INTRODUCTION
Breast cancer is one of the main causes of death worldwide (Plummer et al., 2016). Some studies showed that breast cancer is associated with steroid hormones as the agonist ligand of the nuclear receptor (NR) (Conzen, 2008; Dhiman et al., 2017). NR plays a vital role in many physiological processes, including immunity and cell proliferation (Choi and Bothwell, 2012). NR can directly bind ligands such as steroids and bile acid. Agonist ligands can induce the structural changes within the NR protein, leading to the activation of DNA targets (Danielian et al., 1992; Makishima et al., 1999). On the other hand, antagonist ligand could have anticancer activities by inhibiting cell proliferation through the inactivation of NR.
Limonoids are the modified triterpenes with diverse structures and several bioactivities, including anticancer (Guthrie et al., 2000; Sophia et al., 2016; Tan and Luo, 2011; Zhang and Xu, 2017). Moreover, in vivo studies resulted that limonoids from Azadirachta indica (Neem) showed promising anticancer activity. Many pieces of evidence indicated that limonoid modulates various signaling molecules and networks in cancer cells (Kowshik et al., 2017; Sophia et al., 2016). Azadirachtin, a limonoid from neem, showed a good inhibition toward NR (Thoh et al., 2013). In addition, limonoid from Chisocheton (Meliaceae) also has potential anticancer activity (Awang et al., 2007; Nagoor et al., 2011). Nevertheless, the mechanism of action of these limonoids as the anticancer agents remained unclear.
Many studies suggested that limonoids inhibited NR. However, the expression of NRs is specific in every tissue and type of cancer. There are 12 NRs expressed in breast cancer, and their role in cancer cells has been reviewed (Conzen, 2008; Dhiman et al., 2017).
Previously, four limonoids from Chisocheton sp. showed a good inhibition to the MCF-7 breast cancer cell line. These compounds, namely, compounds 5–8, were further explored by computational methods to predict their mechanism of action at the atomic-level. This work aimed to investigate the possible NR as the molecular target of the antibreast cancer activity of limonoids from Chisocheton sp. using molecular docking and molecular dynamics (MD) simulation.
METHODS
Molecular docking
The crystal structure of NR that expressed in breast tissue was retrieved from Protein Data Bank with PDB ID: 1A28 (Progesterone Receptor, PR), 1H9U (Retinoid X Receptor Beta, RXRβ), 5MKU (Retinoid X Receptor Alpha, RXRα), 1SJ0 (Estrogen Receptor Alpha, ERα), 1QKM (Estrogen Receptor Beta, ERβ), 2LBD (Retinoic Acid Receptor Gamma, RARγ), 1T7R (Androgen Receptor, AR), 3IPQ (Liver X Receptor Alpha, LXR), 4P6W (Glucocorticoid Receptor, GR), 5UNJ (Liver Receptor Homolog 1, LRH1), and 5HJS (Liver X Receptor Alpha, LXRα). The structure of the co-crystal ligand was separated from the receptor using BIOVIA Discovery Studio Visualizer 2017 (Accelrys), including the removal of water molecules and hetero-atoms.
The 3D structures of compounds 5–8 were modeled by Biovia Draw 2018 and geometrically optimized by BIOVIA Discovery Studio Visualizer 2017. The Polar Surface Area (PSA) and LogP were calculated by Biovia Draw 2018. Furthermore, the 3D structures were converted into the PDBQT format by adding the atom type and Gasteiger charges using AutoDock Tools 1.5.6. Docking of all ligands was performed with AutoDock Vina using 24 exhaustiveness of processor. The grid box was centered in the binding site of each NR in size of 25 Å × 25 Å × 28 Å. The docking process was automatized by the PaDEL-ADV program. The free energy of binding from each docking pose was further rescored by AutoDock 4.2.
Molecular dynamics simulation
The docking pose was further refined by MD simulations. The simulations were performed using AMBER16. The temperature was controlled by a Langevin thermostat, and the pressure was preserved using the Berendsen coupling method. Periodic boundary conditions and the particle mesh Ewald method were applied with a nonbonded interaction cut-off of 10 Å. The MD system was gradually heated to 300 K for 150 ps in constant volume condition using harmonic restraints of 5 kcal/molÅ2 on the backbone atoms. Furthermore, 6 ns of constant pressure equilibration was performed, where harmonic restraints on the backbone were slowly decreased by 1 kcal/molÅ2 until it reached 0. Then, 10 ns of the production run in the constant pressure ensemble was performed.
