INTRODUCTION
In the modern medical treatments, the infectious diseases caused by the microbial pathogens can be cured with a wide variety of antibiotics. However, a rapid increase of bacterial strains with the capability of antibiotics resistance widespread both in the clinical and veterinary medicine has triggered worldwide efforts in developing new alternative compounds to deal with such the problem (Aslam et al., 2018). Vibrio cholerae, the Gram-negative bacteria, is an important human pathogen causing the diarrhea disease (Jers et al., 2018) and can be isolated from a variety area of extra-intestinal infections (e.g., fresh wound, ear, sputum, urine, and cerebrospinal fluid) (Morris and Black, 1985). Klebsiella spp., the non-motile Gram-negative bacteria, was reported as one of the major bacterial groups which can be found in the nosocomial infection in Thailand (Danchaivijitr et al., 2007). Either the infection with the well-known Gram-positive bacterium Bacillus subtilis or yeast strain Candida albicans can cause the septicemia in an immunocompromised individual leading to the end-organ damage and finally the death within a few days (Lei et al., 2019). Generally, the treatment of these infections relies on penicillin antibiotics (Zwart et al., 2000). Even the successfulness in bactericidal property, the side effects of penicillin causing allergic symptoms in some patients need to be concerned. In order to avoid such effects, macrolide antibiotics are alternatively selected to treat the patients instead; however, the problem has arisen with the case of increasing of macrolide-resistant bacterial strains which minimizes drug’s efficiency (Fair and Tor, 2014).
To establish an alternative compound to be used in common, antimicrobial peptides (AMPs) have been interested since 1980s due to their potential as the novel antibiotic compounds. With broad-spectrum, antimicrobial activity and specificity in bacteria over eukaryotic cells or patients, it makes them as the attractive candidates for novel drug compounds. AMPs are typically rich in hydrophobic residues, including leucine, isoleucine, valine, phenylalanine, and tryptophan, and their net positive charge properties are due to an excess of cationic amino acids in the sequence. When expose to the phospholipid membranes, it allows the AMPs to adopt into four diverse amphipathic structures, which are α-helix, β-sheet, extended conformation, and looped peptides containing disulfide bridges, respectively (Hancock and Haney, 2013). Among several AMPs, Brucin is a potentially specific antibacterial peptide for Streptococcus pyogenes that comprises 11 amino acids with the sequence of HTLCMDGGATY and an extended random coil primary structure (Sornwatana et al., 2013; 2018). This peptide possesses a neutral charge at physiological pH and hydrophobicity ratio of 36%.
Several methods and rational design of new peptides aimed to create the novel molecules with the improvement of antimicrobial activity and low toxicity to human. In this work, Brucin derivatives were obtained from the sequence template method and designed by the computational prediction. By this method, it would seek to minimize the size, add more selectivity, and/or increase activity of the new synthesized peptide (Vincent et al., 2019; William et al., 2012). Eventually, such procedures provide these two benefits; an increase in the hydrophobicity of the peptide is also a concomitant increase the antimicrobial activity (Ingrid et al., 2016), and the adsorption of AMPs on-to the microbial membrane is conducted by an electrostatic interaction (Shi et al., 2018). To increase the stability of the peptides against the proteolytic degradation, two molecules of Brucin derivatives were structurally designed from the linear to the cyclic by adding glutamine at the N-terminal of Brucin as there is evidence suggested that the N-terminal glutamine can promote cyclization within the short polypeptides (Liu et al., 2011; Neta et al., 2007). Moreover, amino acid substitution at the C-terminus with tyrosine to tryptophan and aspartate to cysteine can increase molecular hydrophobicity and polarity with disulfide bonding ability, respectively. Final step with the cytotoxicity assays, these modified Brucin derivatives were used to test its bacterial inhibitory activity against our laboratory pathogenic strains, including Gram-negative and Gram-positive bacteria, and yeast, respectively. The derivative that showed the most powerful antimicrobial activity against the numbers of selected pathogens was named as Brucyclin and designated as the novel antimicrobial cyclic peptide.
