INTRODUCTION
Cancer is a diversified disease, mainly recognized by unrestrained cell division that can lead toward mortality by acquiring metastatic form. Cancer has been declared the second most deadly disease that makes one out of eight deaths worldwide. According to the latest global cancer data released by the International Agency for Research on Cancer, nearly18.1 million new cases and 9.6 million deaths were reported in the year 2018 (WHO, 2018). Approximately 13.2 million cancer-related deaths and 21.4 million new cancer cases are expected by 2030 (Siegel et al., 2019). Unfortunately, all currently available chemotherapeutic agents suffer from undesirable off-target effects and frequently develop resistance. Thus, the development of potent and effective novel anti-neoplastic drugs is one of the most intensely persuaded goals of existing medicinal chemistry.
Epidermal growth factor receptor (EGFR) is a transmembrane tyrosine kinase receptor that belongs to the human epidermal growth factor receptor family. EGFR mediates the pathways of cell proliferation, growth, survival, and also involved in drug resistance. Pertaining to such involvement in cancer progression, EGFR has become an imperative target to discover novel chemotherapeutic agents (Sigismund et al., 2018). EGFR is highly expressed in many types of cancer cells; a study reveals that EGFR is over-expressed in 14%–91% of breast cancer cases (Fox et al., 2007; Klijn et al., 1992). Advantageously, EGFR provides a larger hydrophobic pocket in which bigger molecules like Erlotinib (Yun et al., 2007) can be docked easily.
Coumarin (2H-1-benzopyran-2-one) is found in the entire plant kingdom, including essential oils such as cinnamon bark oil and cassia leaf oil, and fruits, green tea, and foods such as chicory (Skehan et al., 1990). Coumarin derivative (Fig. 1), i.e., 7,8 dihydroxycoumarin (daphnetin) has been reported to potentially inhibit EGFR tyrosine kinase (Yang et al., 1999) and effectively bind in a hydrophobic pocket of EGFR with good docking score (Kumar et al., 2018). Moreover, Coumarin hybrids (Fig. 1) containing 1,2,3-triazole ring have been reported to inhibit cancer cells with good IC50 values (Pingaew et al., 2014; Singh et al., 2016; 2017).
Uracil (pyrimidine 2,4 dione) is a common nucleobase present in the nucleic acid of RNA. In RNA, Uracil makes two hydrogen bonds with adenine and in DNA sequencing thymine replaces Uracil nucleobase. Owing to have DNA intercalation properties, Uracils impart anti-cancer activity (Fadda and Pome 2011). Analogous to Coumarin, Uracil hybrids (Fig. 2) containing 1,2,3-triazole moiety in their structure have also been proved to actively inhibit various cancer cells with lesser IC50 values (Kumar et al., 2012; Ruddarraju et al., 2016). Concerning to EGFR inhibition, 5-Fluorouracil (5-FU) is reported to form a complex with epidermal growth factor (EGF) protein. This 5FU-EGF complex when docked with EGFR protein results in more affirmative binding (Kanagathara and Kavitha, 2015). Furthermore, 5-FU in combination with oxaliplatin also found to increase EGFR phosphorylation in human colorectal cancer cell lines (Schaeybroeck et al., 2005).
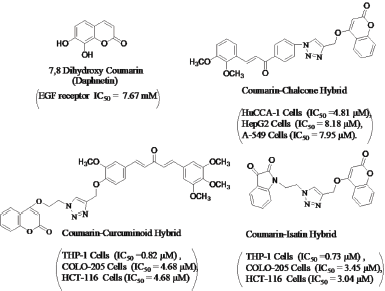 | Figure 1. Anti-cancer profile of Daphnetin and some Coumarin hybrids bearing the triazole ring. [Click here to view] |
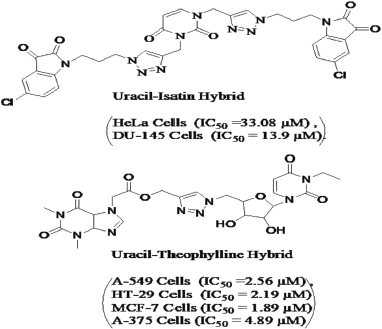 | Figure 2. Anti-cancer profile of some Uracil hybrids bearing the triazole ring. [Click here to view] |
Furthermore, incorporation of 1,2,3-triazole ring between two pharmacophores is evidently a useful tool to produces bifunctional hybrid molecules with improved efficacy toward cancer target (Huber et al., 2009; Jagasia et al., 2009; Zhang et al., 2009). EGFR docking studies of some hybrids confirmed that the role of the triazole ring is in making water-mediated H-bond with Thr766, Lys721, Arg817, and Met769 of target’s amino acid residues (Banerji et al., 2018; Nguyena et al., 2018).
