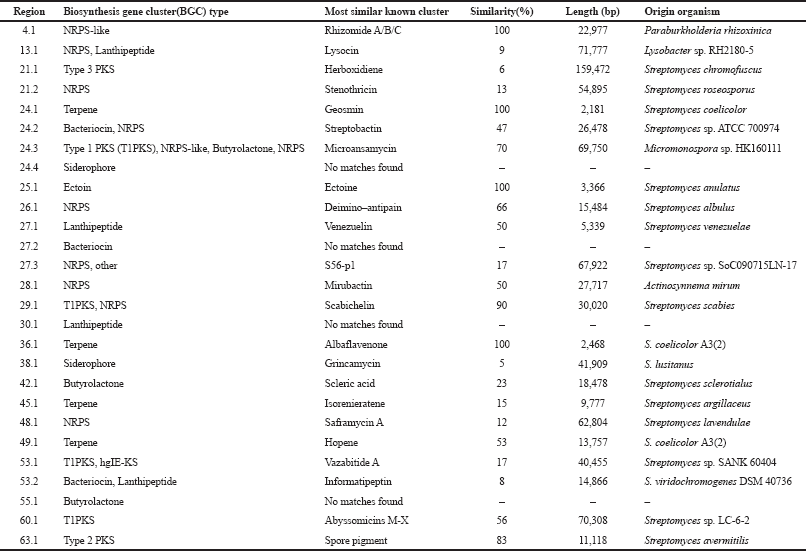 | Supplementary Table S1. Genome mining analysis of Streptomyces sp. GMY01 genome using antiSMASH 5.0. [Click here to view] |
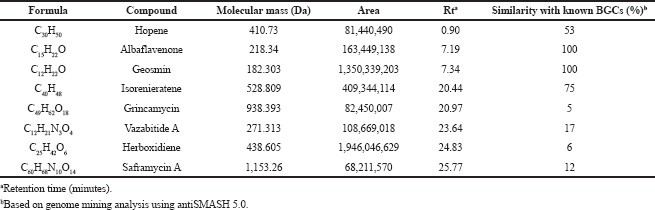 | Supplementary Table S2. Metabolite profile from targeted LC-HRMS of the ethyl acetate extract produced by Streptomyces GMY01. [Click here to view] |