INTRODUCTION
Gastritis is typically defined as episodes of abdominal pain brought on by an inflammation of the submucosa and mucosa of the stomach. The gastric mucosa may enlarge due to this inflammation, releasing the mucosal epithelium on the surface and bringing on more stomach inflammation. In addition, patients with gastritis may experience increased stomach acid, which could worsen lining damage [1]. Several factors, such as the Helicobacter pylori infection, non-steroidal anti-inflammatory drugs, corticosteroids, and alcohol, can cause gastritis. The typical symptoms were upper abdominal pain, nausea, and vomiting [2,3].
Gastritis is a prevalent condition that has had a serious influence on the lives of many people [2]. Current treatments for gastritis include antacids, H-2 blockers, proton pump inhibitors, and antibiotics. However, these drugs have also been linked to several notable adverse effects, including diarrhea, constipation, and abdominal pain [3]. Therefore, researchers engaged in developing a novel treatment for this illness. Recent research shows that medicinal herbs are an effective alternative to treating gastritis [2].
Cinnamomum burmannii has been traditionally added to oral remedies for nausea and epigastric pain associated with digestive diseases. The usage of this plant for gastritis by neutralizing stomach acid and anti-nausea and anti-vomiting was also stated in an Acuan Sediaan Herbal written by the Indonesian Food and Drug Authority (BPOM) [4]. DLBS2411 is a bioactive fraction from C. burmannii and is believed to have anti-ulcer properties through numerous preclinical and clinical research. Several in vitro studies have also identified the proton pump down-regulator and mucosal protector as potential DLBS2411 mechanisms of action for gastritis [5]. However, DLBS2411 was an herbal remedy containing several substances that may create pathways on various targets to treat gastritis. Considering this, the network pharmacology approach is ideal for identifying compounds related to DLBS2411 for treating gastritis and their potential target proteins and signaling pathways. This approach emphasizes the idea of network targeting by numerous compounds and assists in the molecular and systems-level overviews of DLBS2411 [6]. This method involves identifying compound- and disease-related genes, constructing a protein–protein interaction (PPI) network, and analyzing and visualizing the network [7]. The network pharmacology approach was suitable for combining with molecular docking to predict the affinity between the compounds (as ligands) and the protein target [8]. In this study, network pharmacology was employed to comprehend the molecular pathways by which DLBS2411 treats gastritis from a systemic perspective. This method is being used for the first time in this study to investigate the molecular mechanisms of DLBS2411 for gastritis.
MATERIAL AND METHODS
This study consisted of three investigational phases, starting with data mining, then a network analysis that included topology and enrichment analysis, and target validation, using molecular docking.
Data mining
Compound data collection from C. burmannii
The chemical constituents of C. burmannii were discovered using the online databases KNApSAcK (http://www.knapsackfamily.com/KNApSAcK/) database [9] and IJAH (http://ijah.apps.cs.ipb.ac.id/) by importing “Cinnamomum burmannii” as the keyword. Due to a lack of data regarding Cinnamomum burmannii in various databases, potential components were further examined via literature using Google search using the keyword of Cinnamomum burmannii compounds, Cinnamomum burmannii components, and Cinnamomum burmannii substances. After removing the duplicates, the SwissADME (http://www.swissadme.ch/) database [10] was used to generate the solubility and gastrointestinal (GI) absorption data, and the PubChem (https://pubchem.ncbi.nlm.nih.gov/) database [11] was used to gather each compound’s chemical information.
Compound data screening related to DLBS2411
The resulting compounds were screened using the solubility of each component. Since DLBS2411 is a water phase fraction, the poorly water-soluble compounds were eliminated, and the chosen compounds were then employed for the remaining analysis.
Identification of the related target proteins of DLBS2411’s compounds
The protein targets of the active compounds were predicted by the SwissTargetPrediction (http://www.swisstargetprediction.ch/) database [12]. The target protein information, including IDs and names, was standardized using UniProt (http://www.uniprot.org/) [13], and only Homo sapiens were permitted. Then, redundant targets were removed to obtain targets associated with compounds.