The foremost cytotoxic potential of the moieties Uracil, Coumarin, and 1,2,3-triazole prompted us to design a scaffold of 28 novel bi-functional N1, N3 Uracil hybrids linked with Coumarin via 1,2,3-triazole using different carbon chain linker (n = 1 − 4) to get the synergistic inhibition on EGFR. Docking study was performed to find out the potential ligand amid all designed hybrids, which have the best affinity toward the EGFR tyrosine kinase domain. Virtual screening (2D and 3D) of all potential hybrids was compared with standard Erlotinib, a well-known EGFR inhibitor. The binding efficacy of the ligands has been estimated on the basis of Glide score (gScore) and Emodel score because Glide uses Emodel to pick the “best” pose of a ligand (pose selection). Various interactions between ligand and protein were also studied and compared with standard using premium Schrödinger software. To explore further, molecular dynamics studies on different parameters were performed using Groningen Machine for Chemicals Simulations software to check the stability of the ligand–protein complex and to observe the h-bonding interactions in motion. We expect that this study will provide a useful approach for anticancer drug discovery.
MATERIALS AND METHODS
In this present study, the initial binding study was carried out using molecular docking at standard precision (SP) using Glide (Schrödinger Suites 2017-2, LLC, Cam-bridge, USA). The hardware used for this study was from DELL brand with Intel Core i7-6700 clocked at 3.40 GHz X 8 processor with 8 GB RAM running on 64-bit CENT OS Linux operating system.
Molecular docking
Preparation of enzyme
The 3-dimensional (3D) crystallographic model of EGFR tyrosine kinase [Protein Data Bank (PDB) ID: 1M17] was retrieved from Protein Data Bank (www.rcsb.org) using the Protein preparation wizard in Maestro. The protein was preprocessed by performing assign bond orders, removing original hydrogens, adding hydrogens, creating zero-order bonds to metals, creating disulfide bonds, converting selenomethionines to methionines, filling in missing side chains using prime, filling in missing loops using prime, deleting water molecules beyond 5 Å from hetero groups, and generating het states using Epik at pH: 7.0 ± 2.0. Then, the protein was refined by doing H-bond assignment by sample water orientations, using PROPKA pH: 7.0, and optimization was carried out. The restrained protein minimization was performed using OPLS3 force field.
Receptor grid generation
Receptor grid of size 20 Å was generated after preparing the protein and by selecting the co-crystallized ligand Uracil–Coumarin hybrid inside the cavity of EGFR tyrosine kinase as the centroid of the active site.
Preparation of ligand
All the molecules were built, 3D optimized, and energy minimized using ligprep and confgen function in Maestro. The library was saved in .maegz format.
Molecular docking
Subsequently, molecular docking was performed using Glide at the SP algorithm to evaluate the protein–ligand interaction and their binding affinities. For our analysis, we checked to add Epik state penalties to docking score and enhanced planarity of conjugated pi groups. The validation for molecular docking was done by re-docking the co-crystalized ligand Uracil–Coumarin hybrid in the grid. The resulting poses and interactions of co-crystalized ligand and library were compared.
Molecular dynamics simulation
Based on docking results, the lowest energy and best-posed complex of molecules A6 were selected for the simulation study using Groningen Machine for Chemicals Simulations 2019 package (Pronk et al., 2013) with OPLS2015 force field. Ligand topology files were generated using PRODRG (Schuttelkopf and van Aalten, 2004). The charge of the system was neutralized by the addition of the ions. The energy minimization of the complex (nsteps ¼ 50,000) was executed using the steepest descent approach (1,000 ps). The particle mesh Ewald method was employed for energy calculation and for electrostatic and Van der Waals interactions. The linear constraint solver (Amiri et al., 2007) algorithm was used for covalent bond constraints. Finally, 50 ns molecular dynamics simulation (MSD) with periodic boundary conditions was conducted for respective protein–ligand complexes with nsteps 5,00,000. The root-mean-square deviation and fluctuation (RMSD/F), intramolecular hydrogen bonds, radius of gyration (ROG) (Rg), and thermodynamic parameters were analyzed using Xmgrace (http://plasma-gate.weizmann.ac.il/Grace/) and VMD 1.9.2 software.
RESULTS AND DISCUSSION
Molecular docking studies
Docking studies against the target protein EGFR (PDB ID: 1M17) that involved in cancer cell proliferation and growth were carried out for 28 novel designed Uracil–Coumarin hybrid compounds (parent structure is given in the following) to check the binding affinity of ligands with the receptors. To validate the applied docking method, corresponding ligands were docked into the tyrosine kinase domain of EGFR using Schrödinger software. The protein structures presented were natural substrates as a co-crystallized ligand, whereas in others, the co-crystallized ligand was a known EGFR inhibitor (i.e., Erlotinib) took as the reference drug (Fig. 3); in both cases, the same docking and scoring validation process were used. Each co-crystallized ligand was previously removed from the respective protein binding site. The predicted docking poses 2D and 3D were compared with the experimental co-crystallized binding pose.
All the data and docking gScore (Glide score is related to ligand binding free energy) and Emodel score (related to electrostatic and van der Waals energies) are provided in Table 1. The hydrogen-bonding interactions, pi-pi stacking interactions, and pi-cation stacking interactions of ligands with the target protein residues were also mentioned. Amid 28 designed hybrid compounds, almost all of them exhibit good interactions with the target protein EGFR tyrosine kinase.
