INTRODUCTION
The term “inflammatory bowel disease” (IBD) refers to a set of idiopathic immune-related gastrointestinal illnesses, in which, ulcerative colitis (UC) and Crohn’s disease (CD) are linked. Different IBD entities and inflammatory phenotypes have different symptoms, which may include diarrhea, rectal bleeding, bowel pain, and weight loss [1,2]. UC is a disease of the colonic mucosa that often begins in the rectum and spreads proximally through all or a portion of the colon [3]. Compared to CD, UC is more common, and typically manifests in young adults between the ages of 15 and 35 years, although it can occur at any age [4]. Diagnosis of UC is typically made through a combination of clinical evaluation, laboratory tests, imaging studies, and endoscopy with biopsy [5]. UC results from a combination of genetic and environmental factors, while the exact cause of UC is unknown. However, it is thought that it happens in people with genetic susceptibility as a result of an abnormal immunological response to the intestinal microbes. Environmental factors may also play a role in the development of UC, including diet, smoking, and exposure to certain infections [6,7]. There is some evidence to suggest that a Western diet, which is high in fat and low in fiber, may increase the risk of UC [8]. The treatment of UC may involve a variety of medications, including aminosalicylates, corticosteroids, immunomodulators, and biologic agents [9]. Aminosalicylates are the first-line therapy for mild to moderate disease and work by reducing inflammation in the colon [10]. Corticosteroids may be used for short-term control of more severe symptoms. Immunomodulators work by suppressing the immune system to reduce inflammation in the colon. Biologic agents are the newer drugs that target specific proteins in the immune system that are involved in inflammation. However, these therapies are associated with a number of side effects such as weight gain, mood swings, acne, skin rash, and allergic reactions. In severe cases of UC, surgery may be necessary to remove the affected portion of the colon [11].
Traditional herbs are a significant source of novel chemicals that are the basis for medications in all major illness areas. According to Parrotta et al. [12], the family Bombacaceae includes Bombax ceiba Linn. The red cotton tree is also known as the Indian kapok tree in English, semal in Hindi, Shalmali in Sanskrit, shimul in Bengali, kondabruga in Telugu, and mullilavu in Malayalam [13]. According to Ayurveda, B. ceiba, a component in numerous formulations, has shown therapeutic benefits. Menorrhagia, diarrhea, dysentery, and wounds are all treated with it. According to reports, gum or plant petioles are used as enemas in UC [14].
MATERIALS AND METHODS
Retrieval of phytocompounds
Reported phytoconstituents from S. malabarica were collated from phytomedicine databases viz., ChEBI, Dr. Dukes DB, PCIDB, and published literature using the keyword “S. malabarica, and Bombax ceiba.” Each compound’s molecular weight, molecular formula, canonical SMILES, and a number of hydrogen bond donors (HBDs) and acceptors (HBA) were obtained from the PubChem chemical database (https://pubchem.ncbi.nlm.nih.gov/) [15].
Drug-likeness and toxicity properties of phytocompounds
The drug-likeness score for each molecule was predicted using MolSoft’s (https://molsoft.com/) “Lipinski’s Rule of Five” model. The MolSoft server calculates an overall drug-likeness score of an unknown compound based on the chemical fingerprints. The training set of MolSoft includes 5,000 World Drug Index marketed drugs and 10,000 carefully selected non-drug compounds. [16]. The toxicity profile of each compound was predicted by the AdverPred online server. The AdverPred server mainly predicts six major side effects of the compounds, i.e., arrhythmia, cardiac failure, hepatotoxicity, myocardial infarction, nephrotoxicity, and myocardial infarction. The advantage of the server is that it has data on compounds with “Boxed warning,” “Warnings and Precautions,” and “Adverse reactions” [17]. The druggability and toxicity profile of phytocompounds are shown in Table 1.
Target identification
Genecards®: The Human Gene Database that contains information on all known and predicted human genes, including genomic, proteomic, transcriptomic, genetic, and functional details with respect to specific diseases. We first used this server to find UC-related targets, and the UniProt protein database was used to gather gene IDs for each protein with a standard reference to the “Homo sapiens” species. Canonical phytocompound smiles were submitted to the SwissTargetPrediction system for target identification with probable scores of >0.1%. Compounds targeting only therapeutic targets implicated in UC and associated infections were gathered from the GeneCards after the target prediction.
Compound-gene enrichment
The set of phytocompounds modulating target/protein(s) were quired into the STRING [https://string-db.org/] database for protein–protein interaction analysis [18]. Furthermore, molecular pathways modulated by a set of genes were predicted by STRING in-build KEGG pathway database.
