INTRODUCTION
For a decade, computational chemistry has significantly developed. Medicinal chemistry is one of the subjects that were affected by this phenomenon, especially in discovering new drugs. Quantitative Structure-Activity Relationship (QSAR) is one of the alternative approaches to find new drugs supposedly more effective and efficient (Arba et al., 2019). The study of QSAR assumes that there is a quantitative relationship between the molecular structure with its biological activity of a molecule (El-Hamamsy, 2017; Modi et al., 2018; Ujihara et al., 1988). The molecular structure is not only describing atoms and molecules bonding but also its physical and chemical properties (Desai et al., 2013).
Anti-inflammatory drugs are one of the agents that most widely used to treat the inflammation as a local protective response caused by damage in tissue. Physical trauma, damaging chemicals, or microbiology substances cause tissue damage. The inflammation process has its functions to destroy, reduce, or localize destructive agents and damaged tissue. Symptoms of inflammation were signed by swelling/edema, redness, heat, pain, and changes in its functions (Agustina et al., 2015).
Noted that mono-carbonyl analogs of curcumin (MACs) (Fig. 1) have antibacterial activity (Wijianto et al., 2019), anticancer and chemo-preventive effects on certain cancers with low toxicity (Anand et al., 2008; Kurnia et al., 2019), antioxidant (Selvam et al., 2005; Yuliarti and Nugroho, 2013), and anti-inflammatory by inhibit cyclooxygenase (Hayun et al., 2019; Yuliarti and Nugroho, 2013). Curcumin had much biological activity, but its therapeutic usage of curcumin limited by low water solubility, chemical, and metabolic stability and relatively weak in vivo bioavailability (Anand et al., 2008). The study of curcumin has modified to find that the analogs had better physical and chemical properties, better biological activity as well. Mono-carbonyl analogs of curcumin with cyclic as central can inhibit the cyclooxygenase enzyme (Sardjiman et al., 1997). Therefore, it is necessary to perform QSAR study for MACs to determine its anti-inflammatory activity leading to found a new anti-inflammatory drug. The aim of this research is to design the new MACs, synthesize the molecules, and assess its activity in cyclooxygenase inhibition for anti-inflammatory activity.
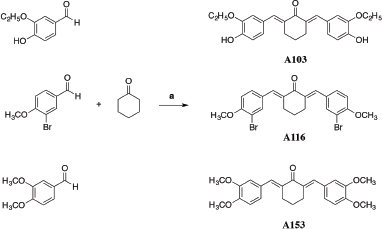 | Scheme 1. Reagent and condition of synthesis: a) THF and HCl; 6-8 h and 60°C (Wijianto et al., 2019). [Click here to view] |
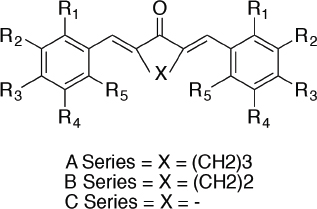 | Figure 1. Structure of mono-carbonyl analogs of curcumin (Wijianto et al., 2019). [Click here to view] |
MATERIALS AND METHODS
Data set
The data set of structure and anti-inflammatory activity used in this study derived from MACs is shown in Table I. Anti-inflammatory activity was determined as pIC50 (negative log of the IC50 in Molar) explaining the ability of each compound in cyclooxygenase inhibition.
Chemical
3,4-dimethoxybenzaldehyde, 3-ethoxy-4-hydroxybenz-aldehyde, 3-Bromo-4-methoxybenzaldehyde, cyclohexanone, tetrahydrofuran (THF), Hydrochloric acid, ethanol, ethyl acetate, sodium hydroxide, methanol, 2-hydroxyl acid), trichloroacetic acid, and arachidonic acid, all obtained from Sigma-Merck. Buffer Tris-HCl (pH 7.4) from Ultrol®. All material compounds used were analytical grade.