In comparison to standard Erlotinib, compounds A6, A2, and A7 (Fig. 4) were found to show better interactions with target and having a docking score of −8.891, −8.712, −8.658 kcal/mol. The hybrid compound A6 exhibits the most effective binding with target protein EGFR tyrosine kinase (PDB ID: 1M17). In terms of affinity, A6 forms four different hydrogen-bonding interactions; out of them, three H-bonds were with Cys773, Met769, Lys721 amino acid residues and one with water (H2O) molecule with gScore −8.891 kcal/mol and Emodel score −100.744. On the other side, reference drug Erlotinib forms three hydrogen bonding interactions; two of them were with Cys773, Met769 and one with (H2O) molecule with gScore −8.538 kcal/mol and Emodel score −80.588.
Virtual 2D and 3D screening comparison between most active hybrid ligand (A6) and standard Erlotinib (Figs. 5 and 6) clearly indicates that A6 forms similar kind of interactions and same mechanistic inhibition as of standard drug follows. One additional hydrogen bonding with Lys721 amino acid residue, extra nitrogen heteroatom, and flexibility provided by the linker makes it a superior ligand for the target EGFR.
Various individual hydrogen bonding interactions with virtual bond length in Å of compound A6 with target protein EGFR tyrosine kinase are depicted in Figure 7, and interactions of reference Erlotinib are also shown in Figure 8 for more focused comparison.
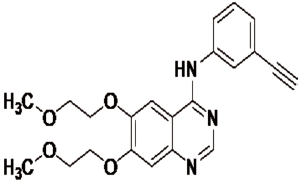 | Figure 3. Structure of reference drug Erlotinib (a well-known EGFR Inhibitor). [Click here to view] |
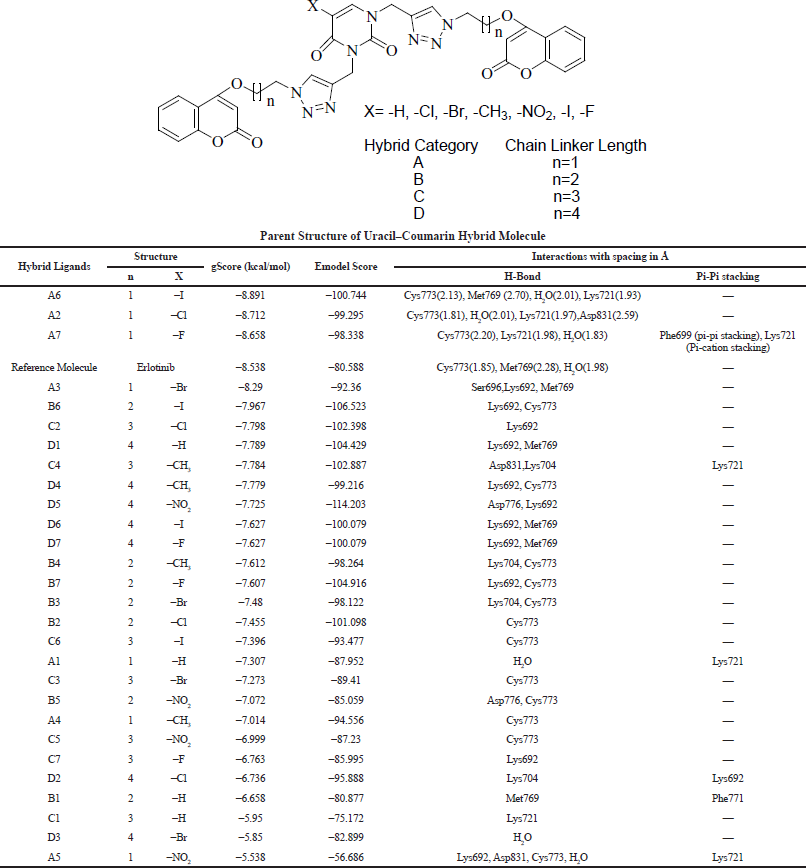 | Table 1. Docking score and interactions of hybrid ligands with EGFR tyrosine kinase. [Click here to view] |
Referring to Table 1, hybrid compounds A2 and A7 were also reported to have good gScore and Emodel score as compared to the reference drugs. The compound A2 with protein EGFR forms four hydrogen-bonding interactions with amino acid residues Cys773, Lys721,Asp831 and one interaction with water H2O molecule. All the interactions of A2 with the target EGFR are shown in Figure 9 in both 2D and 3D dimensions.
Hybrid compound A7 was also found to have hydrogen bonding interactions with Cys773, Lys721, and water (H2O). Moreover, compound A7 also has pi-pi stacking and pi-cation stacking interactions with Phe699 and Lys721 residues, respectively. All the interactions are collectively mentioned in Figure 10, whereas additional interactions make it an effective binder in comparison to standard Erlotinib.
