INTRODUCTION
Biotechnology mediation in the aquatic habitat has unbolted fresh horizons to isolate new organisms that are a powerful source of bioactive compounds. Actinobacterium is an effective ingredient of microbial populations and establishes balanced and continuous populations in multiple marine habitat ecosystems, and is a significant source of new secondary bioactive metabolites (Cumsille et al., 2017; Das et al., 2006; Girão et al., 2019; Mohammadipanah and Wink, 2016 ; Ramesh and William, 2012). The uncaged marine sediments contain a broad range of novel microorganisms, showing a varied and unexploited repository of microorganisms, is essential in obtaining a complex array of innovative metabolic substances and income producing a discovery platform for the pharmaceutical industry (Baltz, 2007; Coutinho et al., 2018; Genitsaris et al., 2016 ; Stach et al., 2003; Sponga et al., 1999). Actinobacterium is an important marine bacterial source for its ability to produce complex commercial lead molecules (Imada, 2005). Actinobacterium lives in distinct extreme settings, is better documented for its capacity to secrete natural compounds with architectural complication and varied biological actions, and is deemed priceless commercially significant prokaryote that is capable of producing antibiotics, anti-tumor agents, immunosuppressive agents, and enzymes (Abdelmohsen et al., 2014; Shuvankar et al., 2015).
Optimization of the physico-chemical parameters along with the media constituents is crucial for high yields of the antimicrobial metabolites (Deka and Jha, 2018; Linda and Rabab, 2017 ; Ripa et al., 2009). Conventional methods for optimization involve changing one independent variable, while other factors are kept constant using one factor at a time that is time-intensive, overpriced for a full total of variables, and cannot evaluate the connected impact of all variables and draw false findings (Marimuthu et al., 2009). Response surface methodology (RSM) investigates and approximates the unknown function using experimentation, modeling, and data analysis (Box and Wilson, 1951; Heinze et al., 2018). RSM is a mixture of statistical and mathematical approach to experiment design, model building, analysis of the interactive impacts of multiple nonpartisan variables against answers (Kishore and Kayastha, 2012).
A mathematical model is set for hypotheses concerning a natural phenomenon translated into mathematical expressions. The experimental data obtained generally confirm the predictions based on these models. Mathematical models not only provide a complete knowledge of dynamic behavior but also optimize and control most of the fermentation processes. Microbiologists recognize the importance of models when created unstructured models mimic bioactive metabolite manufacturing profiles and describe the kinetic interactions between substrate, product, and biomass. Batch mode operated responses are hard to model due to the time and features of cell processes, which often result in nonlinearities. In unstructured models that provide a precise description of whole fermentation, only complete cell levels are considered. Parameter estimation is a critical step for model verification, and consequently, these mathematical models are effectively implemented to study the manufacturing of a multitude of bioproducts (Rama Krishna et al., 2016; Ushakiranmayi et al., 2016).
Liquid chromatography coupled with mass spectrometry is a powerful device for the evaluation of active constituents of actinobacteria because of its high sensitivity, selectivity, and rapid screening ability. The improvement of LC-MS/MS analytical technique requires optimization of both chromatographic and mass spectrometric conditions (Ouakhssase et al., 2019). Once the approach is advanced, it must be proven for selectivity, sensitivity, linearity, accuracy, and precision. In the present study, the morphologically distinct potent actinobacterial isolate, VSM 3, was selected from 57 isolated actinobacterial strains and was classified as Saccharothrix flava (KU507600) by polyphasic taxonomy. The present manuscript has been reported for RSM-based optimization and interactive effects of the variables to produce enhanced bioactive metabolites by VSM-3, unstructured kinetic modeling and elucidation of chemotype analysis of the strain by LC-Q-TOF-MS analysis.
MATERIALS AND METHODS
Chemicals and reagents
The medium components of Starch casein nitrate agar medium and nutritional requirements such as sucrose and biopeptone were obtained from Merck, India, while the solvent, ethyl acetate, and other chemicals/reagents of analytical grade were obtained commercially.