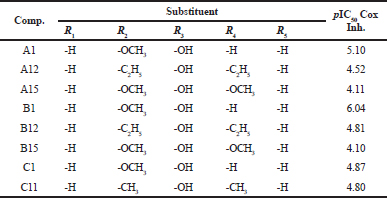 | Table 1. Data sets of compounds with anti-inflammatory activity [Click here to view] |
Instrument
Quantum mechanical calculations and descriptors were performed using the molecular operating environment (MOE) version 2018.0101 licensed. The QSAR model was calculated using the BuildQSAR open source program. BUCHI Melting Point B-540 with temperature gradient at 5°C/minute was used as a melting point test. The purity of compounds was measured using HPLC Elite La-Chrome®. JEOL® spectrophotometer of 500 MHz was used to measure 1H-NMR Spectrum.
Procedure
Molecular modeling
Molecular modeling studies were carried out using the MOE 2018.01.01 software. All the structures of MACs were depicted in 2D in the “builder” menu of MOE. Optimization of the structure of compounds was performed using MOPAC (semi-empirical quantum chemistry algorithms) in MOE. Semi-empirical of the AM-1 method with gradient 0.01 was used to determine the quantum chemical algorithm.
Calculation of descriptors
The optimized structures were calculated using single-point calculations to obtain the value of descriptors. All descriptors were provided by all molecular descriptors that available in 2D and i3D MOE databases. Molecular descriptors data were used, such as highest occupied unoccupied molecular orbitals energy, lowest unoccupied molecular orbitals energy, hydration energy, partition coefficient, polarizability, molecular volume and mass, total energy, the heat of formation, molar refractivity, molecular surface area, amphiphilic, hydrophobic volume, globularity and dipole moment.
Statistical analysis of cyclooxygenase inhibition activity of test compounds (expressed as pIC50) was performed using the multilinear regression (MLR) analysis and the GA-MLRA technique as applied in the BuildQSAR program. The best equation model obtained from the MLR analysis was used to predict the IC50 value. The best equation model was selected by considering the value of the statistical parameters. This approach allows to select the best models with several following characteristics, such as high regression coefficient (r), high Fisher coefficient (F), and low standard deviation (s) (Miladiyah et al., 2018; Sudarmanto and Oetari, 2007; Wijianto et al., 2019). The best equation model was validated using the leave-one-out cross-validation to determine its ability and robustness in predicting, which is confirmed by the Q2 cross-validation coefficient and quadratic prediction errors (sPRESS).
Docking studies
In silico study was conducted to known anti-inflammatory activity through inhibition of cyclooxygenase-1 (COX-1) and cyclooxygenase-2 (COX-2) using test proteins with ID.1EQH and 3PGH that available in the protein data bank. Protein validation and docking process were performed using the alpha PMI method on placement and induced fit on refinement scoring by GBVI/WSA dG as protocols. Protein validated was selected based on the RMSD (Root Mean Square Deviation) value.
Synthesis of 2, 6-bis-(3'-ethoxy, 4’-hydroxybenzylidene)-cyclohexanone
About 6.018 mmol of 3-ethoxy-4-hydroxybenzaldehyde, 3.009 mmol of cyclohexanone, 2 ml of THF, and 0.2 ml of concentrated hydrochloric acid were mixed and stirred at room temperature for 2 hours. Then, the temperature was raised to 50oC–60oC, and stirring was continued for 8 hours until the reaction was completed. Crude products were washed out by adding ethanol and cold water with a ratio of 1:1. The mixture filtered then re-washed out by ethanol and cold water with ratio 3: 2 until pH 7–8 reached out. Powdery residue in Buchner dried in the oven. Recrystallization performed by dissolving the residue in ethanol, and then water was added until a precipitate was formed. Thin-layer chromatography (TLC) and melting point tests were conducted to determine the purity of the product qualitatively.