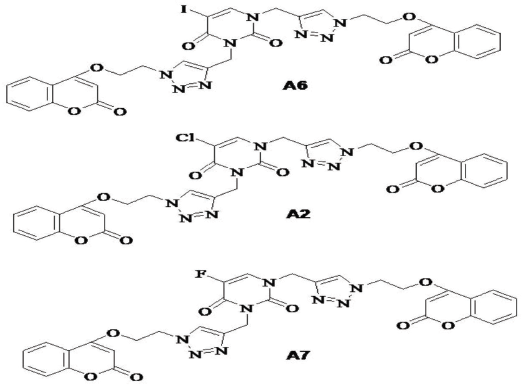 | Figure 4. Structure of most effective hybrid ligands A6, A2, and A7. [Click here to view] |
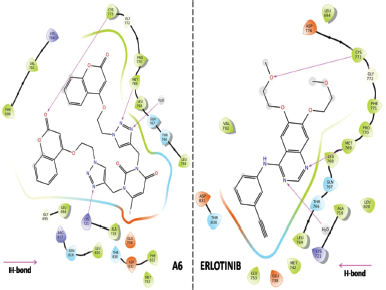 | Figure 5. Two-dimensional (2D) binding interaction comparison between A6 hybrid and Erlotinib (standard). [Click here to view] |
Hence, on the basis of docking scores of hybrid compounds in Table 1, it can be suggested that among the library of hybrid ligands, Uracil–Coumarin hybrids having two carbon chain length (n = 1) with halogen substituent(X = −I, −Cl, −F) at Uracil moiety A6, A2, and A7 were found to have the best affinity for target EGFR. Although A3 with bromine substituent (X = Br) is found to form prominent interactions with target with comparable docking score (−8.29 kcal/mol) to the reference Erlotinib (−8.538 kcal/mol). Being in the same category, compound A5 showed interaction with poor binding score, which justifies its least affinity toward EGFR and ineffective binding too. Compounds A1, A4, and A5 with substituents other than halogens (X = −H –CH3, and −NO2) were found to have a lower affinity and poor docking scores as well.
Molecular dynamic simulations study
MSD is very important to understand structure-to-function relationships in motion and other adaptive conformational changes by the proteins (Gelpí et al., 2015). Complying to this, an MSD study was performed to evaluate the dynamic behavior of the top-scoring ligand A6 in complex with EGFR tyrosine kinase (PDB ID: 1M17) for a period of 50 ns. The study was analyzed on the basis of parameters such as protein-ligand RMSD, root-mean-square fluctuation (RMSF), ROG, thermodynamic parameters, and protein–ligand interactions formed.
RMSD and RMSF analysis
Analyzing the protein RMSD can give insights into a structural conformation that protein undergoes throughout the simulation. The plots for protein Cα versus time (50 ns) for the simulation are shown in Figure 11, which shows that complex attained an equilibrium value of 0.85 nm at around 30 ns. At Initial stage, a sharp increase in the deviation value from 0.2 to 0.8 nm occurs in the first 3 ns pointing toward a large conformational change in the protein during that period.
Analyzing the ligand backbone RMSD from the plot in Figure 12 indicates how stable the ligand is with respect to the protein and its binding pocket. The plot for ligand RMSD (nm) versus time (50 ns) shows a plateau value of 0.3 nm for the ligand A6 at around 10 ns. The deviation value observed was within limits, which is quite stable for binding of the ligand with the protein.
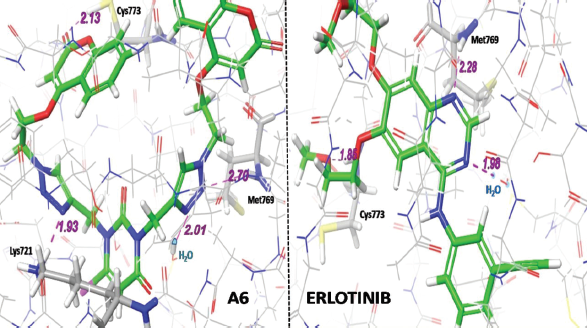 | Figure 6. Three-dimensional (3D) binding interaction comparison between A6 hybrid and Erlotinib (standard). [Click here to view] |
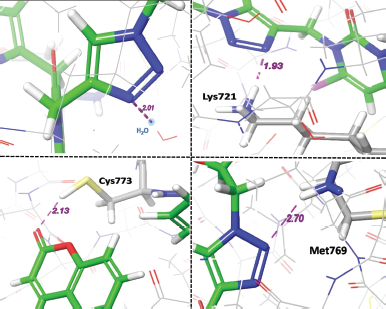 | Figure 7. Interactions of compound A6 with target EGFR tyrosine kinase. [Click here to view] |
The protein RMSF is useful for characterizing local changes along the protein chain. The plot for protein RMSF (nm) versus residue number index shown in Figure 13 describes fluctuations in the range of 0.1–1.4 nm for the complex. The fluctuations in both the terminal ends were higher. The catalytic residue values are no higher than 0.3 nm and are suitable for stable binding.
The ligand RMSF is useful for characterizing adaptive changes in the ligand atom positions (Dong et al., 2018) and can give insights into how ligand fragments interact with the protein and their entropic role in the binding event. Ligand with their atomic index number is shown in Figure 14.