Isolation and identification
The marine silt specimens were gathered from the Bay of Bengal of north coastal Andhra Pradesh, India, and were dried at ambient temperature. The atmospheric dried silt samples were exposed to (6% w/v) peptone pre-treatment and (0.05 %) sodium lauryl sulfate at a temperature of 50°C and for 10 min (Kyung and Yong, 1996) to confine the unsolicited impurities such as molds and microbes. The example of pre-pickled sediment (1 g) was dissolved in a solution of 100 ml of quarter force ringer’s solution, homogenized by vortexing. Starch casein nitrate agar solution composed of 1% soluble Starch, 0.03% Casein, 0.2% K2HPO4, 0.2% KNO3, 0.2% NaCl, 0.005% MgSO4, 0.002% CaCO3, 0.001% FeSO4, and 2% agar supplemented with Nalidixic acid (50 μgml-1) and Secnidazole (50 μg ml−1), was used for serial (100 μl of 10−4) dilution and incubated at 30ºC for three weeks. Morphologically particular strain was specifically isolated and kept up by subcultured on yeast extract malt extract dextrose (YMD) agar medium at 4ºC for further examination. The strain was identified as S. flava VSM-3 by polyphasic taxonomy and deposited in GenBank (Accession number: KU507600).
RSM experimental design
RSM designs are connected for displaying and investigation of the issues in which the reaction of premium is impacted by a few factors and their association (Sandrina et al., 2016) to produce the antimicrobial metabolites by VSM-3. From the preliminary experiments, time of incubation, pH, temperature of medium, concentrations of sucrose, and biopeptone were selected due to their influence for the bioactive metabolite production, and its antimicrobial activity against five responses, namely (measured as Zone of inhibition) Xanthomonas campestris (MTCC 2286), Pseudomonas solanacearum (NCIM 5103), Botrytis cinerea (MTCC 2104), Curvularia maculans, and Penicillium expansum. The most significant factors (A, B, C, D, and E) at their ideal levels were identified for maximal response (inhibition zone against the responses). Low and high scopes of all factors are utilized in RSM in genuine and the coded structure was given in Table 1. In total, 50 (32 factorial points, 10 axial points, and 8 replicates) experiments were performed (Table 2). All the variables’ central-coded value was considered as zero. The data obtained from experiments were fitted into the second-order polynomial model through the RSM.
Analysis of variance (ANOVA)
Statistical analysis of the model was attempted to estimate the ANOVA. Model adequacy of the responses was verified by calculating the determination coefficient (R2), adjusted determination coefficient (adj. R2), variation coefficient (CV), and the Fisher’s test value (F-value). The 3-D surface reaction plots were utilized to break down the connection between independent variables and the responses, and these reaction plots address the dependent variable in capacity of two independent variables, where the third variable is kept steady (Coded terms).
 | Table 1. Coded and actual factors and their experimental ranges. [Click here to view] |
 | Table 2. Central composite factor experimental design along with experimental and predicted values. [Click here to view] |
Unstructured model development
Production of the bioactive metabolites by strain VSM-3 is influenced by the growth rate of biomass and limiting substrate concentration utilization. To simulate the cell (S. flava VSM-3) proliferation and bioactive metabolite synthesis, Logistic (L) and Luedeking–Piret models (LP) were applied. Since the rate of specific bioactive metabolite production and cell growth can be calculated from kinetic expressions. As per the experimental data obtained from batch shake-flask for bioactive metabolite production, our previously developed models (L, Logistic Incorporated LP (LILP) and Logistic incorporated modified LP (LIMLP) were used to determine the process parameters (Ushakiranmayi et al., 2016).
Fermentation and extraction
Spores of potential strain S. flava VSM-3 were scrapped from Starch casein nitrate agar and transferred into 100 ml of YMD seed medium consists of yeast extract 0.4%, malt extract 1%, and dextrose 0.4% and kept in an orbital shaker at 120 rpm, 48h at 35°C. 100 ml of optimized production medium (composed of 0.5 % Sucrose, 0.25 % Biopeptone, 3% NaCl at pH 7.0) was inoculated with 10% inoculum and incubated on an orbital shaker at 120 rpm, 6 days at 35°C. The fermented broth collected at the end of the incubation period was centrifuged, and the culture filtrate obtained was extracted with ethyl acetate. Separating funnel was used to separate the ethyl acetate phase containing bioactive metabolite from the aqueous phase. The pooled solvent extracts were concentrated in a vacuum drier and the crude extract was subjected to LC-Q-TOF-MS for the elucidation of chemical composition analysis for S. flava, as per the method described by Maya and Benjamin (2016).