MATERIALS AND METHODS
Synthesis of modified Brucin derivatives
The two molecules of Brucin derivatives were purchased and synthesized by using the solid phase method (GL Biochem Shanghai Ltd., China). After the synthesis process, high performance liquid chromatography column with OSD-C18 reverse-phase was used to purify the synthetic peptides. Based on the previous suggestion, these derivatde (Neta et al., 2007). The cyclization was then established by the disulfide bond linkage as similar as the plant defensin-like peptides. These two derivatives were designated as QQ and QW with the amino acid sequence of QHTLCMCGGATYQ and QHTLCMCGGATW, respectively.
Pathogenic strains used in the experiment
The antimicrobial activity assay of modified Brucin derivatives was performed against 30 strains of bacteria and 1 strain of yeast (Table 1). The strain samples used in this study were kindly provided by National Institute of Health, Ministry of Public Health, Thailand and Department of Biotechnology, Faculty of Science, Ramkhamhaeng University.
Determination of antimicrobial peptides by minimum inhibitory concentration (MIC) assay
A minimum inhibitory concentration (MIC) value for the synthetic peptides was determined by the agar dilution method (Barry, 1976). The synthetic peptide was first prepared in dimethyl sulfoxide (Carlo Erba), thus its two-fold dilution was mixed together with a sterile melted Muller–Hinton agar to the final volume of 20 ml, immediately poured on the plate. After the agar hardened, 10 μl of 104 CFU/ml of each microorganism strain was spotted onto the mixed agar plates in triplicate, and then incubated at 37°C for 24 hours. The MIC value was determined by the lowest concentration of peptide which could inhibit the visible growth of the cells on the plate. The growth number of the peptide-treated cells was compared to one of the control cells without treatment. Two bacterial strains, E. coli ATCC25922 and S. aureus ATCC25923, were used as the standard references.
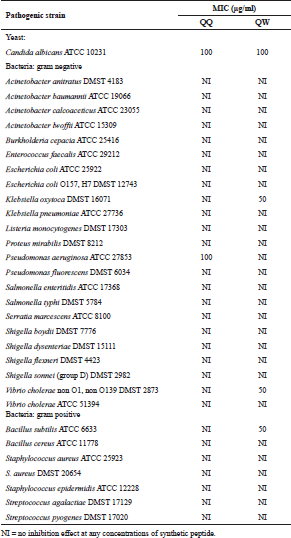 | Table 1. MIC values of synthetic peptides determined by agar-dilution method. [Click here to view] |
Determination of antimicrobial activity by inhibitory concentration (IC50)
The half-maximal inhibitory concentration (IC50) values, in this work, were determined by the method broth microdilution recommended by National Committee for Clinical and Laboratory Standard Institute (CLSI) (1997). To evaluate the IC50 value of the synthetic peptides against tested strains, the peptides were serially diluted two folds with the Muller–Hinton broth to vary the concentration at 25, 50, and 100 μg/ml. For each concentration so far, the 100 μl aliquot was transferred to the 96-well microliter plates (flat-shaped well), and then about 5 μl of 106 CFU/ml of each microorganism strain was inoculated into each well. Culture without peptide treatment was used as a control. For the positive control, the cultures treated with chloramphenicol and penicillin G, were also included. Eventually, the 96-well plates were incubated at 37°C for 24 hours with shaking at 180 rpm. Cell density representing for the microbial growth was determined by the microplate-reader (MRX, Magellan Biosciences) at 600 nm. The assay for each strain was done in three experimental replicates. In data analysis, the absorbance or optical density (OD)600nm value at the lowest concentration of synthetic peptide on each treatment that allows the cells able to grow 50% compared to a control was defined as the IC50. The percentage of growth inhibition was calculated with the following formula:
Structural analysis of synthetic peptides