The plot for ligand RMSF (nm) versus ligand atom index shown in Figure 15 describes fluctuations in the range of 0.05–0.33 nm for ligand A6. The central Uracil fragment acts as the core and holds the two triazole-linked coumarins like two arms attached to the core. The Uracil fragment has the least fluctuation which is nearly 0.1 nm. The triazoles attached tend to fluctuate between 0.1 and 0.2 nm, and the coumarins present at the extended length tend to fluctuate above 0.2 nm. The higher value of RMSF (nm) for coumarin fragments may be due to the three atom long linker that attaches them to triazoles.
H-bond analysis
The H-bond interactions formed during the Molecular Dynamics simulations up to 50 ns were also examined. The plot in Figure 16 represents the number of H-bond contacts formed between the ligands and the protein. Ligand A6 shows one stable H-bond contact during the simulation, although occasionally two H-bond contacts, during the period of 26–42 ns, were also formed. From the discussion, it can be summarised that A6 has an efficient binding inside the cavity of EGFR tyrosine kinase.

The H-bonding interaction fraction shown in Figure 17 conveys that stable H-bonding occurs with residue MET769 for around 38% of duration, followed by supplementary H-bond interactions with residues ARG817, LYS704, and TYR703 with interaction for a duration of 8.34%, 6.30%, and 3.90%, respectively.
Results also suggested that irrespective of substituent X on Uracil moiety, the carbon chain linker (n = 1 − 4) played an important role in making an effective binding pose between ligand and the target. Table 1 clearly suggested that as the carbon linker chain increase gScore of the ligand varies continuously from stable −8.891 kcal/mol to unstable −5.53 kcal/mol. The variation in docking scores clearly indicates that as the linker chain length increases, the flexibility of the molecule increases too, which makes the molecule either unfit to bind in the tyrosine kinase domain of the EGFR or the ligand is unable to make an effective binding pose with protein residues. Evidently, molecular dynamics studies of most effective ligands A6 provided additional proof of forming a stable complex with target EGFR. Ligand A6 forms plateau phase within 10 ns, which retains affirmative up to 50 ns time fraction with little RMSD of 0.3 nm. Both the studies suggested that bigger linker chain hybrids have the property of flexibility and their larger size keep the ligand out of range of interactions, which led to weaker binding interactions and poor docking score. Contrary to this, hybrid molecules with the shortest linker form more affirmative binding probably due to their less flexibility, higher residue interactions in comparison to standard and their fit size and fit size according to the pocket of the kinase domain of EGFR were proved to be the best ligands.