RESULTS AND DISCUSSION
RSM optimization studies and model adequacy for the bioactive metabolite production
Central Composite Design (CCD) module of RSM was involved to assess the interaction among significant variables and to determine their optimal values. Using CCD reduces the experimental run numbers and escalates the resulting efficiency to be obtained. It has been considered as an efficient statistical experimental tool used to design the experiments (Bipasha et al., 2015). Table 2 tabulates the comparative responses of predicted and experimental runs values. Optimized concentration of the bioactive compound synthesis by S. flava VSM-3 and its effect against experimental responses (growth inhibition as the response) were found to be 20, 19, 19, 18, and 17 mm for X. campestris, P. solanacearum, B. cinerea, C. maculans, and P. expansum, respectively, that was identified at 6 days incubation time, 7 pH, 35°C temperature, 0.5% sucrose concentration, and 0.25% biopeptone concentration.
Table 3 tabulates the best outcome quadratic model with the sequential model sum of squares (suggested) and lack of fit tests (showing degrees of freedom; mean square, F-value, and p-value). The results show 99% of confidence (since the Prob > F value of the model was found to be <0.00001, for all the five responses), which implies that the model is significant. ANOVA statistics have shown a 99% confidence level, when models were checked for their adequacy. Furthermore, ANOVA results based on the bioactive metabolite produced by S. flava VSM-3 confirm that the model has a satisfactory level coefficient of determination (R2) against all the responses (Table 4).
Independent variables interaction and their impact on bioactive metabolite response
Figures 1–5 illustrate the 3-D plots for significant interactive effects of the independent variables such as incubation time, pH, temperature, sucrose, and biopeptone concentrations against the bioactive metabolite responses. 3-D plots (showing response (on z-axis against any two independent variables by keeping the third variable constant) were generated from CCD of RSM to analyze the effect of these variables’ interactions.
 | Table 3. Sequential model fitting for all the responses (in terms of inhibition zone from the bioactive metabolite production). [Click here to view] |
 | Table 4. Results of Analysis of variance for predicting the model adequacy. [Click here to view] |
 | Figure 1. Response surface plots comprised of 3-D views and contours showing interactive effects of selective variables on Zone of Inhibition (mm) of the bioactive compound production by S. flava VSM 3 against X. campestris. [Click here to view] |
 | Figure 2. Response surface plots comprised of 3-D views and contours showing interactive effects of selective variables on Zone of Inhibition (mm) of the bioactive compound production by S. flava VSM 3 against P. solanacearum. [Click here to view] |
The least square method of nonlinear regression (in Microsoft Excel solver 2010) was attempted to fit the experimental data with unstructured models. Experimental versus model-predicted inhibition zones were compared in terms of the bioactive metabolite responses by Kirby Bauer disk diffusion method against the test pathogens X. campestris, P. solanacearum, B. cinerea, C. maculans, and P. expansums strains against the time, were shown in Figure 6 (a)–(e). Figure 6(f) shows the experimental and model-predicted comparative profiles of S. flava VSM-3 substrate utilization. All the profiles show very good fit between model and experiment values. Table 5 lists the evaluated biokinetic parameters used in the mathematical model equations, which indicate high R2 values L, LILP, and LIMLP models fitting to the experimental data revealing good precision of these models. Inhibition zones of agar diffusion tests were also found to be matching with experimental and model data (Table 6). In this way, these developed mathematical models found to be a good representation of bioactive metabolite production kinetics of S. flava VSM-3 in submerged shake-flask fermentations (Ushakiranmayi et al., 2016).