RESULTS AND DISCUSSION
Eleven NRs were selected as the candidate of the receptor of limonoid since they are specifically expressed in breast tissue (Conzen, 2008; Dhiman et al., 2017). The docking method was validated with the positive control docking, by re-docking the co-crystal ligand of each receptor into the binding site using AutoDock Vina. The positive control docking of all complex showed a successful prediction, indicated by the low root-mean-square deviation (RMSD) value between the docking- and co-crystal poses of ligand (<2 Å). Furthermore, compounds 5–8 from Chisocheton were docked to all of the NRs using the same parameter with that of positive control docking. The docking result was filtered based on the similarity of poses with the respective co-crystal ligand. It is shown that the docking pose of Chisocheton limonoids to theRXRβ, RXRα, ERα, ERβ, RARγ, AR, LXR, LRH1, and LXRα were different from that of the co-crystal ligand. Interestingly, the docking pose of Chicocheton limonoids was similar to that of PR and GR co-crystal ligands (Fig. 2). For this reason, the docking complex of Chisocheton limonoid with PR and GR was selected for further investigation using MD simulation.
The docking result showed that the carbonyl group at C-3 of all the limonoid compounds formed hydrogen bonds with Arg776 and Arg611 of PR and GR, respectively, similar to that of the co-crystal ligand. The Phe778 and Phe623 of PR and GR stabilized the A- and B-rings of limonoids and formed the hydrophobic interactions with a methyl group at C-4 of the compounds. Moreover, the C-ring of limonoids was stabilized by Leu718 of PR and Leu563 of GR. Met801 of PR formed hydrogen bond with the OH-group at the C-6 of compound 7. However, the ester group at C-6 and C-7 of compounds 5 and 6 formed weak interactions with the receptor. The ester group of limonoids sterically interacted with Leu797 of PR and Gln647 of GR. To improve the binding prediction, the docking score resulting from AutoDock Vina was re-scored using AutoDock 4.2 (Table 1).
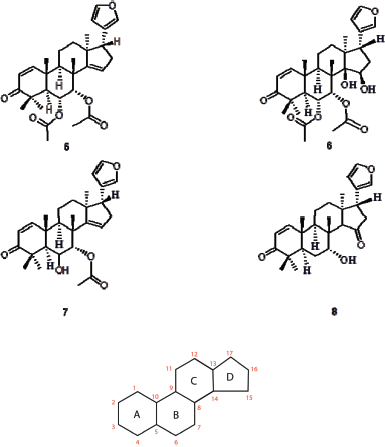 | Figure 1. Structure of compounds 5–8 isolated from Chisocheton. The numbering of the atom and the labeling of the ring are provided. (Laphookieo et al., 2008; Maneerat et al., 2008) [Click here to view] |
PR is one of the oncogenic proteins, which modulate cancer cells to proliferate, whereas GR utilizes endocrine hormones as their endogenous ligands that play as a tumor suppressor. The GR has been shown to contribute to the progression and survival of breast cancer by activating proliferative and resistance genes (Conzen, 2008; Dhiman et al., 2017). Since both PR and GR are expressed in the breast cancer cell, then these NR share necessary for initiating transcription of certain oncogenes. The structure and sequence of PR and GR are homologs with 55.2% of identity and 83.3% of similarity.
Moreover, the ligand-binding domain (LBD) of PR shares a similarity with that of GR. There are two distinct conformations related to their function, which are agonist- and antagonist-form. In the antagonist-form, Helix-12 of PR and GR shows high flexibility, preventing the helix from covering the LBD. On the other hand, Helix-12 in agonist-form covers the LBD (Kauppi et al., 2003; Lusher et al., 2011; Williams and Sigler, 1998; Zheng et al., 2016). It is noted that both PR and GR structures (1A28 and 4P6W) used in this study were complexed with agonist ligand (Williams and Sigler, 1998; Zhang and Xu, 2017).