The peptide sequence was compared to the other peptides corresponding to an antimicrobial property using the Antimicrobial Peptide Database as described in Wang et al. (2009). Next, the sequence was submitted to predict its secondary structure by the Garnier–Osguthorpe–Robson IV Secondary Structure Prediction Method (Garnier et al., 1996). For the hard evidence, the circular dichroism (CD) spectroscopy was applied to determine the secondary structure and folding properties of the synthesized peptide. After the chemical synthesis, the peptide concentration of 0.2 mg/ml was dissolved in distilled water (DW) or 10% sodium dodecyl sulfate (SDS), representing the anionic micelles forming (Sangtanoo et al., 2014). Later on, the peptide was transferred to the quartz cuvette (1-mm path length), the spectra were scanned at the room temperature by JASCO CD spectrophotometer model J-815. The scanning speed was set at 50 nm/minute ranging from 190 to 260 nm, and each spectrum was the average of three replicates. The graph of CD spectra was then produced with ellipticity θ as a function of the wavelength (λ). The ellipticity, which is proportional to the difference between the absorbance of left and right circularly polarized light of peaks, reflects the thermodynamic stability for peptide structure. Peak position and the sharp of a curve represent the conformation of the synthetic peptides, in which, a typical form possesses a positive peak in between 260 and 270 nm, and a negative peak between 210 and 220 nm (Lu et al., 2012).
Statistical analysis
The antimicrobial activity of Brucyclin peptide in between the microbial candidates and the control groups were analyzed by two-way analysis of variance method. The p-value of <0.05 was considered to be significant differences in between data set. Statistical analyses were done by the program GraphPad Prism 5.04 (GraphPad Software, Inc.).
RESULTS
In this study, two Brucin derivatives, QQ (QHTLCMCGGATYQ) and QW (QHTLCMCGGATW), were chemically synthesized, and their antibacterial activity was hence determined. As shown in Table 1, antimicrobial activity for the synthetic peptides against human pathogenic microorganisms assigned by MIC assay was observed. From the results, we found that the QW peptide showed the most powerful inhibitory effect, and the concentration between 50 and 100 μg/ml was sufficient to inhibit cell growth against, at least, four bacterial strains; V. cholera non O1 and non O139 Department of Medical Sciences Culture Collection (DMST) 2873, K. oxytoca DMST 16071, and B. subtillis ATCC 6633, and yeast C. albicans The American Type Culture Collection (ATCC) 10231. Although the number of growth inhibition of the other, QQ, was quite lower than that of the QW, the QQ at 100 μg/ml concentration did inhibit only in P. aeruginosa ATCC 27853 and C. albicans ATCC 10231, respectively. Hence, the QW peptide with molecular mass of m/z1290 was subjected as a candidate for further analyses, and later so called Brucylin. The chemical structure of Brucyclin and the liquid chromatography-mass spectrometry data were shown in Figures 5 and 6, respectively, and there was no α-helix found in the secondary structure of Brucyclin as confirmed in CD spectrum analysis (Fig. 4). To evaluate the potential effect of Brucyclin, the bacterial growth inhibition by the peptide was carried out at the different time points as shown in Figure 1. Showing that, Brucyclin substantially inhibited cell growth by 50% begun at 13 hours after the treatment. For the antimicrobial activity assay, Brucyclin was evaluated by the microdilution as described above. Suggesting that Brucyclin at the concentration between 50 and 100 μg/ml could inhibit by 50% cell growth (IC50) in the three bacterial strains and yeast as shown in Figure 2, while the microbial colony count exhibited the same trend and was also shown in Figure 3.