 | Figure 3. Response surface plots comprised of 3-D views and contours showing interactive effects of selective variables on Zone of Inhibition (mm) of the bioactive compound production by S. flava VSM 3 against B. cinerea. [Click here to view] |
.png) | Figure 4. Response surface plots comprised of 3-D views and contours showing interactive effects of selective variables on Zone of Inhibition (mm) of the bioactive compound production by S. flava VSM 3 against C. maculans. [Click here to view] |
Chemotype analysis by ESI-LC-MS/MS
The chemical composition of S. flava VSM-3 was elucidated for the first time by liquid chromatography quadrupole time-of-flight mass spectrometry analysis and the compounds’ spectra was shown in Figure 7. Mass fragmentation analysis confirms the identity of the compounds. Positive ion mode detected a total of seven active metabolites from Saccharothrix flava ethyl acetate extract. The compounds identified were 7-Deazaadenosine, 5-Hydroxy-9-Methylstreptimidone, Amiclenomycin, Dihydroabikoviromycin, Epopromycin A, OAP Silane 55, and MKN-003B. Negative ion mode did not yield any known compounds (Fig. 8).
RSM designs are applied for modeling and analysis of the problems in which the response of interest is influenced by several variables and their interaction (Sandrina et al., 2016) to produce the antimicrobial metabolite by VSM-3. The model’s variability suggested that R2 should be close to 1, or at least 0.80, to be a good fit model (Xiangli et al., 2008). Adjusted R2 is used to determine the model adequacy (in number of model terms) (Onsekizoglua et al., 2010). The desirable Coefficient of variation (CV) should be below 10% and adequate precision must be above 4. Non-significance of the lack fit indicates that the model is a good fit (Mojtaba et al., 2014).
Logistic (L) model-based parameters, μmax, X0, and Xmax were calculated from the growth kinetic profile of the data obtained from the shake-flask culture of S. flava VSM-3 used in this study and LILP model yields α and β (non-growth associated product parameters). Comparatively, α value greater than β concludes bioactive metabolite synthesis associated by S. flava VSM-3 growth (Table 5). In addition, the simulated LIMLP model parameters, γ and η, have good fit with experimental data confirms that this model well represents metabolite synthesis is done from substrate consumption in S. flava VSM-3 fermentation (Ushakiranmayi et al., 2016).
Seven potent bioactive compounds were identified in the VSM 3 strain’s LC-Q-TOF-MS analysis. Among the active compounds, 7-Deazaadenosine (Tubercidin), a nucleoside antibiotic, exhibited potent cytotoxic activity (Biabani et al., 2002), while 5-Hydroxy-9-Methylstreptimidone, a glutarimide metabolite, was reported for its herbicidal activity (Chatterjee et al., 1995). Amiclenomycin, a free amino acid, was reported bioactive against Mycobacteria and has part of di- and tri-peptides that show antibiotic activities (Kern et al., 1985). The compound Dihydroabikoviromycin was formerly mentioned for its genotoxic activity (Holmalahti et al., 1998), while Epopromycin A, an analog of eponeomycin showed herbicidal activity and potent cytotoxicity (Tsuchiya et al., 1997). The compound OAP Silane55 (S-520) was reported for its antibacterial activity, especially in opposition to gram positive bacteria (Shoji et al., 1970), while the compound MKN-003B, a butenolide confirmed moderate to weak antifungal activity (Wang et al., 2014).