Network analysis
Cytoscape is a software program that facilitates the construction of network models to represent interactions and provides functionality for annotating and evaluating connections or relationships within a given dataset. This dataset may include a wide range of entities, including genes, proteins, chemicals, and enzymes. The datasheet of phytocompounds targeting protein molecules and protein molecules’ association with molecular pathways was created from the STRING and KEGG databases. Using these datasheets, the “compound-gene-pathway network” was built by employing Cytoscape ver. 3.6.1. The network was assessed with the “Network Analyzer” command, and the “network was treated as direct.” To express the degree of connections, the edge count and map node size from “low values to small sizes” and map node color from “low values to bright colors” were used between the compound and gene [19].
Docking study
AutoDock Vina was used for the docking study. Autodock Vina is much faster and more accurate compared to AutoDock 4.0. Each compound’s 3D structures were obtained in the structural data file (*.sdf) format from the PubChem database. Compounds were then converted to pdb format using Discovery Studio Visualizer (DSV) version 2019. From the network analysis, we prioritized the PTGS2 target as a therapeutic target of UC that is potentially associated with inflammation. PTGS2 X-ray crystallographic structure was obtained in pdb format (PDB ID: 5IKT) from the RCSB PDB. The compound “Tolfenamic Acid” is used as a reference compound. The protein was loaded into the PyRx 0.8v software and converted into AutoDock molecules. The grid box was set to the maximum with center_x = 164, y = 186.46, z = 191.95; size_x = 24.04, y = 19.24, size_z = 21.23; and spacing 1 Å and 100 exhaustiveness was set to the system. As a result, nine different docked conformations were obtained, conformation with the least RMSD was selected and the complex was generated. The intermolecular interaction between the ligand and protein with complex was visualized by DSV version 2019 [20–22].
 | Table 1. Druggability of compounds. [Click here to view] |
 | Table 2. Toxicity profile of phytocompounds. [Click here to view] |
RESULTS
Phytocompounds druggability
10 phytocompounds were reported to be present in the S. malabarica / B. ceiba. Among 10 bioactives, 7 exhibited positive DLS scores, and Rutin had the highest score of 0.91 (Table 1). Astragalin and Isoquercitrin were also found to be the next best druggable compounds which scored a DLS score of 0.67 and 0.68, respectively. Out of 10 compounds, beta-Amyrin, Lupeol, and Olean-12-en-3-one were found to have no side effects. Cellotriose and Magniferin were found to have p-value of > 0.5 for Nephrotoxicity. Whereas the remaining compounds exhibited p-value < 0.5 for side effects (Table 2).
Targets prediction
From the GeneCards, about 2,830 genes were obtained for UC of which 489 were prioritized based on geneCards inferred functionality scores cut-off >50. From the TTD database, 21 therapeutic targets of UC were collated. Finally, the probable protein targets of 10 compounds were predicted by SwissTargetPrediction. 10 compounds were predicted to hit 297 targets. Out of 297 targets, 39 targets matched with the further peer-interpretation of probable targets with the approved therapeutic targets GeneCards and TTD database UC targets.
Gene set enrichment and network analysis
Pathway enrichment study revealed that 10 phytocompounds affected 125 molecular pathways; among which 14 were linked to UC, i.e., “VEGF, IL-17, Neurotrophin, JAK-STAT, cAMP, TNF, TGF-beta, and NF-kappa B, MAPK, PI3K-Akt signaling pathway, Inflammatory bowel disease (IBD), Leukocyte transendothelial migration, Gastric acid secretion, and Apoptosis” (Table 3). The network analysis showed MAPK3, TNF, and PTGS2 protein targets to score the highest edge count. MAPK3 enriched to be involved in VEGF, IL-17, Neurotrophin, cAMP, TNF, TGF-beta, MAPK, PI3K-Akt signaling pathway, and Apoptosis; TNF was enriched to be involved in IL-17, TNF, TGF-beta, NF-kappa B signaling pathways, IBD; whereas, IL-17, TNF, and NF-kappa B signaling pathways (Fig. 1).