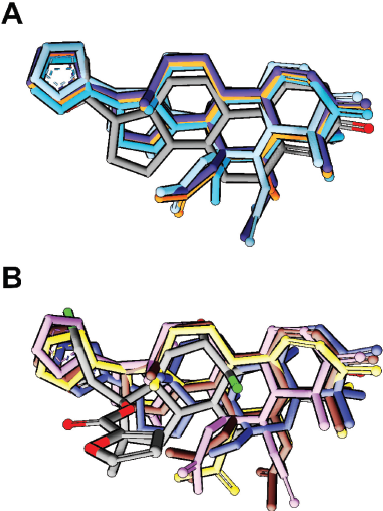 | Figure 2. Binding mode of all compounds in (A) PR and (B) GR. The docking pose of all compounds showed a similar orientation with the co-crystal ligand (gray color). [Click here to view] |
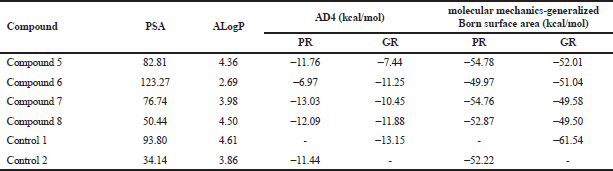 | Table 1. Docking and MD binding affinity. [Click here to view] |
In the MD simulations of the PR system, the backbone RMSD of the receptor of all the systems increased quickly in the first nanosecond and then went stable at 0.5–2.0 Å. This result indicated that the initial phase of the system was still looking for its relaxed conformation by decreasing the steric constraint between residues. The plot of RMSD also suggested no major changes in the flexibility of the protein structure, whereas the root-mean-square fluctuation (RMSF) plot suggested only a slight movement of Thr706 at the loop-1 in the complex system of PR with compound 8.
It is worth noting that the increasing fluctuation of Helix-12 and its previous loop was observed (Fig. 3). A sterical clash between ligand and Met909 at Helix-12 of PR was one of the key factors in initiating the antagonist response (Lusher et al., 2011; Zheng et al., 2016). However, unlike the RU-482, a well-known antagonist compound for PR and GR that can interact with Helix-12, all the limonoids (compounds 5 and 8) do not have bulky groups at C-11 to drive away from the Met909 of Helix-12. Nevertheless, compounds 5 and 6 have ester group at C-7, which is large enough to force the movement of limonoids toward Helix-12 (Fig. 5).
Different from the PR, Helix-12 in GR does not have a bulky side chain like methionine. Therefore, there are no sterical clashes observed between Helix-12 of GR and the limonoids. This observation was supported by the RMSF plot of the GR system, i.e., low fluctuation of the Helix-12 region. The RMSD plot of GR showed that in compound 7, the structural changes of the receptor were high (Fig. 4), whereas in compound 5 system, the ligand was not formed a hydrogen bond with Arg776 ( Fig. 6).
The substituents of compounds 5 and 6 are less hydrophobic than that of compounds 7 and 8. Their hydrophobicity was indicated by the PSA and LogP values. Since the binding site of PR and GR was hydrophobic, then compounds 7 and 8 were predicted to have better activity than the others (Williams and Sigler, 1998).
The conformational change throughout simulation was visually inspected. There was no major change in the conformation of PR and GR complexes. In the PR system, compound 7 slightly makes the receptor structure smaller than the other systems (Fig. 7). While in the GR system, compound 7 increased the fluctuations of the receptor compared to the other compounds.
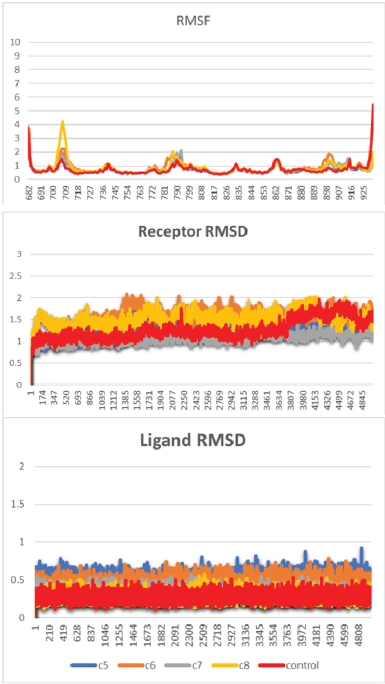 | Figure 3. RMSF and RMSD of the MD simulation of the PR system. [Click here to view] |
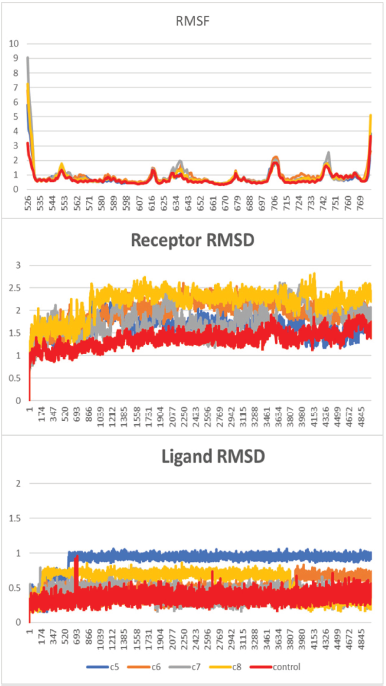 | Figure 4. RMSF and RMSD of the MD simulation of the GR system. [Click here to view] |