Identification of the related target proteins of gastritis
Protein targets associated with gastritis were provided via DisGeNET (https://www.disgenet.org/home/) [14], GeneCards (https://www.genecards.org/) [15], OMIM (https://omim.org/) [16], GenBank (https://www.ncbi.nlm.nih.gov/genbank/) [17], and PharmGKB (https://www.pharmgkb.org/) with “gastritis” as the keyword. All the targets were limited to “homo sapiens.” The UniProt database (www.uniprot.org) [13] was used to standardize the proteins to their standard protein symbol with a “homo sapiens” restriction. After that, duplicate targets were eliminated to identify targets associated with gastritis.
Identification of the overlap target proteins
The intersection of the DLBS2411 and gastritis targets was identified using an online Venn diagram website (http://bioinformatics.psb.ugent.be/webtools/Venn/) by importing the lists of compound-target and gastritis-target to the website. The overlapped targets were served as potential DLBS2411 targets for gastritis and used for network analysis.
Identification of the details related to gastritis
The MalaCards (https://www.malacards.org/) database [18], an integrated collection of human diseases and related annotations, was used to assess the relevant information regarding gastritis. Information on related genes, gene ontologies, and signaling pathways for gastritis was assembled using the keyword “Gastritis.” The list of protein-coding genes retrieved was further considered as a protein symbol. After that, the list of proteins was examined using an online Venn diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) to determine which proteins from MalaCards were also found in the list of proteins retrieved from Section “Identification of the overlap target proteins” (overlapping proteins of this section and Section “Identification of the overlap target proteins”).
Network analysis
Construction and analysis of PPI network
The STRING 11.0 database (https://cn.string-db.org/) [19] created a PPI network of overlapping targets. The parameter was adjusted to moderate confidence (0.400), and the organism was restricted to Homo sapiens. After that, the “.tsv” (tab-separated values) file format that can be opened in Excel and Cytoscape, which represents the constructed network, was downloaded. The TSV file was then imported into Cytoscape 3.9.1 [20], and the Analyze Network tool was used to analyze the topology of the interaction network by calculating the degree centrality (DC)—the topology study aimed to identify the network’s hub proteins. Hub proteins were chosen from the nodes with DC values greater than twice the median.
Gene ontology (GO) and pathway enrichment
DAVID database (https://david.ncifcrf.gov/tools.jsp) [21] was used to conduct enrichment analysis on GO (biological processes, cellular components, and molecular functions) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways to obtain insight through the potential mechanism of DLBS2411 on gastritis. The enrichment study used the proteins from section “Identification of the details related to gastritis” and the hub proteins from Section “Construction and analysis of PPI network”. This list of proteins was further considered as core target proteins. The study was restricted to “Homo sapiens” and was screened using a p-value and false discovery rate (FDR) of less than 0.05.
Target validation
Selection data for molecular docking analysis
A molecular docking study was conducted to validate the interaction between the substance and its target protein. This confirmation test was conducted only on a few numbers of the hub targets. Only the hub proteins listed among the MalaCards proteins were chosen. Following that, a list of the substances associated with these particular proteins was compiled, and only chemicals that had the greatest impact on the targeted proteins were the focus of the molecular docking investigation.
Preparation data for molecular docking analysis
The 2-D structures of the molecule ligands (structure data file files of the chosen compounds) were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) [11]. The chosen proteins’ 3-D structures were obtained by first looking up their UniProt IDs in the UniProt database and then downloading the PDB format file from the Research Collaboratory for Structural Bioinformatics (RCSB) protein data bank (PDB) database (https://www.rcsb.org/) [22], which was connected directly to the UniProt database. After that, the UCSF Chimera v.1.17.1 program (https://www.cgl.ucsf.edu/chimera/download.html) [23] was used to remove the original ligands and water molecules from the receptor protein structure.