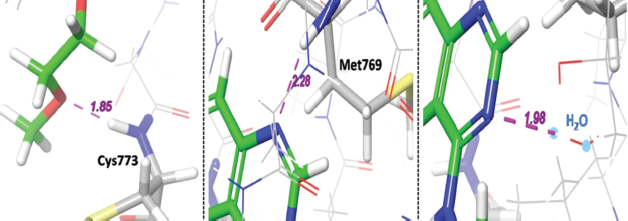 | Figure 8. Interactions of reference Erlotinib with target EGFR tyrosine kinase. [Click here to view] |
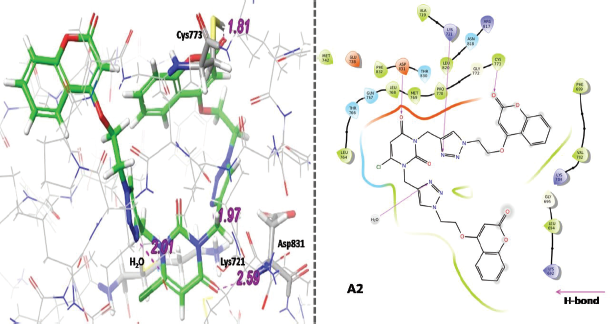 | Figure 9. Three-dimensional (3D) and two-dimensional (2D) binding interaction of A2 with target EGFR. [Click here to view] |
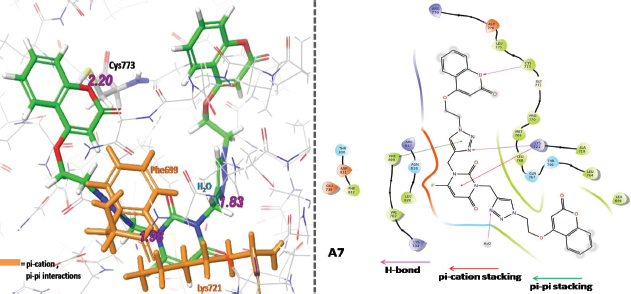 | Figure 10. Three-dimensional (3D) and two-dimensional (2D) binding interaction of A7 with target EGFR. [Click here to view] |
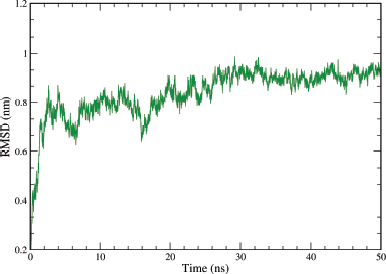 | Figure 11. Graphical representation of the plots showing protein CαRMSD (nm) versus time (50 ns) for A6 ligand-complex of EGFR tyrosine kinase (PDB ID: 1M17). [Click here to view] |
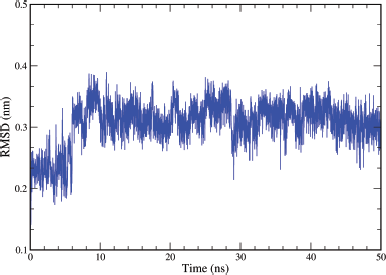 | Figure 12. Graphical representation of the plots showing ligand backbone RMSD (nm) versus time (50 ns) for A6 ligand complex of EGFR tyrosine kinase (PDB ID: 1M17). [Click here to view] |
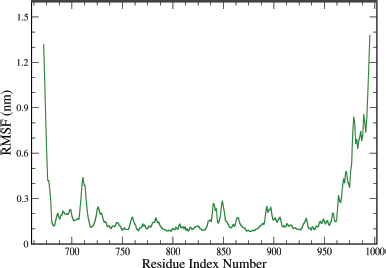 | Figure 13. Graphical representation of the plots showing the protein RMSF (nm) versus residue index number of protein for A6 ligand complex of EGFR tyrosine kinase (PDB ID: 1M17). [Click here to view] |
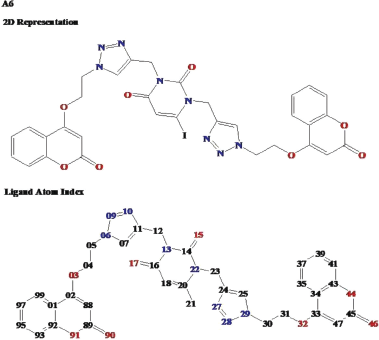 | Figure 14. 2D representation and ligand atom index representation of ligand A6. The ligand atom index numbers are written in short for the last two digits, the numbers 88–99 stand for 3288–3299 and 01–47 stand for 3301–3347, respectively. [Click here to view] |
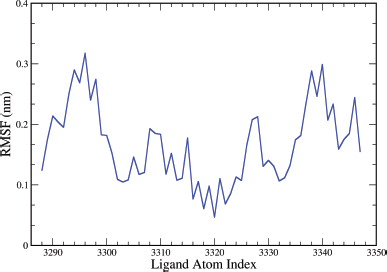 | Figure 15. Graphical representation of the plots showing the ligand RMSF (nm) versus ligand atom index of ligand for A6 ligand complex of EGFR tyrosine kinase (PDB ID: 1M17). [Click here to view] |
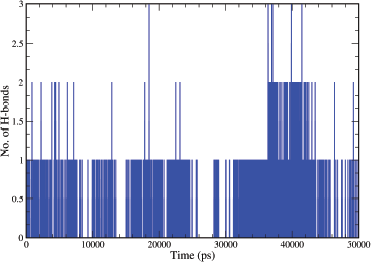 | Figure 16. Pictorial representation of the number of H-bond contacts formed by ligand A6 with EGFR tyrosine kinase (PDB ID: 1M17). [Click here to view] |
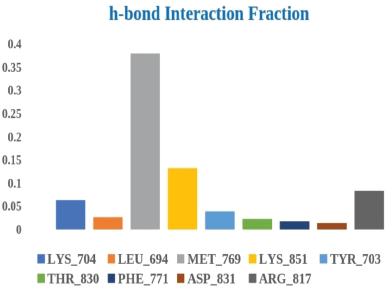 | Figure 17. Histogram representation of the H-bond interaction fraction of ligand A6 with EGFR tyrosine kinase (PDB ID: 1M17). [Click here to view] |
Proposed synthetic route for designed hybrids
Hybridization of active pharmacophores is a modern approach to synthesized multiple-targeted anti-cancer compounds (kucuksayan and Ozben, 2016). Generally, 1,2,3-triazole tethered hybrids having two active moieties can be easily synthesized via click chemistry approach (Nepali et al., 2013). Various N1,N3 bifunctional Uracil-1,2,3-triazole linked hybrids have been reported to have an anti-cancer profile (Lal and Yadav, 2018); and all kinds of 1,4-disubstituted 1,2,3-triazole hybrids have been synthesized via copper(I)-catalyzed cycloaddition reaction between organic azides and terminal alkynes (Gregorić et al., 2017; Lal and Yadav, 2018). Following these existing procedures, Uracil–Coumarin hybrids tethered via 1,2,3-triazole ring could be synthesized by the reaction mentioned in Scheme 1.