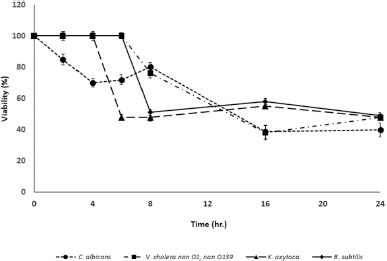 | Figure 1. The bacterial growth inhibition by the Brucyclin peptide was determined at different time point, data presented the mean ± standard deviation (SD) of three independent assays, p < 0.0001. [Click here to view] |
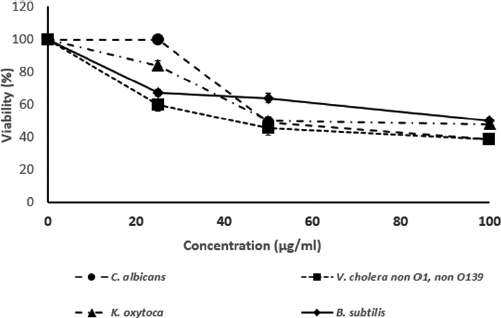 | Figure 2. The bacterial growth inhibition (IC50) at various concentration of Brucyclin peptide, Data presented the mean ± SD of three independent assays, p < 0.0001. [Click here to view] |
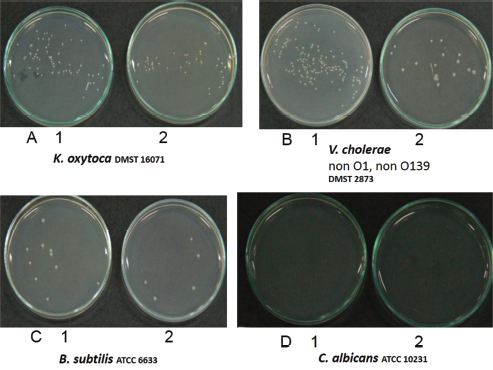 | Figure 3. The microbial colony count representing 50% growth inhibition (IC50), IC50 = a half maximal inhibitory concentration. A1–D1 = control (untreated), A2–D2 = Brucyclin treated, A1 = 88 CFU, A2 = 43 CFU, B1 = 142 CFU, B2 = 17 CFU, C1 = 10 CFU, C2 = 5 CFU, D1 = 37 CFU, D2 = 18 CFU. [Click here to view] |
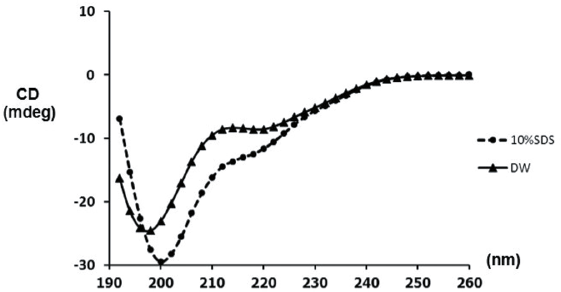 | Figure 4. CD spectra of Brucyclin peptide. The peptide (0.2 mg/ml) was prepared in DW and 10% SDS. [Click here to view] |
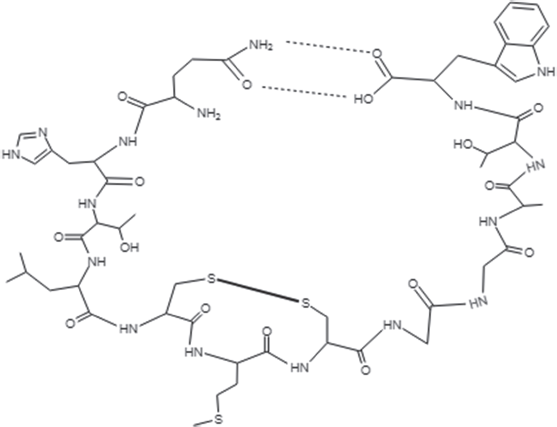 | Figure 5. Chemical structure of the cyclic peptide, Brucyclin. [Click here to view] |
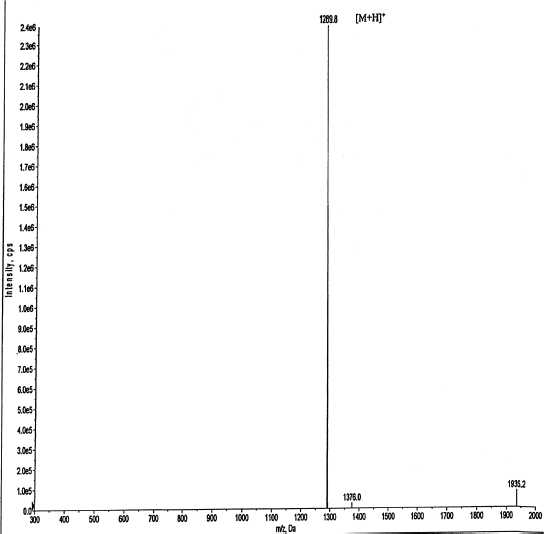 | Figure 6. LC-MS analysis of cyclic peptide showed a molecular mass of m/z 1290 with confirmed the molecular mass of Brucyclin. [Click here to view] |
DISCUSSION
Cyclic peptides are more stable against proteolytic degradation in the cell host according to lacking of free N- or C-terminus and reduced conformational deliverance. Moreover, the cyclic structures are more penetrable than the linear as they are able to form intra-molecular hydrogen bonds and transverse the lipid bilayer making them the greater membrane permeability (Hruby, 2016). More than thousands of AMPs are known to have great variety in both primary and secondary structure; however, most of them share some general structural characteristics (Kumar et al., 2018). Their general character is the cationic amphipathic molecule at physiological pH, consisting of lesser than 40 amino acid residues. Key features of the peptides are answerable for antimicrobial activity depending on different forms of amphipathic structure, hydrophobic ratio, and the cationic parts (Cantisani et al., 2014; Jenssen et al., 2006). The amphipathic characters in some peptides are shown to be more impact than their native secondary structures (Godballe et al., 2011; Marcia et al., 2019).