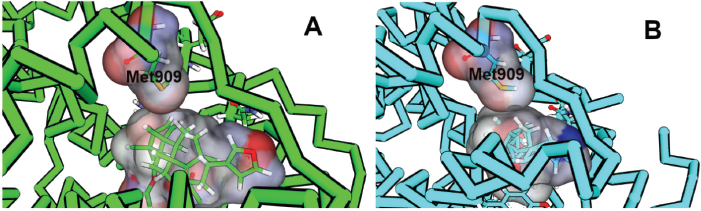 | Figure 5. The sterical clashes between Met909 of PR and limonoids. (A) Compound 5 showed a smaller gap with Met909 (part of Helix-12) than (B) compound 7. [Click here to view] |
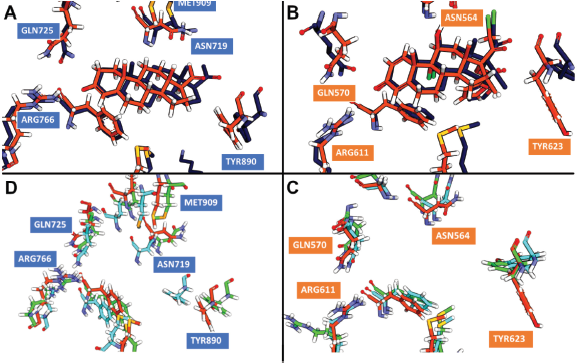 | Figure 6. The average structures from 50 ns of MD trajectory. The average structure of co-crystal ligand in (A) GR and (B) PR showed no difference between the initial structure (orange color) and the last structure (dark blue color) from MD simulation. (C) GR simulations showed no major deviation of binding residues, except Arg611 in compound 5 system (green color). (D) PR simulations showed the movement of binding residues closer to compound 7 (cyan). [Click here to view] |
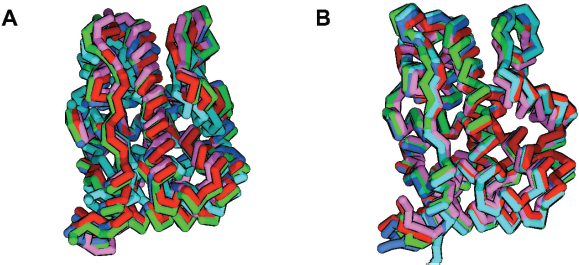 | Figure 7. The average structure of all simulations visualized in carbon alpha stick representation. (A) Compound 7 shrank the volume of PR binding site (cyan). (B) GR system is not affected by limonoid binding. [Click here to view] |
From the MD simulations, all limonoids were observed to form a direct hydrogen bond with Asn719 and Asn564 of PR and GR via an oxygen atom of furan. Previously, it is known that the inhibitor of PR always requires the presence of a water molecule to mediate the interaction with Asn719. This result suggested that the water molecule around Asn719 is not always needed for the inhibition of PR. In addition, Asn564 of GR, which is equivalent to Asn719 of PR, was appeared to be an important residue in mediating the agonist response (Lusher et al., 2011; Williams and Sigler, 1998; Zheng et al., 2016). This study should be further verified by the cell-based assay with T47D that expressed PR at a high level (Dhiman et al., 2017).
CONCLUSION
This study proposes a novel hypothesis on the molecular target of limonoids from Chisocheton sp., which are PR and GR. These compounds formed similar docking poses with the co-crystal ligands of PR and GR within a range of free energy of binding from −6.97 to −13.03 kcal/mol. Interestingly, MD simulations indicated a slight fluctuation of Helix-12 only on the PR system. Therefore, within the limitation of this study, it is suggested that the ligand antagonist activity was specifically only on PR, not GR. This finding warrants further development of limonoids as antibreast cancer, including a future work with a cell-based assay using a T47D cancer cell line, which expressed a high level of PR.
ACKNOWLEDGMENTS
The work of Nurlelasari was supported by grant PDUPT 2018, and the work of Desi Harneti was supported by the Internal Grant of Universitas Padjadjaran.
AUTHOR CONTRIBUTIONS STATEMENT
Nurlelasari Nurlelasari, Ade Rizqi Ridwan Firdaus, Desi Harneti, Nenden Indrayati, and Muhammad Yusuf conceived the experiment(s); Ade Rizqi Ridwan Firdaus, and Muhammad Yusuf conducted the experiment(s); and Ade Rizqi Ridwan Firdaus and Umi Baroroh analyzed the results. All authors reviewed the manuscript.