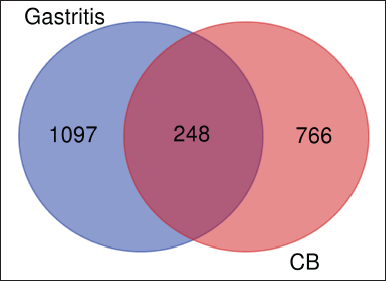 | Figure 1. Venn diagram of the target of DLBS2411 compounds and gastritis. [Click here to view] |
 | Table 1. List of 49 closely related genes to gastritis was collected from the MalaCards database. [Click here to view] |
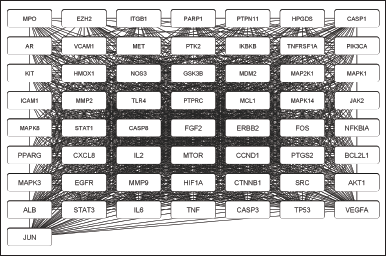 | Figure 2. PPI network of hub proteins. [Click here to view] |
Molecular docking
The CBDock2 website (https://cadd.labshare.cn/cb-dock2/) [24,25] was used to compute the centers and sizes and forecast the binding regions of potential target proteins to obtain the best pose with the least binding energy. CB-Dock2 displayed the binding modes in an interactive 3-D graphic in a Vina score-based order. According to the molecular docking principle, the most stable ligand structure corresponds to the energy value with the lowest Vina score [26]. According to Song et al. [8] and Lin et al. [27], a high ligand-receptor binding activity can be assumed if the minimum binding energy is less than −5.0.
RESULT AND DISCUSSION
Data mining
Cinnamomum burmannii was found to include a total of 146 chemical components that were found in the KNApSAcK, IJAH (accessed on February 5, 2023), and previously published literature. The poorly water-soluble chemicals were removed to leave only the compounds associated with DLBS2411, leaving 130 compounds. DLBS2411 was a water phase fraction created by liquid–liquid extraction from C. burmannii [28].
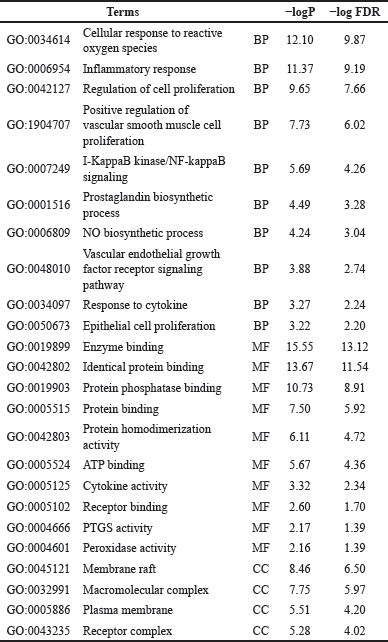 | Table 2. Top GO of 65 proteins. [Click here to view] |
The targets were retrieved from the Swiss Target Prediction database (accessed on April 7, 2023) using the SMILES of the components as mentioned earlier, and after deleting duplicates, 1,014 possible targets were identified. The GeneCards, DisGeNET, GenBank, PharmGKB, and OMIM databases were also searched for 1,345 observed therapeutic targets for gastritis (accessed on April 8, 2023).
The intersection of DLBS2411-target proteins with gastritis-related proteins led to the discovery of 248 shared target proteins (Fig. 1). These 248 proteins were chosen as overlapped targets for further investigation. On the other hand, the MalaCards database (accessed on May 6, 2023) was used to assemble the most closely related information to gastritis. This information included 49 genes (shown in Table 1). The 49 genes consisted of 45 protein-coding genes (further considered as a protein symbol) and 4 RNA genes.
 | Table 3. Top 10 KEGG pathways of 65 proteins. [Click here to view] |
Network analysis
A PPI network with 248 overlapping targets was built in the STRING database with a confidence level of 0.4 on April 18, 2023. This network contains 248 nodes and 4,619 edges. The constructed network was evaluated using the Analyze Network tools in Cytoscape for DC analysis to identify the hub proteins in this network. According to research by Yu et al. [3], nodes were considered hubs when their degree was higher than double the median of all the other nodes in the network. This criterion was utilized as the initial cutoff point, yielding 57 hub proteins.
Figure 2 shows the network of the hub proteins, which included 57 nodes and 1,279 edges. The target proteins were sorted according to their degree of centrality, from lowest to highest. Each node had between 28 and 56 edges, which shows that different proteins in the network interact with one another.