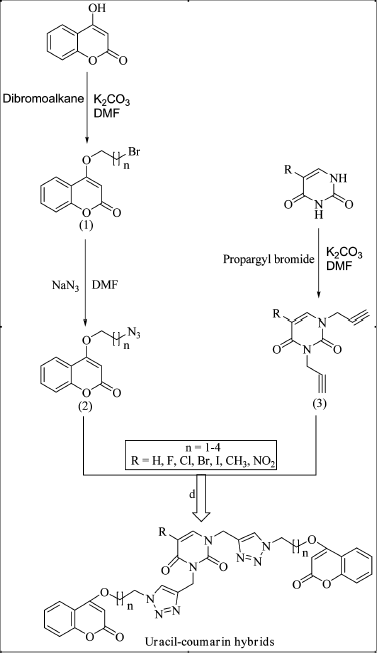 | Scheme 1. Proposed synthetic scheme of 1,2,3-triazole tethered Uracil–Coumarin hybrid. [Click here to view] |
CONCLUSION
This study emphasizes on the potential interactions between 1,2,3-triazole tethered Uracil–Coumarin hybrids and target protein EGFR tyrosine kinase. Docking results clarified that all hybrid ligands have good H-bonding interactions with the target and few of them also showed pi-pi stacking interactions too. It was also deduced from the results that short carbon chain linker (n = 1) between triazole and Coumarin ring and a halogen substituent (X = −I, −Cl, −F, −Br) at Uracil moiety are two essential parameters for effective binding and to fit properly in the active tyrosine kinase pocket of EGFR. In the library of designed hybrids (A1–D7), compounds A6, A2, and A7 are reported to have good binding interactions as compared to standard Erlotinib. Hybrid compound A6 was found to have the highest binding affinity and form a perfect binding pose with EGFR, justified by gScore and Emodel score. Furthermore, the molecular dynamic study of the most effective ligand A6 has proved the stability of ligand–protein complex with little deviation and fluctuation up to 50 ns. Hence, it can be concluded that hybrid ligand A6 can serve as the lead compound in the designing of a novel anticancer molecule. Hopefully, the results may help the medicinal chemist in designing and development of a new hybrid drug having a better anti-cancer profile.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
- Mohit Sanduja: Conceived the presented idea, literature search, designed the molecules, found the target for the study, and planned for their computational analysis, and also took the lead in writing of manuscript according to the review of literature and results obtained.
- Jyoti Gupta: Supervised the project. Investigated and contributed to finalizing the manuscript.
- Ravi Rawat: Performed the computational studies of the designed molecules and derived the results, and also participated in the writing of the manuscript.
- Uttam Singh: Performed the computational MSD studies and derived the graphs of the study and helped in drafting the manuscript.
- Saurabh M. Verma: Participated as a reviewer and helped in overall direction and planning.
ACKNOWLEDGMENT
The authors would like to thank the Department of Pharmaceutical Sciences and Technology, Birla Institute of Technology, Mesra, for the constant support in molecular docking and dynamics studies.
REFERENCES
Amiri S, Sansom MSP, Biggin PC. Molecular dynamics studies of AChBP with nicotine and carbamylcholine: the role of water in the binding pocket. Protein Eng Des Sel, 2007; 20(7):353–9. CrossRef
Banerji B, Chandrasekhar K, Sreenath K, Roy S, Nag S, Saha KD. Synthesis of triazole-substituted quinazoline hybrids for anticancer activity and a lead compound as the EGFR blocker and ROS inducer agent. ACS Omega, 2018; 3:16134−42. CrossRef
Dong YW, Liao ML, Meng XL, Somero GN. Structural flexibility and protein adaptation to temperature: molecular dynamics analysis of malate dehydrogenases of marine molluscs. Proc Natl Acad Sci USA, 2018; 115(6):1274–9. CrossRef
Fadda E, Pome R. On the molecular basis of uracil recognition in DNA: comparative study of T-A versus U-A structure, dynamics, and open base-pair kinetics. Nucleic Acids Res, 2011; 39(2):767–80. CrossRef
Fox SB, Smith K, Hollyer J, Greenall M, Hastrich D, Lexes AL. Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell, 2007; 11:217–27. CrossRef
Gelpí JL, Hospital A, Goñi JR, Orozco M. Molecular dynamics simulations: advances and applications. Adv Appl Bioinforma Chem, 2015; 8(1):37–47. CrossRef
Gregorić T, Sedić M, GrbÄić P, Tomljenović Paravić A, Kraljević Pavelić S, Cetina M, Vianello R, Raić-Malić S. Novel pyrimidine-2,4-dione–1,2,3-triazole and furo[2,3-d]pyrimidine-2-one–1,2,3-triazole hybrids as potential anti-cancer agents: synthesis, computational and X-ray analysis and biological evaluation. Eur J Med Chem, 2017; 125:1247–67. CrossRef
Huber D, Hubner H, Gmeiner P. 1,1′-disubstituted ferrocenes as molecular hinges in mono- and bivalent dopamine receptor ligands. J Med Chem, 2009; 52:6860–70. CrossRef
Jagasia R, Holub JM, Bollinger M, Kirshenbaum K, Finn MG. Peptide cyclization and cyclodimerization by CuI-mediated azide−alkyne cycloaddition. J Org Chem, 2009; 74:2964–74. CrossRef
Kanagathara N, Kavitha K. Molecular docking studies on EGF – 5 fluorouracil. Asian J Pharm Sci Clin Res, 2015; 8(2):463–5.