REFERENCES
Awang K, Lim CS, Mohamad K, Morita H, Hirasawa Y, Takeya K, Thoison O, Hadi AH. Erythrocarpines A–E, new cytotoxic limonoids from Chisocheton erythrocarpus. Bioorg Med Chem, 2007; 15:5997–6002. CrossRef
Choi J-M, Bothwell ALM. The nuclear receptor PPARs as important regulators of T-cell functions and autoimmune diseases. Mol Cells, 2012; 33:217–22. CrossRef
Conzen SD. Nuclear receptors and breast cancer. Mol Endocrinol, 2008; 22:2215–28. CrossRef
Danielian PS, White R, Lees JA, Parker MG. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J , 1992; 11:1025–33. CrossRef
Dhiman VK, Bolt MJ, White KP. Nuclear receptors in cancer uncovering new and evolving roles through genomic analysis. Nat Publ Gr, 2017; doi:10.1038/nrg.2017.102 CrossRef
Guthri N, Morley K, Hasegawa S, Manners GD, Vandenberg T. Inhibition of human breast cancer cells by citrus limonoids. In Citrus Limonoids. ACS Symposium Series. Washington DC, United State: American Chemical Society, 2000. CrossRef
Kauppi B, Jakob C, Färnegårdh M, Yang J, Ahola H, Alarcon M, Calles K, Engström O, Harlan J, Muchmore S, Ramqvist AK. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J Biol Chem, 2003; 278:22748–54. CrossRef
Kowshik J, Mishra R, Sophia J, Rautray S, Anbarasu K, Reddy GD, Dixit M, Mahalingam S, Nagini S. Nimbolide upregulates RECK by targeting miR-21 and HIF-1 α in cell lines and in a hamster oral carcinogenesis model. Sci Rep, 2017; 7:2045. CrossRef
Laphookie S, Maneerat W, Koysomboon S, Kiattansakul R, Chantrapromma K, Syers JK. A novel limonoid from the seed of Chisocheton siamensis. Can J Chem, 2008; 86:205–8. CrossRef
Lusher SJ, Raaijmakers HC, Vu-Pham D, Dechering K, Lam TW, Brown AR, Hamilton NM, Nimz O, Bosch R, McGuire R, Oubrie A, de Vlieg J. Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators. J Biol Chem, 2011; 286(40):35079–86. CrossRef
Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science, 1999; 284:1362–5. CrossRef
Maneerat W, Laphookie S, Koysomboon S, Chantrapromma K. Antimalarial, antimycobacterial and cytotoxic limonoid from Chisocheton siamensis. Phytomed, 2008; 15(12):1130–34. CrossRef
Nagoor NH, Shah Jehan Muttiah N, Soon Lim C, In LLA, Mohammad K, Awang K. Regulation of apoptotic effects by Erythrocarpine E, a Cytotoxic limonoid from chisocheton erythrocarpus in HSC-4 Human Oral Cancer Cells. PLoS One, 2011; 6:e23661. CrossRef
Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Heal, 2016; 4:609–16 . CrossRef
Sophia J, Kiran Kishore T. K, Kowshik J, Mishra R, Nagini S. Nimbolide, a neem limonoid inhibits Phosphatidyl Inositol-3 Kinase to activate glycogen synthase kinase-3β in a hamster model of oral oncogenesis. Sci Rep, 2016; 6:1–13. CrossRef
Tan Q-G, Luo X-D. Meliaceous Limonoids: chemistry and biological activities. Chem Rev, 2011; 111:7437–522. CrossRef
Thoh M, Babajan B, Raghavendra PB, Sureshkumar C, Manna SK. Azadirachtin interacts with retinoic acid receptors and inhibits retinoic acid-mediated biological responses [retracted in: J Biol Chem. 2013 Mar 22; 288(12):8563]. J Biol Chem, 2011; 286(6):4690–702. CrossRef
Williams SP, Sigler PB. Atomic structure of progesterone complexed with its receptor. Nature, 1998; 393:392–96. CrossRef
Zhang Y, Xu H. Recent progress in the chemistry and biology of limonoids. RSC Adv, 2017; 7:35191–220. CrossRef
Zheng L, Lin VC, Mu Y. Exploring flexibility of progesterone receptor ligand binding domain using molecular dynamics. PLoS One, 2016; 11:e0165824. CrossRef