Docking study
From the network analysis, prioritized PTGS2 protein target as a therapeutic target and has a core role in inflammation. Within the network, Astragalin, Isoquercitrin, and Rutin were identified to hit PTGS2. From the docking analysis, it is inferred that Tolfenamic Acid possesses the lowest BE of −9.1 kcal/mol via forming 11 intermolecular interactions. It formed 1 H-bond with Tyr385…O- and 10 non-H-bond interaction with Leu352, Ala527 (3), Val116, Val349 (3), Leu531 (2). All eleven interactions were with active site residues of PTGS2. Among three phytocompounds, Rutin scored the lowest BE of −7.9 kcal/mol via forming 22 intermolecular interactions, i.e., 6 H-Bonds (Ser530…O-, Ser530….OH, Val523….OH, Leu352….OH, His90…O, Tyr385…O-) and 14 non-H-bonds (Phe209, Phe381, Phe210, Phe205, Tyr385, Leu359, Leu531, Ala527 (2), Val349 (2), Ser353 (2), Val523 (2), Ser530). Among 22 interactions, 20 were with active site residues. Interestingly, the other two compounds, Astragalin and Isoquercitrin also formed interactions with active site residues of PTGS2. Astragalin formed 14 and Isoquercitrin formed 9 interactions with active site residues of PTGS2. Table 4 and Figure 2 represents the intermolecular interaction of compounds with PTGS2
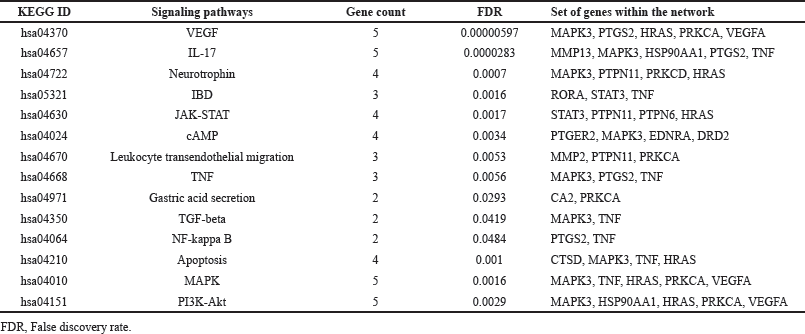 | Table 3. Pathway enrichment study of protein targets in UC regulated by compounds. [Click here to view] |
 | Figure 1. Phytochemical, target protein, and pathway interactions. [Click here to view] |
DISCUSSION
The current study deals with exploring the effect of B. ceiba on UC by multiple computational studies viz “target identification, gene set enrichment analysis, network pharmacology coupled with molecular docking studies.” Identification of potential therapeutic targets is the key to good drug design. Since traditional plants are the starting point for the discovery of new drugs, it stands to reason that a novel treatment strategy, supported by polypharmacology and bioinformatics approaches, could be developed by using multiple compounds found in herbs with a traditional claim against a specific disease with minimal side effects. The influence of medications on numerous protein targets altering disease pathways is the focus of polypharmacology and bioinformatics. A good therapeutic approach aimed at the potential therapeutic targets will not cause adverse effects by altering the physiological function of the target in healthy tissues. In the current study, initially, we retrieved phytocompounds present in B. ceiba using curated herb databases and scientific reports.
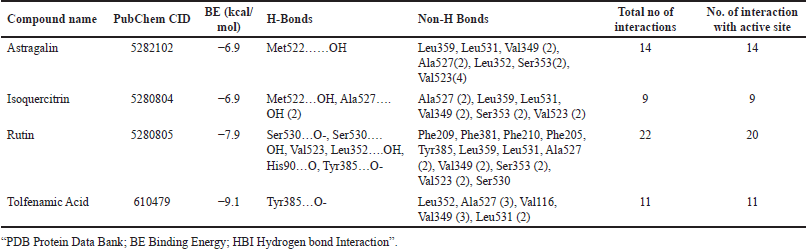 | Table 4. Binding of compounds with cyclooxygenase 2 protein. [Click here to view] |
 | Figure 2. Binding mode of compounds with PTGS2. a) and c) 2D representation of interaction of Rutin and Tolfenamic acid with PTGS2, respectively. b) and d) 3D representation of Rutin and Tolfenamic acid, respectively, within the PTGS2 binding pocket. [Click here to view] |
We used phytocompounds with drug-like properties to forecast their druggable characteristics. We also used SwissTargetPrediction to identify potential targets for each phytocompound with drug-like properties and selected those compounds that modulated solely therapeutic targets related to UC. The pathways affected by B. ceiba phytocompounds were inferred using gene set enrichment and network analysis. To learn more about the molecular mechanism of action and pharmacological basis of B. ceiba with respect to certain proteins and their association with disease-modifying capacity, a network pharmacology study was conducted. This can be scientifically studied by network construction using the Cytoscape tool (topological network) with ease since it can link to external public databases and assures the diverse interpretations
We inferred 8 bioactives of B. ceiba to trigger 19 therapeutic protein targets linked with UC, i.e., MAPK3, PTGS2, HRAS, PRKCA, VEGFA, MMP13, HSP90AA1, TNF, PTPN11, PRKCD, RORA, STAT3, PTPN6, PTGER2, EDNRA, DRD2, MMP2, CA2, and CTSD. Furthermore, these 19 targets were linked with 125 molecular pathways, of which 14 were related to UC pathogenesis, i.e., VEGF, IL-17, Neurotrophin, JAK-STAT, cAMP, TNF, TGF-beta, and NF-kappa B, MAPK, PI3K-Akt signaling pathway, IBD, Leukocyte transendothelial migration, Gastric acid secretion, and Apoptosis. The network analysis showed MAPK3, TNF, and PTGS2 protein targets to score the highest edge count. MAPK3 enriched to be involved in VEGF, IL-17, Neurotrophin, cAMP, TNF, TGF-beta, MAPK, PI3K-Akt signaling pathway, and Apoptosis; TNF was enriched to involve in “IL-17, TNF, TGF-beta, NF-kappa B signaling pathways, IBD; whereas, IL-17, TNF, and NF-kappa B signaling pathways.”