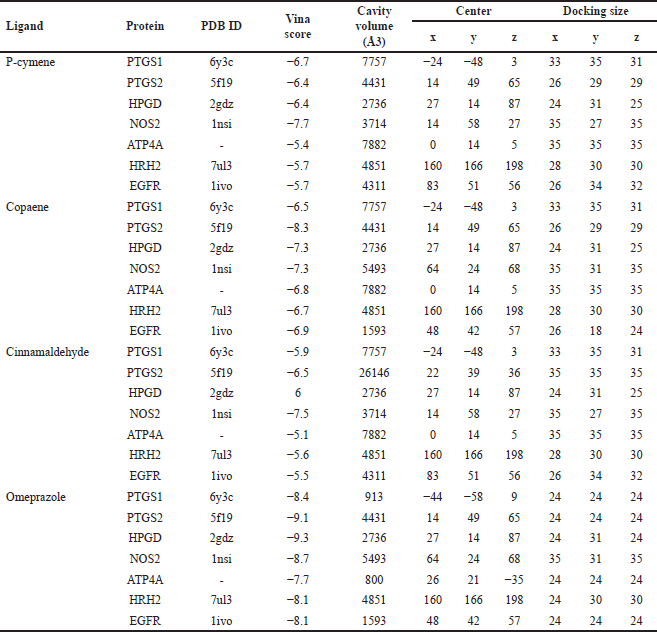 | Table 4. The most suitable CB-Dock pose for each putative target protein and selected compound of DLBS2411 based on Vina score. [Click here to view] |
Eight of the 14 proteins from MalaCards that were collected were not included as hub proteins. However, these eight proteins had to be considered in a subsequent enrichment investigation because of their potential impact on gastritis. The hub proteins in this study were screened based on centrality analysis, which identifies the most important nodes in a network based on the connections and relationships between the nodes. It does not consider the specific characteristics or functions of the nodes themselves, whereby the specific functions and characteristics of the genes or proteins are often more important than their connections to other genes [29].
A total of 65 proteins from MalaCards (a combination of hub targets and MalaCards) were subsequently added to the DAVID database for KEGG pathway and GO enrichment, which consists of three parts: biological process, cellular component, and molecular function. 532 GO-BP, 94 GO-MF, and 57 GO-CC analytical data were gathered, but only 251 BP, 48 MF, and 27 CC were significant according to the p-value and FDR rate less than 0.05. Because the p-value and FDR values were too low, the p-value and FDR values were converted into −logP and −logFDR. Table 2 presents the top ten results of the GO analysis. Of 159 pathways, 151 were found to be significant (p-value and FDR < 0.05) due to the enrichment of the KEGG pathway. The top 10 KEGG pathways are listed in Table 3.
Target validation
This investigation selected seven target proteins—PTGS1, PTGS2, 15-hydroxyprostaglandin dehydrogenase (HPGD), NOS2, APT4A, histamine H2 receptors (HRH2), and epidermal growth factor receptor (EGFR)—for molecular docking. All protein structures were retrieved from the RCSB PDB database except for the ATPase H+/K+ transporting subunit alpha (ATP4A) protein because the ATP4A protein for humans was not provided in this database. The ATP4A homology structure was downloaded from EMBL-EBI [30]. The compounds used in this validation experiment were p-cymene, copaene, and cinnamaldehyde. All these active ingredients could easily bind to and enter all the targeted proteins, as indicated by the vina score of less than −5 (Table 4). Table 4 contains the findings of the molecular docking investigation.
 | Figure 3. NF-Kappa B pathway. [Click here to view] |
DISCUSSION
Gastritis is typically defined as episodes of abdominal pain brought on by an inflammation of the submucosa and mucosa of the stomach. In addition, patients with gastritis may experience increased stomach acid, which could worsen lining damage [1]. Gastritis is treated with antibiotics, proton pump inhibitors, H-2 blockers, and antacids. However, these drugs have several apparent side effects, including constipation, diarrhea, and abdominal pain [3].
The bioactive fraction DLBS2411 from C. burmannii is proven to have anti-ulcer properties. Its effects have been demonstrated in several preclinical and clinical studies. Several in vitro and in vivo investigations have also suggested mucosal protectors and proton pump down-regulators as potential modes of action for DLBS2411 [5]. Network pharmacology was employed in this study to investigate the molecular mechanisms of DLBS2411’s ability to treat gastritis. This approach supports the systems perspective and molecular level overview of DLBS2411 and is a concept of network target by many substances.
Most of the chemicals in DLBS2411 were shown to be active in their effects on gastritis, with the components p-cymene, copaene, and cinnamaldehyde being particularly notable. Significant target proteins affected by these compounds include PTGS1, PTGS2, HPGD, NOS2, ATP4A, histamine receptor H2, and EGFR.