 | Figure 5. Response surface plots comprised of 3-D views and contours showing interactive effects of selective variables on Zone of Inhibition (mm) of the bioactive compound production by S. flava VSM 3 against P. expansum. [Click here to view] |
 | Figure 6. Comparison of experimental and model predicted kinetics (a)–(e): for zone of inhibition (mm); (f): for biomass growth (g/l), substrate utilization (g/l). [Click here to view] |
 | Table 5. Estimated kinetic parameters using L, LILP, and LIMLP model equations. [Click here to view] |
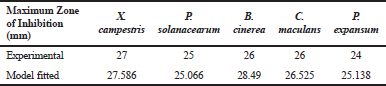 | Table 6. Maximum zones of inhibition (mm) comparison for experimental with model results. [Click here to view] |
 | Figure 7. Positive ion (+) ESI-MS/MS analysis of ethyl acetate extract of strain VSM-3. [Click here to view] |
Till date, Madumycin antibiotic was synthesized by Actinomadura flava (later renamed as Nocardiopsis flava) INA 2171 strain with maximum quantities of madumycin I and II by fifth day of cultivation (Gauze and Sveshnikova, 1985; Gauze et al., 1974; Kochetkova et al., 1976). Nocardiopsis flava has been mentioned to be transferred to the genus Saccharothrix on chemotaxonomic traits basis (Labeda and Lechevalier, 1989). The current work also found that only after 6th day of fermentation yielded maximum bioactive metabolite from S. flava. Zitouni et al. (2005) have reported that Saccharothrix and Nocardiopsis species were active against Gram positive bacteria and yeast. In the present study, bioactive compounds synthesized from S. flava VSM-3 exhibited good antimicrobial activity against gram-positive bacteria and Gram-negative plant pathogenic bacteria like P. solanacearum, agriculturally important fungi like B. cinerea. Hence, the strain S. flava VSM-3 collected from shore samples of Bay of Bengal, located in Andhra Pradesh, India, is novel actinomycete which enabled the production of antibacterial and antifungal compounds.
 | Figure 8. Negative ion (-) ESI-MS/MS analysis of ethyl acetate extract of strain VSM-3. [Click here to view] |
CONCLUSION
Central composite design (of RSM) was successfully employed to determine the optimizing conditions and to analyze the independent and interactive effects of the process variables, namely, incubation time, pH, temperature, concentrations of sucrose, and bio peptone. The results indicated that the process variables have a significant effect on bioactive metabolites production by S. flava VSM-3 and their effects counter to the five responses (inhibition zones). Statistical summary of the model explained that developed model was adequate and precise with the experimental data that were in near concurrence with the predicted values. Further unstructured models, applied in the study, had given a superior guess of kinetic profiles of bioactive metabolite synthesis by S. flava VSM-3 in submerged shake flask fermentations. The values of predicted (model) and experimental responses (zones of inhibition) for five responses were almost similar, which concludes that the developed kinetic model fits the data accurately. The bioactive compounds obtained from strain VSM-3 exhibited significant bioactivity especially against agriculturally important bacteria and fungi, and reveals that the strain is a promising producer of antimicrobial compounds. The actinobacterium’s ethyl acetate extract was subjected to LC-QTOF-MS, which revealed the presence of significant bioactive compounds also supported that the strain VSM 3, and it was most potent for further pharmacological studies. The study also supports that north coastal Bay of Bengal of Andhra Pradesh, a potent ecological niche with inimitable strain diversity remained to be explored for bioactive compounds.
ACKNOWLEDGMENT
Ushakiranmayi M, the first author, greatly acknowledges the University grants commission (U.G.C), New Delhi, Government of India, for granting financial aid to execute this research.
ABBREVIATIONS
RSM Response surface methodology
CONFLICT OF INTEREST
The authors declared that they have no conflict of interest.
REFERENCES
Abdelmohsen UR, Bayer K, Hentschel U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat Prod Rep, 2014; 31:381–9.
Baltz RH. Antimicrobials from actinomycetes: back to the future. Microbe, 2007; 2:125–31.
Biabani MF, Gunasekera SP, Longley RE, Wright AE, Pomponi SA, Tubercidin. A cytotoxic agent from the marine sponge Caulospongiabi flabellate. Pharm Biol, 2002; 40:302–3.
Bipasha D, Ananda Prasad R, Sangita B, Sudip C, Chiranjib B. Lactose hydrolysis by ß-galactosidase enzyme: optimization using response surface methodology. Ecotoxicol Environ Saf, 2015; 121:244–52.
Box GEP, Wilson KB. On the experimental attainment of optimum conditions. J Roy Statist Soc Ser B, 1951; 13:1–45.
Chatterjee S, Vijaya Kumar EKS, Chatterjee S, Blumbach J, Ganguly BN. 5-Hydroxy-9-Methyl Streptimidone, a new Glutarimide from Streptomyces sp. HIL Y-9065403. J Antibiot, 1995; 48:271–3.
Coutinho FH, Gregoracci GB, Walter JM, Thompson CC, Thompson FL. Metagenomics sheds light on the ecology of marine microbes and their viruses. Trends Microbiol, 2018; 26(11):955–65.