Synthesis of 2, 6-bis-(3'-Bromo, 4'-methoxybenzylidene)-cyclohexanone
About 4.650 mmol of 3-Bromo-4-methoxybenzaldehyde, 2.325 mmol of cyclohexanone, 2.0 mL of THF, and 0.25 ml of concentrated Hydrochloric acid were mixed and stirred up to 2 hours at room temperature. Then, the temperature was raised to 50–60oC, and stirring was continued for 8 hours until the reaction was completed. The crude product was washed out with ethanol and cold water (1:1) then filtered by Buchner. The second washed out with ethanol and cold water (3:2) up to pH 7–8 reached out, then filtered and dried in the oven. Recrystallization conducted by dissolving the residue in acetone, and then cold water was added until a precipitate was formed. TLC and melting point examination were performed to determine the purity of the product qualitatively.
Synthesis of 2, 6-bis-(3', 4'-dimethoxybenzyllidene)-cyclohexanone
About 6.018 mmol of 3, 4-dimethoxybenzaldehyde, 3.009 mmol of cyclohexanone, 2.0 ml of THF, and 0.25 ml of concentrated hydrochloric acid were mixed and stirred at room temperature for 2 hours. Then, the temperature was raised to 50oC–60oC, and stirring was continued for 8 hours until the reaction was completed. The crude product was washed out by ethanol: cold water (1:1) then filtered by Buchner. Second, washed out for residue was performed by different ratios of ethanol and cold water (3:2) until pH 7–8 reached out. The residue was filtered and dried in the oven. Recrystallization performed by dissolving the residue in acetone, and then cold water was added until a precipitate formed. TLC and melting point tests carried out to find out the purity of the product qualitatively.
Purity test by high-performance liquid chromatography (HPLC)
Analysis of purity was performed using HPLC Elite La-Chrome®, and for comparison, we used aldehydes as each starting material. Synthesis products and starting material were analyzed at 100 ppm concentration. Measurements were determined at UV wavelength of 350 nm and a mobile phase of 80:20 acetonitrile and water with a flow rate of 1 ml/ minute, 100 psi pressure, and 20 μl volume injection in column C18.
From the results of the purity test conducted by HPLC, each compound can be separated appropriately using the system. The test results showed that the synthesized compound was pure, which was confirmed using the respective starting material as seen in Figure 2 and Table 2.
2, 6-bis-(3′-ethoxy, 4′-hydroxybenzylidene)- cyclohexanone (A103)
Greenish yellow powder, yield 57.84%, mp 156.1°C–157.8°C; Rf = 0.3, ethyl acetate:CHCl3(1:20); IR γ (cm−1) (KBr): 3,368.86 (-OH hydrogen bonded); 2,979.35 (=C-H stretching aromatik); 2,881.23 (=C-H stretching alkena); 1,658.85 (C=C stretching alkena); 1595,93 (C=O stretching αβ,α’β’-unsat); 1,433.48 (C=C stretching aromatik); 1,399.68 (C-H bending alifatik); 1,288.36 (C-CO-C coupled stretching and bending); 1,213.24 (C-O-C stretching); 1,161.49 (C-OH stretching); MS (EI-MS, m/z )394 [M++2H]; 61 (base line); 1 H-NMR (500 MHz, ppm, DMSO-d6): δ 1.384 (3H, t, C9′, CH3 aliphatic), δ 1.727 (1H, qui, H4, CH2 cyclohexanone); δ 2.885 (2H, qui, H3,5, CH2 cyclohexanone); δ 4.065 (2H, k, H8′, CH2 aliphatic); δ 6.878 (1H, d, J = 8Hz H5′, Ar-CH); δ 7.040 (1H, dd, J1 = 10 Hz J2 = 1.5Hz, H6′, Ar-CH); δ 7.100 (1H, s, H2′, Ar-CH); δ 7.551 (1H, s, H7′, =C-H alkena); δ 9.437 (1H, s, Ar-OH).