The cyclooxygenase (COX) enzymes, produced by the PTGS1 and PTGS2 genes, catalyze the conversion of arachidonate to prostaglandin. Prostaglandin-endoperoxide synthase (PTGS) has two isozymes: PTGS1 (COX-1), which is constitutive, and PTGS2 (COX-2), which is induced [15]. The COX-2 pathway significantly mediates PGE2 production during inflammation [31]. Prostaglandin E2 (PGE2), which is produced and stored in the gastric and duodenal mucosa, is a potent inhibitor of gastric acid and pepsin production and an inducer of gastric mucus and gastroduodenal bicarbonate secretion [32]. This finding was supported by an earlier in vitro study that showed DLBS2411 treatment increased COX-2 gene expression. The earlier study also demonstrated that increased COX-2 expression affected increased mucin 5AC (MUC5AC) gene expression [28]. MUC5AC serves as a selective diffusion barrier for HCl and is one of the main components of the surface-protecting layer in the gastric mucosa. MUC5AC also defends the stomach epithelium from H. pylori. The glycan structures on MUC5AC, Leb, and sialyl Lex, serve as ligands for the bacteria, competing with the ligands on the epithelial cell surface [33].
In addition, in line with previous investigations that indicated that DLBS2411 treatment decreased the gene expression level of the HPGD gene [28], our research showed the effects of DLBS2411 compounds on HPGD protein. The HPGD gene generates 15-hydroxyprostaglandin dehydrogenase (15-PGDH), a crucial enzyme in the biological inactivation of PGE2 [34]. The prior study also showed that DLBS2411 increases the production of COX-2 and PGE2 by inhibiting the expression of the 15-PGDH gene in the stomach cell [28].
NOS2 genes (iNOS proteins) generate nitric oxide (NO), a messenger molecule with a variety of functions throughout the body [15]. According to earlier studies, DLBS2411 upregulated NO production, enhancing gastric blood flow and sustaining gastric microcirculation, which raises the possibility that DLBS2411 might improve mucosal blood flow through NO production [28]. The research by Wulandari et al. [28] demonstrated that DLBS2411’s impact on COX-2 and NO generation occurred via the NF-Kappa B pathway, whereby an increase in NF-Kappa B results in an increase in COX-2 expression and NO production (Fig. 3).
The ATP4A gene encodes a catalytic alpha subunit of the gastric H+, K+-ATPase, the gastric proton pumps responsible for maintaining an acidic environment in the stomach. They are typically found in the parietal cells of the stomach mucosa and use the energy generated during the hydrolysis of ATP to transport the ions H+ and K+ against concentration gradients [15]. This result was supported by an earlier investigation that showed that treatment with DLBS2411 decreased the expression of the H+/K+ ATPase messenger RNA on rat stomach parietal cells and human embryonic kidney 293 cells in vitro and ex vivo in a dose-dependent manner. The stomach H+/K+ ATPase activity was inhibited by DLBS2411 at various pH levels, indicating that it also functioned as a competitive inhibitor [35]. The current work also discovered HRH2 genes (HRH2 receptors). This protein regulates stomach acid production and controls intestinal and GI motility. These results require additional in vitro and in vivo studies to support them because there are no preclinical investigations of DLBS2411 as an H2 blocker.
One of the most important factors in gastric tissue repair and cell regeneration is the interaction of EGF and its receptor (EGFR) in the gastric epithelium basal or bilateral membrane. While normal epithelial cells rarely express EGFR, it is abundantly detectable in perfused epithelial cells and damaged gastric barriers. In addition, it was shown that EGFR was more highly expressed in the surface mucosal layer of the stomach, suggesting that this factor may impact the growth and healing of damaged gastric epithelial cells. Therefore, it is crucial for developing and repairing the gastric mucosa [3].
The molecular docking study has supported the findings of this network pharmacology investigation. Each of the DLBS2411 compounds used in this investigation was found to have strong binding capabilities with all of the anticipated target proteins, as evidenced by Vina scores that were less than −5. This data suggests that copaene, cinnamaldehyde, and p-cymene had stable ligand structures on the target proteins. In this molecular docking investigation, omeprazole, the reference drug, was utilized. Omeprazole had a low Vina score, which indicated high binding affinity to all expected proteins. The binding energies of copaene, cinnamaldehyde, and p-cymene were slightly higher than those of omeprazole. However, more research is necessary to fully comprehend the atoms’ and molecules’ motions and interactions (a molecular dynamics simulation).