Cumsille A, Undabarrena A, González V, Claverías F, Rojas C, Cámara B. Biodiversity of actinobacteria from the South Pacific and the assessment of Streptomyces chemical diversity with metabolic profiling. Mar Drugs, 2017; 15(9):286.
Das S, Lyla PS, Khan SA. Marine microbial diversity and ecology: importance and future perspectives. Curr Sci, 2006; 90:1325–35.
Deka D, Jha DK. Optimization of culture parameters for improved production of bioactive metabolite by endophytic Geosmithia pallida (KU693285) isolated from Brucea mollis Wall ex. Kurz, an endangered medicinal plant. J Pure Appl Microbiol, 2018; 12(3):1205–13.
Gauze GF, Sveshnikova MA. In validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol, 1985; 35:223–5.
Gauze GF, Maksimova TS, Okkhovatova OL, Sveshnikova MA, Kochetkova GV, Ilchenko, GB. Production of Madumycin, an antibacterial antibiotic by Actinomaduraflava sp. Nov. Antibiotiki, 1974; 19(9): 771–5.
Genitsaris S, Monchy S, Denonfoux J, Ferreira S, Kormas KA, Sime-Ngando T, Viscogliosi E, Christaki U. Marine microbial community structure assessed from combined metagenomic analysis and ribosomal amplicon deep-sequencing. Mar Biol Res, 2016;12(1):30–42.
Girão M, Ribeiro I, Ribeiro T, Azevedo IC, Pereira F, Urbatzka R, Leão PN, Carvalho MF. Actinobacteria isolated from Laminaria ochroleuca: a source of new bioactive compounds. Front Microbiol, 2019; 10:683.
Heinze G, Wallisch C, Dunkler D. Variable selection–a review and recommendations for the practicing statistician. Biometric J, 2018; 60(3):431–49.
Holmalahti J, Raatikainen O, Von WA, Laatsch H, Spohr A, Lyngberg OK, Nielsen J. Production of dihydroabikoviromycin by Streptomyces anulatus: production parameters and chemical characterization of genotoxicity. J Appl Microbiol, 1998; 85(1): 61–8.
Imada C. Enzyme inhibitors and other bioactive compounds from marine actinomycetes. Antonie Van Leeuwenhoek, 2005; 87(1):59–63.
Kern A, Kabatek U, Jung G, Werner RG, Poetsch M, Zähner H. Amiclenomycin peptides-isolation and structure elucidation of new biotin antimetabolites. Liebigs Annalen der Chemie, 1985; 1985(5):877–92.
Kishore D, Kayastha AM. Optimization of immobilization conditions for chick pea β-galactosidase (CpGAL) to alkylamine glass using response surface methodology and its applications in lactose hydrolysis. Food Chem, 2012; 134:1650–7.
Kochetkova GV, Maksimova TS, Ilchenko GB, Olkhovatova OL. Madumycin biosynthesis by an Actinomaduraflava culture under conditions of varying aeration. Antibiotiki,1976; 4:296–8.
Kyung MY, Yong KP. A new method for the isolation of actinomycetes from soil. Biotechnol Techn, 1996; 10:541–6.
Labeda DP, Lechevalier MP. Amendment of the genus Saccharothrix and description of Saccharothrix espanesis sp. nov., Saccharothrix cryophilus sp. nov. and Saccharothrix mutabilis comb. nov. Int J Syst Bacteriol. 1989; 39:426–31.
Linda HG, Rabab O. Optimization of medium composition for antibacterial metabolite production from Streptomyces sp. Asian J Pharm Clin Res, 2017;10:381–5.
Marimuthu J, Ye-Wang Z, In-Won K, Jung-Kul L. Enhanced saccharification of alkali-treated rice straw by cellulase from Trameteshirsuta and statistical optimization of hydrolysis conditions by RSM. Bioresour Technol, 2009; 100:5155–61.
Maya R, Benjamin S. Penicillium verruculosum Strain BS3 Produces Aurantioclavine and Rugulosuvine B Alkaloids. Electronic J Biol, 2016; 12(4):484–9.