2, 6-bis-(3′-bromo, 4′-methoxybenzylidene)-cyclohexanone (A116)
Yellow crystal, yield 62.76%, mp 182.1oC–182.5oC, Rf = 0.58, ethyl acetate : n-hexane (1:9); IR γ (cm−1) (KBr): 2,939.52 (=C-H stretching aromatik); 2,839.22 (-C-H stretching alkena); 1,658.78 (C=C stretching alkena); 1,589.34 (C=O stretching αβ,α′β′-unsat); 1,496.76 (C=C stretching aromatik); 1,288.45 (C-CO-C coupled stretching and bending); 1,157.29 (C-O-C stretching); 551.41 (C-Br stretching); MS (EI-MS, m/z): 492 [M+] (100%); 1H-NMR (500 MHz, DMSO-d6): δ 1.726 (1H, qui, H4, CH2 cyclohexanone); δ 2.866 (2H, t, H3,5, CH2 cyclohexanone); δ 3.899 (3H, s, H8’, O-CH3); δ 7.200 (1H, d, J = 8.5 Hz H5’, Ar-CH); δ 7.539 (1H, s, H7’ =CH alkena); δ 7.577 (1H, dd, J1 = 9, J2 = 2.5 Hz, H6’Ar-CH); δ 7.772 (1H, d, J = 2 Hz C2’Ar-CH).
2, 6-bis-(3′, 4′-dimethoxybenzyllidene)-cyclohexanone (A153)
Yellow powder, yield 42.02%, mp 149.7oC–151.6oC, Rf = 0.23, DCM : n-hexane (2:1); IR γ (cm-1) (KBr): 2,931.42 (=C-H stretching aromatik); 2,837.33 (-C-H stretching alkena); 1,650.70 (C=C stretching alkena); 1,595.85 (C=O stretching αβ,α′β′-unsat); 1,450.24 (C=C stretching aromatik); 1,334.92 (C-H bending alifatik); 1,285.56 (C-CO-C coupled stretching and bending); 1,139.51 (C-O-C stretching).; MS (EI-MS, m/z): 394 [M++2H]; 131 (base line); 1H-NMR (500 MHz, DMSO-d6): δ 1.732 (1H, qui, H4, CH2 cyclohexanone); δ 2.912 (2H, qui, H3,5, CH2 cyclohexanone), δ 3.830 (6H, s, H8’,9’, O-CH3); δ 7.043 (1H, d, J=8.5 Hz, H6’, Ar-CH); δ 7.139 (2H, s, H2’,5’, Ar-CH); δ 7.702 (1H, s, H7’, =CH alkena).
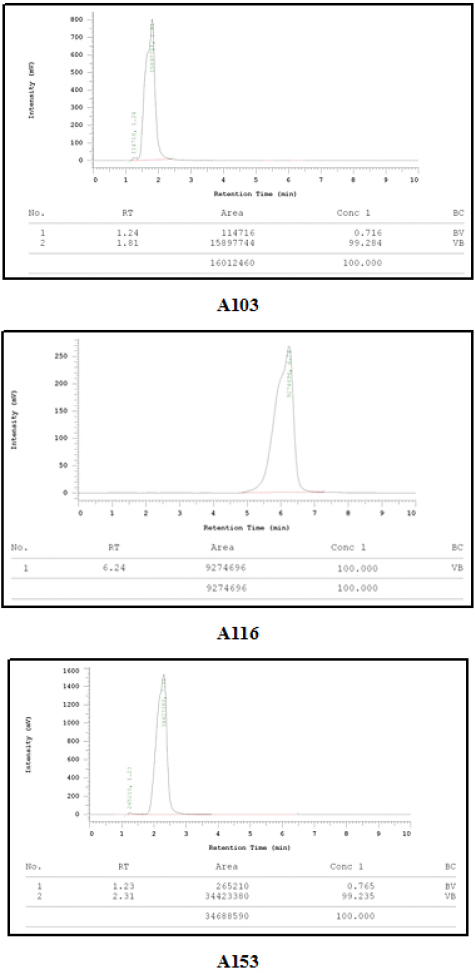 | Figure 2. Chromatogram of the purity test results. [Click here to view] |
In vitro anti-inflammatory assay
Several stages were performed for in vitro anti-inflammatory assay started by the platelet-rich plasma (PRP) preparation from fresh human blood. The non-enzymatic method was used in this study by measuring the level of malondialdehyde (MDA) in fluorescence spectrophotometer.