Patients with IBD have been found to have elevated levels of expression of MAPKs, which are intracellular serine/threonine-specific kinases that regulate the transcription and stimulation of several transcription factors governing genes involved in IBD in responses to pro-inflammatory cytokines. It has been found that VEGF, a key regulator of angiogenesis in UC, is upregulated in the sera of UC patients and animals with experimental UC. The JAK-STAT signaling plays an essential role in the pathophysiology of UC. Both normal physiological processes and inflammatory responses are mediated by the JAK-STAT pathway. TNF is a proinflammatory mediator that plays an important role in the pathogenesis of IBD. Serum, feces, and mucosal biopsies from IBD patients have been shown to have significantly higher amounts of TNF.
Furthermore, based on the network analysis and well-known therapeutic targets of UC, we chose PTGS2, also known as cyclooxygenase 2 (COX2) from the network. Astragalin, Isoquercitrin, and Rutin were predicted to modulate PTGS2, and their molecular docking study was carried out using AutoDock vina. The results were compared with the Tolfenamic acid, a known inhibitor of PTGS2. Astragalin, Isoquercitrin, and Rutin formed 14, 9, and 22 interactions with PTGS2, in which 14, 9, and 20 interactions were with active site residues of PTGS2, respectively. While Tolfenamic Acid formed 11 interactions with PTGS2 and all interactions were with active site residues. Tolfenamic Acid scored the lowest BE of −9.1 kcal/mol and Rutin scored −7.9 kcal/mol. However, among three phytocompounds, Rutin BE was found to have the lowest and maximum number of interactions with active site residues. The previous reports also suggest Rutin as a potent inhibitor or down regulator of COX2/PTGS2 [23–25] and could be the better lead molecule against UC along with other molecules present in the B. ceiba via multicompound, multiprotein, and multipathway interaction mechanism.
CONCLUSION
This work reports the molecular mechanisms of phytocompounds reported in the petiole of B. ceiba leaves for the management of UC via the use of multiple compounds, multiple proteins, and multiple pathways concept. The identified compounds exhibited druggability with minimal adverse effects when predicted in MolSoft and AdverPred servers. Furthermore, gene set enrichment analysis revealed phytocompounds to modulate key pathways mainly IBD, leukocyte transendothelial migration, gastric acid secretion, and apoptosis. VEGF, IL-17, Neurotrophin, JAK-STAT, cAMP, TNF, TGF-beta, and NF-kappa B were the signaling pathways that are linked to UC. A molecular docking study validated astragalin, isoquercitrin, and rutin to interact with the active site of PTGS2. Hence, the use of a paste made from the leaf petioles of B. ceiba (as described in AYUSH), could be one of the most cost-effective therapeutic natural resources for the management of UC.