Mohammadipanah F, Wink J. Actinobacteria from arid and desert habitats: diversity and biological activity. Front Microbiol, 2016; 6:1541.
Mojtaba HM, Ahmad R, Davoud SB, Seyyed AM, Shadi, B. Optimization of ultrasonic-assisted extraction of phenolic compounds from bovine pennyroyal (Phlomis schema parviflorum) leaves using response surface methodology. Ind Crop Prod, 2014; 57:195–202.
Onsekizoglua P, Bahcecib KS, Acara J. The use of factorial design for modelling membrane distillation. J Membr Sci, 2010; 349:225–30.
Ouakhssase A, Chahid A, Choubbane H, Aitmazirt A, Addi EA. Optimization and validation of a liquid chromatography/tandem mass spectrometry (LC-MS/MS) method for the determination of aflatoxins in maize. Heliyon, 2019; 5(5):e01565; doi:10.1016/j.heliyon.2019.e01565
Rama Krishna GVS, Ushakiranmayi M, Vijayalakshmi M, Sudhakar P. Model-based kinetic parameters estimation in batch pullulan fermentation using Jaggery as substrate. J Chem Pharma Res, 2016; 8:217–24.
Ramesh S, William A. Marine actinomycetes: An ongoing source of novel bioactive metabolites. Microbiol Res, 2012; 167:571–80.
Ripa FA, Nikkon F, Zaman S, Khondkar P. Optimal conditions for antimicrobial metabolites production from a new Streptomyces sp. RUPA-08PR isolated from Bangladeshi soil. Mycobiology, 2009; 37:211–4.
Sandrina AH, Patrícia D, Prieto MA, Lillian B, Alírio R, Maria FB. Optimization of ultrasound-assisted extraction to obtain mycosterols from AgaricusbisporusL with response surface methodology and comparison with conventional Soxhlet extraction. Food Chem, 2016; 197:1054–63.
Shoji J, Kozuki S, Mayama M, Shimaoka N. A new peptide antibiotic complex S-520. I. Isolation and characterization. J Antibiot, 1970; 23:429–31.
Shuvankar B, Savita K, Sabu T, Nimmy A. Halophilic and halotolerant actinomycetes from a marine saltern of Goa, India producing anti-bacterial metabolites. J Biosci Bioengg, 2015; 119:323–30.
Sponga F, Cavaletti L, Lazzarini A, Borghi A, Ciciliato I, Losi D. Biodiversity and potentials of marine derived microorganisms. J Biotechnol, 1999; 70:65–9.
Stach JE, Maldonado LA, Masson DG, Ward AC, Goodfellow M, Bullm AT. Statistical approaches for estimating actinobacteria diversity in marine sediments. Appl Environ Microbiol, 2003; 69:6189–200.
Tsuchiya K, Kobayashi S, Nishikiori T, Nakagawa T, Tatsuta K. Epopromycins, Novel cell wall synthesis inhibitors of plant protoplast produced by Streptomyces sp. NK04000. J Antibiotics, 1997; 50:261–3.
Ushakiranmayi M, Vijayalakshmi M, Sudhakar P, Rama Krishna GVS, SatishBabu R. Optimization of culture conditions by response surface methodology and unstructured kinetic modeling for bioactive metabolite production by NocardiopsislitoralisVSM-8. 3 Biotech, 2016; 6:219; doi:10.1007/s13205-016-0535-2
Wang T, Jiang Y, Ma KX, Li YQ, Huang R., Xie XS, Wu SH. Two new butenolides produced by an actinomycete Streptomyces sp. Che Biodiverse, 2014; 11:929–33.
Xiangli F, Wei W, Chen Y, Jin W, Xu N. Optimization of preparation conditions for polydimethylsiloxane (PDMS)/ceramic composite pervaporation membranes using response surface methodology. J Memb Sci, 2008; 311(1-2):23–33.
Zitouni A, Boudjella H, Lamari L, Mathieu F, Lebrihi A, Sabaou N. Nocardiopsis and Saccharothrix genera in Saharan soils in Algeria: Isolation, biological activities and partial characterization of antibiotics. Res Microbiol, 2005; 156: 984–93.