About 0.5 mL of plasma (PRP) and 5 μl of 3.13–50.0 μM sample [prepared by diluted in Dimethyl sulfoxide (DMSO)] are placed into Eppendorf tube then pre-incubated for 5 minutes at 37oC. About 100 μl of the fresh arachidonic acid solution was added then incubated for 30 minutes at 37oC. The reaction was stopped using 1 ml of TBA reagent in cold temperature. The mixture was heated at 80oC for 15 minutes and centrifuged at 3,000 rpm for 15 minutes. The absorbance was determined using fluorescence spectrophotometry in 553 nm wavelength for emission and 510 nm wavelength for excitation. DMSO was used as a control.
RESULTS AND DISCUSSION
QSAR study provides the best equation model to design new curcumin analogs of mono-ketone as an anti-inflammatory. QSAR studies were conducted several lead compounds that have reported having cyclooxygenase inhibitory activity (Sardjiman et al., 1997). As a result, 80 descriptors were selected that founded in 2D and i3D molecular database (Table 3).
Geometry optimization was performed toward optimized structure using the AM-1 semi-empirical method. Based on the previous study, the best analysis results on the QSAR series of curcumin analog compounds as antioxidants are shown in the AM-1 method compared to the PM-3 method (Sudarmanto and Oetari, 2007). In the previous study, AM-1 was used to predict curcumin compounds and their derivatives as class μ GST inhibitors as well (Istyastono et al., 2003).
Model 1 (Table 4) shows the best statistical parameter values. It shows the highest values of r and F by 0.999 and 501.319, respectively, and the smallest standard deviation (s) value of 0.042 compared to other models. The cross-validation value also shows a good statistical parameter, where the 0.991 Q2 value is the highest compared to other models and has the lowest sPRESS value of 0.077. Tables 5 and 6 show new MACs compounds with anti-inflammatory activity obtained from the best equation model.
Based on Table 5 results, we can observe that the equation we used is to provide good prediction value. Good prediction value determines by the small number of the residual value from each compound. Correlation analysis between observations over calculation value is shown in Figure 3. The best equation provided by model 1 is also used to predict three new MACs compounds.
In vitro evaluation of new mono-ketone curcumin analog compound
Three compounds, A103; A116; and A153, were used to determine their anti-inflammatory activity dissolved in DMSO. These compounds were obtained based on our QSAR studies, which have the best predictive values to be analyzed for in vitro anti-inflammatory assay. In this study, aspirin was used as a positive control.
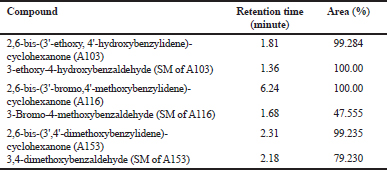 | Table 2. Area and retention time of HPLC purity test results. [Click here to view] |
 | Table 3. List of descriptors used in the QSAR study. [Click here to view] |
As a standard gold method, anti-inflammatory assay with cyclooxygenase inhibition activity in human blood samples was used in our study. PRP isolated from human blood was used as a source of cyclooxygenase and arachidonic acid as the substrate was used to stimulate the inflammation. TBA was used to form Malondialdehyde-TBA (MDA-TBA) complex.
Five concentrations examined from each compound show inhibition activity of COX (Fig. 4). There was a correlation between increasing of concentration and the percentage of cyclooxygenase inhibition. We have verified that the highest concentration will provide cyclooxygenase inhibition activity. We found that A116 at 3.125 μg/mL concentration has the best inhibition percentage values (23.77 ± 3.18). The ability of A116 to inhibit cyclooxygenase is higher than positive control aspirin as well (10.79 ± 5.34). The presence of halogen and methoxy groups on the benzene structure exerts a resonant and induction effect, which causes more potent cyclooxygenase inhibition activity.