Klijn JG, Berns PM, Schmitz PI, Foekens JA. The clinical significance of epidermal growth factor receptor (EGF-R) in human breast cancer: a review on 5232 patients. Endocr Rev, 1992; 13(1):3–17. CrossRef
Kucuksayan E, Ozben T. Hybrid compounds as multitarget directed anticancer agents. Curr Top Med Chem, 2016; 17(8):907–18. CrossRef
Kumar A, Sunita P, Jha S, Pattanayak SP. 7,8-dihydroxycoumarin exerts antitumor potential on DMBA-induced mammary carcinogenesis by inhibiting ERα, PR, EGFR, and IGF1R: involvement of MAPK1/2-JNK1/2-Akt pathway. J Physiol Biochem, 2018; 74(2):223–34. CrossRef
Kumar K, Sagar S, Easu L, Kaur M, Kumar V. Synthesis of novel 1H-1,2,3-triazole tethered C-5 substituted uracil–isatin conjugates and their cytotoxic evaluation. Eur J Med Chem, 2012; 58:153–9. CrossRef
Lal K, Yadav P. Recent advancements in 1,4-disubstituted 1H-1,2,3-triazoles as potential anticancer agents. Anticancer Agents Med Chem, 2016; 18(1):21–37. CrossRef
Nepali K, Singh J, Sharma S, Saxena AK, Bedi PMS. Synthesis of 1,2,3-triazole tethered bifunctional hybrids by click chemistry and their cytotoxic studies. Med Chem Res, 2013; 22(7):3160–9. CrossRef
Nguyena TV, Van Dinh T, Pham-The H, Quang HN, Thi NN, Thi TA, Thi PH, Le Thi TA, Nguyen HT, Thanh PN, Le Duc T. Design, synthesis and evaluation of novel hybrids between 4-Anilinoquinazolines and substituted triazoles as potent Cytotoxic agents. Biorganic Med Chem Lett, 2018; 28(23–24):3741–7. CrossRef
Pingaew R, Saekee A, Mandi P, Nantasenamat C, Prachayasittikul S, Ruchirawat S, Prachayasittikul V. Synthesis, biological evaluation and molecular docking of novel chalcone-coumarin hybrids as anticancer and antimalarial agents. Eur J Med Chem, 2014; 85(6):65–76. CrossRef
Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, Van Der Spoel D, Hess B. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation tool kit. Bioinformatics, 2013; 29(7):845–54. CrossRef
Ruddarraju RR, Murugulla AC, Kotla R, Chandra Babu Tirumalasetty M, Wudayagiri R, Donthabakthuni S, Maroju R, Baburao K, Parasa LS. Design, synthesis, anticancer, antimicrobial activities and molecular docking studies of theophylline containing acetylenes and theophylline containing 1,2,3-triazoles with variant nucleoside derivatives. Eur J Med Chem, 2016; 123:379–96. CrossRef
Schaeybroeck SV, McCaul AK, Kelly D, Longley D, Galligan L, Cutsem EV, Johnston P. Epidermal growth factor receptor activity determines response of colorectal cancer cells to Gefitinib alone and in combination with chemotherapy. Clin Cancer Res, 2005; 11(20):7480–9. CrossRef
Schuttelkopf AW, van Aalten DM. PRODRUG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr, 2004; 60:1355–63. CrossRef
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: A Cancer J Clin, 2019; 69(1):7–34. Available via http://doi.wiley.com/10.3322/caac.21551
Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Mol Oncol, 2018; 12(1):3–20. CrossRef
Singh H, Kumar M, Nepali K, Gupta MK, Saxena AK, Sharma S, Bedi PMS. Triazole tethered C5-curcuminoid-coumarin based molecular hybrids as novel antitubulin agents: design, synthesis, biological investigation and docking studies. Eur J Med Chem, 2016; 116(30):102–115. CrossRef
Singh H, Singh JV, Gupta MK, Saxena AK, Sharma S, Nepali K, Bedi PMS. Triazole tethered isatin-coumarin based molecular hybrids as novel antitubulin agents: design, synthesis, biological investigation and docking studies. Bioorg Med Chem Lett, 2017; 27(17):3974–9. CrossRef
Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. New colorimetric cytotoxicity assay for anticancer-drug screening. J Nat Cancer Institute, 1990; 83(13):1107–12. CrossRef
WHO (World Health Organization). Latest global cancer data: cancer burden rises to 18.1 million new cases and 9.6 million deaths in 2018. Int Agency Res Cancer, 2018: 13–5. Available via: http://gco.iarc.fr/ (accessed 24 August 2019).
Yang EB, Zhao YN, Zhang K, Mack P. Daphnetin, one of coumarin derivatives, is a protein kinase inhibitor. Biochem Biophys Res Commun, 1999; 260(3):682–5. CrossRef
Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell, 2007; 11(3):217–27. CrossRef
Zhang H, Zhang J, Cai WX, Yu LP, Zhen XC, Zhang A. Click’ D1 receptor agonists with a 5-HT1A receptor pharmacophore producing D2 receptor activity. Bioorg Med Chem, 2009; 17:4873–80. CrossRef