Based on such advantages, in this study, a rationally designed peptides, Brucyclin (QHTLCMCGGATW), was derived from the plant antimicrobial peptide Brucin by cyclization and disulfide bond formation of the original peptide. Brucyclin is the small cyclic peptide (~1,290 Da) possessing a net neutral charge at physiological pH. The secondary structure analysis showed that it is an amphipathic molecule, which may be capable for the antibacterial activity. AMPs can be found in wide variety of plant species (Barbosa Pelegrini et al., 2011; Nawrot et al., 2014). Some peptides exhibit specificity toward only Gram-negative or Gram-positive bacteria but most of them have broad-spectrum antibacterial activities. Some plant AMPs inhibit the growth of Gram-positive bacteria and those from poke weed, Phytolacca americana (Lui et al., 2000) and Macadamia integrifolia kernels (Marcus et al., 1999). General mechanism for most AMPs in action mainly generates membrane collapse by the interaction between charged molecules and the bacterial membrane with the electrostatic property resulting membrane pores formation, rapidly lysis, and cell death, respectively (Barbosa Pelegrini et al., 2011; Hollmann et al., 2018; Jenssen et al., 2006). The pore formation mechanisms have been reviewed for plant antimicrobial peptides. From the amino acid sequence of Brucyclin and its broad-spectrum inhibitory activity to bacteria, it could be postulated that the hydrophobic and hydrophilic moieties of Brucyclin may segregate into distinct patches and interact with either phospholipids or teichoic acid, and specific receptors on the bacterial cell surface, allowing it to form pores in the cytoplasmic membrane, leading to the membrane disruption. Mode of its antibacterial action on bacterial cell wall and the relative importance of amino acid residues of Brucyclin are likely to clarify in the future. Its structure consists of hydrophobic, hydrophilic, and positive and negative charge 50% of amino acid residues, which may improve an antimicrobial activity of the peptide. The inhibitory activity of Brucyclin toward bacteria (Table 1) and its resistance to the intestinal enzyme hydrolysis imply the potential application use of the peptide, Brucyclin, as a drug candidate for treatment of the diseases. However, some clinical trails should be performed for evaluation of its safety, bioavailability, and metabolic stability in vivo.