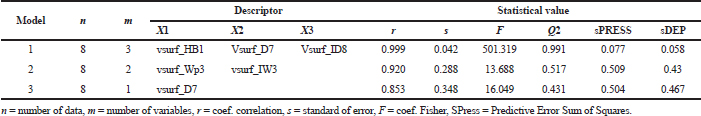 | Table 4. Parameter statistical of MLR result. [Click here to view] |
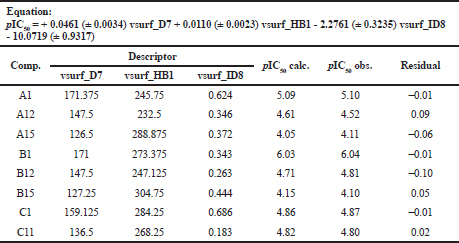 | Table 5. The best equation model to calculated calculation-value of cyclooxygenase inhibition. [Click here to view] |
.png) | Table 6. Descriptor value of new design mono-carbonyl analogs of curcumin with anti-inflammatory activity. [Click here to view] |
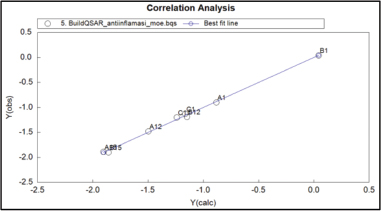 | Figure 3. Correlation analysis between calculation and observation QSAR. [Click here to view] |
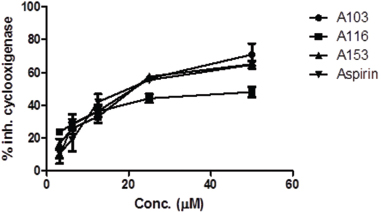 | Figure 4. Percentage of cyclooxygenase inhibition. [Click here to view] |
The percentages of cyclooxygenase inhibition were used to determine the IC50 that indicates the ability of a compound to inhibit 50% cyclooxygenase. The smallest IC50 value indicates the best cyclooxygenase inhibition ability. IC50 values from each compound are shown in Table 7. Efforts to compare the results of in vitro tests of new compounds with existing mono-carbonyl curcumin compounds were not carried out because they only used IC50 value of cyclooxygenase inhibition data in the process of designing compounds in silico can be seen in Table 7.
Result of study docking
In silico studies computationally were conducted over COX-1 and COX-2 receptor to determine the cyclooxygenase enzyme inhibition of new analogs of curcumin mono-ketone obtaining from QSAR study and those previously had been proven in vitro assay. MOE 2018.01.01 was used as a tool. All proteins downloaded then used in this docking study and then prepared in the 'sequence editor' menu of the software. The proteins are containing one strand of amino acid residue without a molecule of water. Autocorrect was performed after the protein was ready to be validated. The validated protein used in docking studies is shown in Figure 5.
The validity of protein was verified by the value of RMSD as a parameter used to evaluate the similarity of two structures. The similarity was measured based on the different distances of similar atoms. RMSD value < 2 indicates that the protein is valid and ready to use for further docking studies. The RMSD value of each protein is shown in Table 8. The results of the docking study show the interaction between the test compounds, whereas the protein target is shown in Figures 6 and 7.
ARG120 and TYR355 are the critical receptors necessary for the activity of cyclooxygenase-1 (COX-1) enzyme inhibition. It explains from the 2D and 3D interaction between native ligand (flurbiprofen) and proteins. For A103, O atom (55, from the ketone) interacts with N atom from ARG120 over H-acceptor. For A116, NH2 from ARG120 and OH groups from TYR355 interact through the H acceptor with O atoms (47, from ketones). For A153, C atom (11, from methoxy in the aromatic ring) interacts with 6-ring (benzene aromatic) of TYR385 over H-pi. For aspirin, O atom (6, from aldehydes) interacts with NH2 from ARG120 and OH from TYR355 over H-acceptor.