The limitations of this study are acknowledged. Data collection for the chemical compounds of C. burmannii was limited to reliable databases and published literature. There might have been substances missing from those sources. The conclusions of this network pharmacology study are just based on computational analysis, thus, are still only predictions and required to be confirmed in the lab using in vivo or in vitro experiments. However, the findings of this study revealed DLBS2411’s bioactive component and its potential target protein for treating gastritis through the proton pump inhibitor and mucosal protector pathways. The outcome of this study needed to be validated by further studies on molecular dynamics and in vitro and in vivo evaluation.
CONCLUSION
In this study, the DLBS2411 compound p-cymene, copaene, and cinnamaldehyde were selected as highlighted compounds for gastritis-treating action, notably on seven main target proteins, including PTGS1, PTGS2, HPGD, NOS2, APT4A, HRH2, and EGFR. These proteins mainly function through cell regeneration, mucosal protection, and proton pump-down regulator pathways. The molecular docking analysis showed that the ligand-receptor binding activity was less than −5, indicating stable ligand-protein complexes. Thus, this work presents new insight into the efficacy of DLBS2411 compounds in treating gastritis, which might guide future research on this bioactive fraction.
ACKNOWLEDGMENTS
The authors would like to thank Emilia Utomo for the manuscript review in this research.
AUTHOR CONTRIBUTIONS
Jeni Rustan developed the methodology, collected and analyzed data, and prepared the manuscript. Raymond R. Tjandrawinata designed the concept, supervised this research, and contributed to the review of the manuscript. Adi Yulandi designed the methodology, supervised this research, and contributed to the review of the manuscript.
FUNDING
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
LIST OF ABBREVIATIONS
PTGS: prostaglandin-endoperoxide synthase; HPGD: 15-hydroxyprostaglandin dehydrogenase; ATP4A: ATPase H+/K+ transporting subunit alpha; EGFR: epidermal growth factor receptor; MUC5AC: mucin 5AC; DC: degree centrality; GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; FDR: false discovery rate.
REFERENCES
1. Rahmawati F. Therapies for gastritis pain. Palembang, Indonesia: University Sriwijaya; 2020. p 2020.
2. Bashir A, Zulfikar S, Bhutto A, Farid N, Ali K, Fatima K. Gastritis: a brief review of herbal treatment and conventional therapy alternatives. Sci Int (Lahore). 2021;33(2):189–92.
3. Yu G, Wang W, Wang X, Xu M, Zhang L, Ding L, et al. Network pharmacology-based strategy to investigate pharmacological mechanisms of Zuojinwan for treatment of gastritis. BMC Complement Altern Med. 2018 Nov 1;18(1):292. CrossRef
4. BPOM RI. Acuan Sediaan herbal. 1st ed. Indonesia: BPOM RI; 2006. Vol. 2.
5. Nailufar F, Tjandrawinata RR. The evaluation of proton pump inhibitor bioactive fraction DLBS2411 from Cinnamomum burmannii (Nees & T. Nees) in animal model of gastric ulceration healing. Am J Pharmacol Toxicol. 2017 Apr;12(4):79–88. CrossRef
6. Wu Y, Zhu Y, Xie N, Wang H, Wang F, Zhou J, et al. A network pharmacology approach to explore active compounds and pharmacological mechanisms of a patented Chinese herbal medicine in the treatment of endometriosis. PLoS One. 2022 Feb 7;17(2):e0263614. CrossRef
7. Noor F, Tahir Ul Qamar M, Ashfaq UA, Albutti A, Alwashmi ASS, Aljasir MA. Network pharmacology approach for medicinal plants: review and assessment. Pharmaceuticals (Basel). 2022 May 4;15(5):572. CrossRef
8. Song S, Zhou J, Li Y, Liu J, Li J, Shu P. Network pharmacology and experimental verification based research into the effect and mechanism of Aucklandiae Radix-Amomi Fructus against gastric cancer. Sci Rep. 2022 Jun 7;12(1):9401. CrossRef
9. Afendi FM, Okada T, Yamazaki M, Hirai-Morita A, Nakamura Y, Nakamura K, et al. KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012 Feb;53(2):e1. CrossRef
10. Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017 Mar 3;7:42717. CrossRef
11. Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2023 update. Nucleic Acids Res. 2023 Jan 6;51(D1):D1373–80. CrossRef
12. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019 Jul;47(W1):W357–664. CrossRef
13. The UniProt Consortium. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res. 2023 Jan;51(D1):D523–31. CrossRef
14. Piñero J, Saüch J, Sanz F, Furlong LI. The DisGeNET cytoscape app: exploring and visualizing disease genomics data. Comput Struct Biotechnol J. 2021 May 11;19:2960–7. CrossRef
15. Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards Suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016 Jun 20;54:1.30.1–33. CrossRef
16. McKusick VA. Mendelian inheritance in man. A catalog of human genes and genetic disorders. 12th ed. Baltimore, MD: Johns Hopkins University Press; 1998.
17. Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2016;44(D1):D67–72. CrossRef
18. Rappaport N, Nativ N, Stelzer G, Twik M, Guan-Golan Y, Stein TI, et al. MalaCards: an integrated compendium for diseases and their annotation. Database (Oxford). 2013 Apr 12;2013:bat018. CrossRef
19. von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005 Jan 1;33(Database issue):D433–7. CrossRef
20. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003 Nov;13(11):2498–504. CrossRef
21. Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(9):60.
22. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235–42.
23. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25(13):1605–12. CrossRef
24. Liu Y, Yang X, Gan J, Chen S, Xiao ZX, Cao Y. CB-Dock2: improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022 Jul 5;50(W1):W159–64.
25. Yang X, Liu Y, Gan J, Xiao ZX, Cao Y. FitDock: protein-ligand docking by template fitting. Brief Bioinform. 2022 May 13;23(3):bbac087.
26. Abbas A, Agusta MK, Dipojono HK, Saputro AG, Rachmawati H, Ismaya WT. Preliminary insight into recognizing of mannose toward LSMT protein: molecular docking and DFT study. J Appl Phys Sci. 2018;4(3):95–100. CrossRef
27. Lin Y, Shen C, Wang F, Fang Z, Shen G. Network pharmacology and molecular docking study on the potential mechanism of Yi-Qi-Huo-Xue-Tong-Luo formula in treating diabetic peripheral neuropathy. J Diabetes Res. 2021 May 28;2021:9941791. CrossRef
28. Wulandari AS, Tandrasasmita OM, Tjandrawinata RR. Bioactive fraction DLBS2411 from Cinnamomum burmannii (Nees and T. Nees) blume as colon and gastroprotector by stimulating MUC5AC and cyclooxygenase-2 gene expression. Int J Pharm Pharm Sci. 2016 Aug 1;8(8):202–7.
29. Kumar N, Mukhtar MS. Ranking plant network nodes based on their centrality measures. Entropy (Basel). 2023 Apr 18;25(4):676. CrossRef
30. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 Aug;596(7873):583–9. CrossRef
31. Kulesza A, Paczek L, Burdzinska A. The role of COX-2 and PGE2 in the regulation of immunomodulation and other functions of mesenchymal stromal cells. Biomedicines. 2023 Feb 3;11(2):445. CrossRef
32. Park SM, Yoo BC, Lee HR, Chung H, Lee YS. Distribution of prostaglandin E2 in gastric and duodenal mucosa: possible role in the pathogenesis of peptic ulcer. Korean J Intern Med. 1992 Jan;7(1):1–8. CrossRef
33. Mejías-Luque R, Cobler L, de Bolós C. MUC5AC (mucin 5AC, oligomeric mucus/gel-forming). Atlas Genet Cytogenet Oncol Haematol. 2010;14:566–9.
34. Tai HH, Tong M, Ding Y. 15-hydroxyprostaglandin dehydrogenase (15-PGDH) and lung cancer. Prostaglandins Other Lipid Mediat. 2007 May;83(3):203–8.
35. Tjandrawinata RR, Nailufar F, Arifin PF. Hydrogen potassium adenosine triphosphatase activity inhibition and downregulation of its expression by bioactive fraction DLBS2411 from Cinnamomum burmannii in gastric parietal cells. Int J Gen Med. 2013 Sep 23;6:807–15. CrossRef