CONCLUSION
In this study, a novel broad-spectrum antimicrobial peptide, Brucyclin, was rationally designed from the plant antibacterial peptide, Brucin. The amphipathic characteristic of Brucyclin suggested for its antibacterial capability and selectivity toward a wide-range of Gram-positive and Gram-negative bacteria as well as yeast. Further experiments on mode of antimicrobial action of the peptide and in vivo clinical trials may also be investigated to success in application of Brucyclin as an antimicrobial drug in the future.
ACKNOWLEDGMENTS
The Higher Education Commission Research Networks was supported this work to T. Sornwatana. The authors also gratefully thank Dr. Tiwa Rotchanapreeda for his helps in reading and editing the manuscript.
CONFLICT OF INTERESTS
No conflicts of interest to declare.
REFERENCES
Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, Nisar MA, Alvi RF, Aslam MA, Qamar MU, Salamat MKF, Baloch Z. Antibiotic resistance: a rundown of global crisis. Infect Drug Resist, 2018; 11:1645–58. CrossRef
Barry AL. Agar dilution techniques. In: Barry AL (ed.). The antimicrobial susceptibility test; principle and practices. Led and Febiges, Philadelphia, PA, p 76, 1976.
Barbosa Pelegrini P, del Sarto RF, Silva ON, Franco OL, Grossi-de-Sa MF. Antibacterial peptides from plants: what they are and how they probably work. Biochem Res Int, 2011; 2011:250349. CrossRef
Cantisani M, Finamore E, Mignogna E, Falanga A, Nicoletti GF, Pedone C, Morelli G, Leone M, Galdiero M, Galdiero S. Structural insights into and activity analysis of the antimicrobial peptide Myxinidin. Antimicrob Agents Chemother, 2014; 58(9):5280–90. CrossRef
Danchaivijitr S, Judaeng T, Sripalakij S, Naksawas K, Plipat T. Prevalence of nosocomial infection in Thailand 2006. J Med Assoc Thai, 2007; 90(8):1524–9.
Dos Santos Cabrera MP, Rangel M, Ruggiero Neto J, Konno K. Chemical and biological characteristics of antimicrobial α-helical peptides found in solitary wasp venoms and their interactions with model membranes. Toxins, 2019; 11(10):559. CrossRef
Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem, 2014; 6:25–64. CrossRef
Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol, 1996; 266:540–53 CrossRef
Godballe T, Nilsson LL, Peterson PD, Jenssen H. Antimicrobial β-peptides and α-peptoids. Chem Biol Drug Des, 2011; 77:107–16. CrossRef
Hancock REW, Haney EF. Peptide design for antimicrobial and immunomodulatory applications. Biopolymers, 2013; 100:572–83. CrossRef
Hollmann A, Martinez M, Maturana P, Semorile LC, Maffia PC. Antimicrobial peptides: interaction with model and biological membranes and synergism with chemical antibiotics. Front Chem, 2018; 204:1–13. CrossRef
Hruby VJ. Design of cyclic peptides with biological activities from biologically active peptides: the case of peptide modulators of melanocortin receptors. Biopolymers, 2016; 106(6):884–8. CrossRef
Ingrid AE, Alysha GE, Angela MK, Johannes Z, Mark ATB, Matthew AC. Contribution of amphipathicity and hydrophobicity to the antimicrobial activity and cytotoxicity of b-Hairpin peptides. ACS Infect Dis, 2016; 2(6):42–50. CrossRef
Jenssen H, Hamill P, Hancock REW Hancock. Peptide antimicrobial agents. Clin Microbiol Rev, 2006; 19:491–511. CrossRef
Jers C, Ravikumar V, Lezyk M, Sultan A, SjÖling A, Wai SN, Mijakovic I. The Global acetylome of human pathogen Vibrio cholera V52 reveals lysine acetylation of major transcriptional regulators. Front Cell Infect Microbiol, 2018; 7:1–13. CrossRef