 | Table 7. IC50 values of COX inhibition. [Click here to view] |
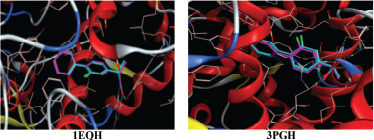 | Figure 5. Appearance of validation proteins test. [Click here to view] |
 | Table 8. RMSD value of protein test. [Click here to view] |
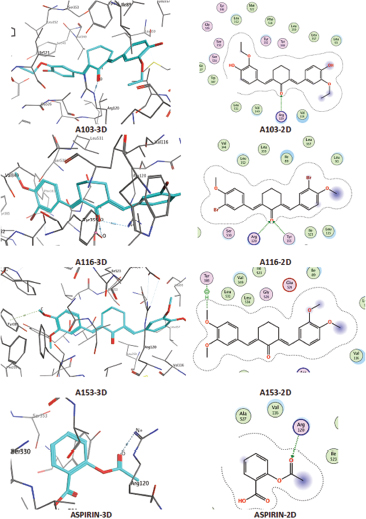 | Figure 6. Interaction of test ligands on 1EQH protein. [Click here to view] |
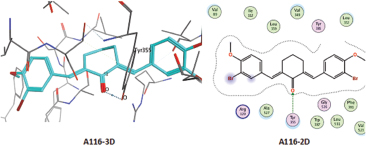 | Figure 7. Interaction of test ligands on 3PGH protein. [Click here to view] |
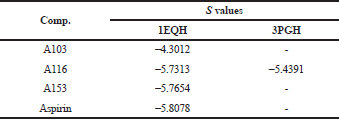 | Table 9. s value of new mono-carbonyl analogs of curcumin. [Click here to view] |
The results of 2D and 3D interactions between native ligands (flurbiprofen) and proteins test known that ARG120 and TYR355 are critical receptors that play an essential role in the cyclooxygenase-2 (COX-2) inhibitory activity of 3PGH proteins. For A116, O atom (47, from ketone) interacts with OH group from TYR355 over H-acceptor. For A103 and A153, in silico study, results show no interaction between these two compounds and 3PGH protein. Thus, it can conclude that A103 and A153 do not have inhibition activity over the COX-2 enzyme. A similar pattern of results obtained for aspirin does not show interaction with the test protein as well. The findings are directly in line with previous findings that aspirin works to inhibit the COX-1 enzyme as irreversible (Kalantzi et al., 2012).
After the docking process, the S value (Table 9) of each test ligand was found, which described the strength of the bond affinity that occurs between the test ligand and the target protein. The smallest S value explains its more excellent affinity bond leading to the better for its biological activity.
CONCLUSION
The results show that the new curcumin analogs of mono-ketone compounds obtained from QSAR study using BuildQSAR have anti-inflammatory activity through their inhibition over cyclooxygenase enzyme, which confirmed by docking studies.
ACKNOWLEDGMENT
The authors are grateful to Medicinal Chemistry Department, Faculty of Pharmacy UGM and Universitas Gadjah Mada, Yogyakarta, Indonesia, for MOE version 2018.01.01 licenses and funding this research.
FUNDING
This research funded by Rekognisi Tugas Akhir (RTA) Program from Universitas Gadjah.
AUTHORS CONTRIBUTIONS
All the authors have contributed equally.
CONFLICT OF INTERESTS
The authors confirm that this article content has no conflicts of interest.
REFERENCES
Agustina R, Indrawati DT, Masruhim MA. Aktivitas ekstrak daun salam (eugenia polyantha) sebagai antiinflamasi pada tikus putih (rattus norvegicus). J Trop Pharm Chem, 2015; 3:120–3. CrossRef
Anand P, Thomas SG, Kunnumakkara AB, Sundaram C, Harikumar KB, Sung B, Tharakan ST, Misra K, Priyadarsini IK, Rajasekharan KN, Aggarwal BB. Biological activities of curcumin and its analogues (Congeners) made by man and Mother Nature. Biochem Pharmacol, 2008; 76:1590–611. CrossRef
Arba M, Azali H, Ombe S, Armid A, Usman I. 3D-QSAR, molecular docking and dynamics simulation of difluorophenol pyridine derivatives as RSK2 inhibitor. J Appl Pharm Sci, 2019; 9(06):001–9. CrossRef
Desai SA, Kumbhar SS, Katti VS, Choudhari PB, Bhatia MS. 3D QSAR and pharmacophore modelling on chalcones as antileishmanial agents potential trypanothione reductase inhibitors. J Appl Pharm Sci, 2013; 3(12):099–102.