ACKNOWLEDGMENTS
The authors thank to the Principal, Shri B. M. Kakanwadi Ayurveda Mahavidyalaya, KLE Academy of Higher Education and Research, Belagavi, for providing necessary facilities to carry out the research.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117(3):514–21. CrossRef
2. Gracie DJ, Ford AC. IBS-like symptoms in patients with ulcerative colitis. Clin Exp Gastroenterol. 2015; 19:101–9. CrossRef
3. Feagan BG, Rutgeerts P, Sands BE, Hanauer S, Colombel JF, Sandborn WJ, et al. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2013;369(8):699–710. CrossRef
4. Lockhart-Mummery HE, Morson BC. Crohn’s disease (regional enteritis) of the large intestine and its distinction from ulcerative colitis. Gut. 1960;1(2):87–105. CrossRef
5. Peeters M, Joossens S, Vermeire S, Vlietinck R, Bossuyt X, Rutgeerts P. Diagnostic value of anti-Saccharomyces cerevisiae and antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease. Am J Gastroenterol. 2001;96(3):730–4. CrossRef
6. Danese S, Fiorino G, Peyrin-Biroulet L. Positioning therapies in ulcerative colitis. Clin Gastroenterol Hepatol. 2020;18(6):1280–90. CrossRef
7. Loftus Jr EV. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126(6):1504–17. CrossRef
8. Yang H, McElree C, Roth MP, Shanahan F, Targan SR, Rotter JI. Familial empirical risks for inflammatory bowel disease: differences between Jews and non-Jews. Gut. 1993;34:517–24. CrossRef
9. Ford AC, Khan KJ, Achkar JP, Moayyedi P. Efficacy of oral vs. topical, or combined oral and topical 5-aminosalicylates, in ulcerative colitis: systematic review and meta-analysis. Am J Gastroenterol. 2012;107(2):167–76. CrossRef
10. Carter MJ, Lobo AJ, Travis SP. Guidelines for the management of inflammatory bowel disease in adults. Gut. 2004;53:1–6. CrossRef
11. Feagan BG, MacDonald JK. Oral 5-aminosalicylic acid for induction of remission in ulcerative colitis. Cochrane Database Syst Rev. 2012;10:CD000543. CrossRef
12. Parrotta JA. Healing plants of peninsular India. Wallingford, UK: CABI publishing; 2001. CrossRef
13. Chaudhary PH, Khadabadi SS. Bombax ceiba Linn.: pharmacognosy, ethnobotany and phyto-pharmacology. Phcog Commun. 2012;2(3):2–9. CrossRef
14. Jain A, Katewa SS. Medicinal plant diversity of Sitamata wildlife sanctuary, Rajasthan, India. J Ethnopharmacol 2005;102:143–57. CrossRef
15. Lagunin AS. Ivanov A. Rudik D. Filimonov V. Poroikov. DIGEP-Pred: web service for in silico prediction of drug-induced gene expression profiles based on structural formula. Bioinformatics. 2013;29:2062–3. CrossRef
16. Lipinski. C.A, Lead- and drug-like compounds: the rule-of-five revolution, Drug Discov Today Technol. 2004;1:337–41. CrossRef
17. Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. CrossRef
18. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The string database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45. CrossRef
19. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. CrossRef
20. Samdani A, Vetrivel U. POAP: A GNU parallel based multithreaded pipeline of open babel and Auto-Dock suite for boosted high throughput virtual screening. Comput Biol Chem. 2018;74:39–48. CrossRef
21. Biradar P, Patil V, Joshi H, Khanal P. Mallapur S. Experimental validation and network pharmacology evaluation to decipher the mechanism of action of Erythrina variegata L. bark against scopolamine-induced memory impairment in rats. Adv Tradit Med. 2020;22:1–14. CrossRef
22. Patil RS, Khatib NA, Patil VS, Suryawanshi SS. Chlorogenic acid may be a potent inhibitor of dimeric SARS-CoV-2 main protease 3CLpro: an in silico study. Tradit Med Res. 2021;6:20. CrossRef
23. Enogieru AB, Haylett W, Hiss D, Ekpo O. Inhibition of γH2AX, COX-2 and regulation of antioxidant enzymes in MPP+-exposed SH-SY5Y cells pre-treated with rutin. Metab Brain Dis. 2021;36(7):2119–30. CrossRef
24. Choi S, Lim TG, Hwang MK, Kim YA, Kim J, Kang NJ, et al. Rutin inhibits B[a]PDE-induced cyclooxygenase-2 expression by targeting EGFR kinase activity. Biochem Pharmacol. 2013;86(10):1468–75. CrossRef
25. Moutinho MS, Aragao S, Carmo D, Casaca F, Silva S, Ribeiro J, et al. Curcumin and rutin down-regulate cyclooxygenase-2 and reduce tumor-associated inflammation in HPV16-transgenic mice. Anticancer Res. 2018;38(3):1461–6. CrossRef