Kumar P, Kizhakkedathu JN, Straus SK. Antimicrobial peptides: diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules, 2018; 8(1):4. CrossRef
Lei S, Zhao H, Pang B, Qu R, Lian Z, Jiang C, Shao D, Huang Q, Jin M, Shi J. Capability of iturin from Bacillus subtilis to inhibit Canida albican in vitro and in vivo. Appl Microbiol Biotechnol, 2019; 103(11):4377–92. CrossRef
Liu YD, Goetze AM, Bass RB, Flynn GC. N-terminal glutamate to pyroglutamate conversion in vivo for human IgG2 antibodies. J Biol Chem, 2011; 286(13):11211–7. CrossRef
Lu L, Dionyssios K, Susan TM, Harvey AR, Ciara B, Emilie MC, Herbert Yu Functional study of risk loci of stem-cell associated gene lin-28B and associations with disease survival out comes in epithelial ovarian cancer. Carcinogenesis, 2012; 33:2119–25. CrossRef
Lui Y, Luo J, Xu C, Ren F, Peng C, Wu G, Zhao J. Purification,characterization, and molecular cloning of gene of a seed-specific antimicrobial protein from pokeweed. Plant Physiol, 2000; 122:1015–24. CrossRef
Marcus JP, Green JL, Goulter KC, Manners JM. A family of antimicrobial peptides is produced by processing of a 7S protein in Macadamia integrifola kernels. Plant J, 1999; 19:699–710. CrossRef
Morris JG Jr, Black RE. Cholera and other vibrioses in the United States. N Engl J Med, 1985; 312(6):343–50. CrossRef
National Committee for Clinical Laboratory Standards Insitute (CLSI). Performance standards for dilution antimicrobial susceptibility testing for bacteria that grow aerobically, NCCLS document M100-58. Clinical Laboratory Standards Institute, Villanova, PA, 1997.
Nawrot R, Barylski J, Nowicki G, Broniarczyk J, Buchwald W, JÓzefiak AG. Plant antimicrobial peptides. Folia Microbiol (Praha), 2014; 59(3):181–96. CrossRef
Neta P, Pu QL, Kilpatrick L, Yang X, stein SE. Dehydration versus deamination of N-terminal Glutamine in collision-induced dissociation of protonated peptides. J Am Soc Mass Spectrom, 2007; 18:27–36. CrossRef
Sangtanoo P, Choowongkomon K, Surat W, Nitisinprasert S, obialKubera A. Antimicrobial peptides of Latobacillus Salivarius K4 isolated from chicken intestine. Science Asia, 2014; 40:135–40. CrossRef
Shi Y, Wan M, Fu L, Zhang S, Wang S, Gao L, fang W. Peptide-lipid interaction sites affect vesicles responses to antimicrobial peptides. Biophys J, 2018; 115(8):1518–29. CrossRef
Sornwatana T, Roytrakul S, Wetprasit N, Ratanapo S. Brucin, an antibacterial peptide derived from fruit protein of Fructus Bruceae, Brucea javanica (L) Merr. Lett Appl Microbiol, 2013; 57(2):129–36. CrossRef
Sornwatana T, Arpornsuwan T, Roytrakul S, Wetprasit N. M-Brucin, an antibacterial peptide against Staphylococcus epidermidis and Streptococcus pyogenes. J App Pharm Sci, 2018; 8(2):27–32.
Vincent F, Justin MJ, Jianfu Z, Gevory G, Amy EK. Tertiary structural motif sequence statistics enable facile prediction and design of peptides that bind anti-apoptic Bfl-1 and Mcl-1. Structure, 2019; 27(4):606–17. CrossRef
Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucl Acids Res, 2009; 37:D933–7. CrossRef
William FP, Osmar NS, Octavio LF. Prediction and rational design of antimicrobial peptide. In: Faraggi E(ed.). Protein Structure, Intech Open Limited, London, UK, 2012.
Zwart S, Sachs AP, Ruijs GJ, Gubbels JW, Hoes AW, de Melker RA. Penicillin for acute sore throat: randomised double blind trial of seven days versus three days treatment or placebo in adults. BMJ, 2000; 320:150.