El-Hamamsy MH. Accessing the anti-proliferating activity of tankyrase-2 inhibitors via 2d, 3d-QSAR and molecular docking: assessment of structure activity relationships. J Appl Pharm Sci, 2017; 7 (12):014–27.
Hayun H, maggadani PB, Kurnia A, Fadlol A, Yuliandi M, Fitriyani I, PUTRI Hadrianti PS. Anti-inflammatory and antioxidant activity of synthesized Mannich base derivatives of (2e,6e)-2-[(4-hydroxy-3-methoxyphenyl)methylidene]-6-(phenyl -methylidene)cyclohexan-1-one. Int J Appl Pharm, 2019; 11:246–50. CrossRef
Istyastono EP, Martono S, Pranowo HD, Tahir I. Quantitative structure-activity relationship analysis of curcumin and its derivatives as GST inhibitors based on computational chemistry calculation. Indones J Chem, 2003; 3:179–86. CrossRef
Kalantzi KI, Tsoumani ME, Goudevenos IA, Tselepis AD. Pharmacodynamic properties of antiplatelet agents: current knowledge and future perspectives. Expert Rev Clin Pharmacol, 2012; 5:319–36. CrossRef
Kurnia A, Saputri FC, Hayun H. Synthesis and anticancer potential of aminomethyl derivatives of methyl-substituted asymmetrical curcumin mono-carbonyl. J Appl Pharm Sci, 2019; 9:18–24. CrossRef
Miladiyah I, Jumina J, Haryana SM, Mustofa M. Biological activity, quantitative structure-activity relationship analysis, and molecular docking of xanthone derivatives as anticancer drugs. Drug Des Dev Ther, 2018; 12:149–58. CrossRef
Modi SJ, Kulkarni VM. 3D-QSAR analysis of pyrimidine derivatives as AXL kinase inhibitors as anticancer agents. J Appl Pharm Sci, 2018; 8(11):015–27. CrossRef
Sardjiman S, Reksohadiprodjo MS, Hakim L, Van der Goot H, Timmerman H. 1, 5-Diphenyl-1, 4-pentadiene-3-ones and cyclic analogues as antioxidative agents. Synthesis and structure-activity relationship. Eur J Med Chem, 1997; 32:625–30. CrossRef
Selvam C, Jachak SM, Thilagavathi R, Chakraborti AK. Design, synthesis, biological evaluation and molecular docking of curcumin analogues as antioxidant, cyclooxygenase inhibitory and anti-inflammatory agents. Bioorg Med Chem Lett, 2005; 15:1793–7. CrossRef
Sudarmanto BA, Oetari RA. Aplikasi deskriptor kimia kuantum dalam analisis QSAR derivat kurkumin sebagai penghambat o-dealkilasi ethoxyresorufin. Maj Farm Indones, 2007; 18:147–53.
Ujihara M, Tsuchida S, Satoh K, Sato K, Urade Y. Biochemical and immunological demonstration of prostaglandin D2, E2, and F2α formation from prostaglandin H2 by various rat glutathione S-transferase isozymes. Arch Biochem Biophys, 1988; 264:428–37. CrossRef
Wijianto B, Ritmaleni R, Purnomo H, Nurrochmad A. In silico and in vitro assay of HGV analogue as antibacterial. Int J Pharm Pharm Sci, 2019; 11:78–85. CrossRef
Yuliarti NN, Nugroho AE. Synthesis and evaluation of antioxidant and antiinflammatory activities of 2-(4’-dimethylamino benzylidene)-6-benzylidene cyclohexanone. Indones J Pharm, 2013; 24:198–205.