INTRODUCTION
Streptomyces species are among the most industrially relevant producers of antibiotics and other bioactive compounds [1]. Some of these compounds, like tetracyclines, have been used due to their efficacy against a wide range of pathogens; and others have also contributed to the development of new medications, as the case of Lipstatin [2,3]. Clavulanic acid (CA) is a C-8 β-lactam antibiotic well-known for its β-lactamase inhibition capacity which is mainly produced by submerged cultures of Streptomyces clavuligerus [4]. It is synthesized through a complex biosynthetic pathway that condenses arginine (C-5) and glyceraldehyde 3-phosphate (C-3) molecules [5,6]. A series of studies in the 1980s identified glycerol as the C-3 precursor through 13C labeling experiments [7–9]. Only by the mid-1990s L-arginine was confirmed as the C-5 precursor over other candidates such as L-glutamic acid and L-glycine [10,11]. L-arginine, initially a C-6 molecule, is condensed with glyceraldehyde 3-phosphate to synthesize N2-(2-carboxyethyl)-arginine (a C-9 molecule). This C-9 structure is conserved in two reaction steps, but the ureido group of the arginine is lost in the formation of the intermediate proclavaminic acid (a C-8 molecule), thus five carbons of L-arginine are retained [5,6]. This C-8 structure is conserved up to the formation of CA. The chemical structure of CA and the β-lactam carbon precedence is shown in Figure 1.
Despite several studies on the optimization of nutritional conditions and metabolic and genomic perturbations, low CA yields persist during fermentation, motivating the application of metabolic engineering strategies like fluxomic and strain engineering, as alternatives for determination of feasible scenarios of production and improving the CA accumulation levels [5,12,13]. In addition, CA is temperature sensitive, which leads to its degradation during fermentation. The latter phenomenon further contributes to the low yields of CA [14,15].
Given the importance of the C-3 and C-5 precursors on CA synthesis, several organic nitrogen sources have exhibited a positive impact on CA production [16,17]. Suitable nitrogen sources could enhance the metabolic activity of the urea and tricarboxylic acid cycle (TCA) cycles, and therefore the CA biosynthesis [18,19]. Furthermore, understanding the assimilation of these types of nitrogen sources, including amino acids, as well as maintaining high and stable C-3 and C-5 fluxes, is a key factor in increasing CA production levels.
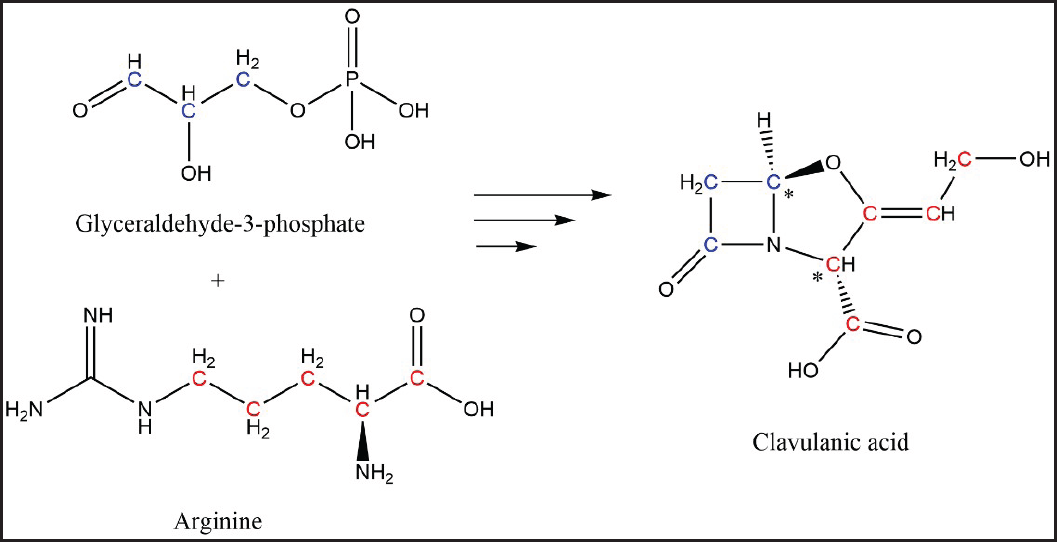 | Figure 1. Clavulanic acid chemical structure. Labeled red and blue atoms correspond to those coming from L-arginine and D-glyceraldehyde-3-phosphate, respectively. *Stereochemical centers on CA structure. [Click here to view] |
Over the last decade, diverse systems biology approaches have been applied for further understanding of Streptomyces metabolic pathways for metabolite production. In fact, some genome-scale constraint-based modeling and in silico simulations have been used to understand the characteristic low CA titers observed in wild-type strains, as well as for the rational selection of potential metabolic targets under different bioprocessing conditions [4,5]. Nevertheless, the knowledge about the role of C-5 amino acid precursors in the regulation of CA production is still low. Some studies have pointed out that amino acids connected with 2-oxoglutarate may contribute to enhancing CA biosynthesis and secretion [20–22].
Specifically, flux balance analysis (FBA) helped to understand S. clavuligerus physiology concerning nutritional conditions [23,24]. In addition, dynamic FBA (dFBA) has been used recently for this purpose [12,25], as dFBA is able to estimate numerically time-dependent concentrations and reactions [26,27]. Therefore, fed-batch and batch operation modes can be evaluated and analyzed employing this approach in contrast to the classical FBA.
In this work, a preliminary literature screening was performed to identify nutritional conditions leading to high CA secretion rates in wild-type S. clavuligerus cultivated in the bioreactor scale. After this initial screening, cultivations were performed at the shake flask scale to confirm the optimal conditions of production referred in the literature and further scaled up to the 5 l bioreactor scale. A genome-scale in silico study of the nutritional conditions attained in each experimental scenario of CA production was performed to get insights into the genome-scale dynamic distribution of the metabolic fluxes during the cultivation, focusing on the metabolism of C-5 precursors for further enhancement of CA biosynthesis in cultivations using with a wild-type S. clavuligerus.
MATERIALS AND METHODS
Dynamic genome-scale simulations
The static optimization approach was used in the dFBA formulation [12,26,27]. In this approach, each time interval assumes a pseudo-steady state, then, the FBA optimization problem is solved according to a dynamic set of hard constraints applied to the uptake of nutrients. The instantaneous uptake flux vector is obtained by solving a nonstructured model comprised of the set of ordinary differential equations presented in the following equations (Equations 1 to 4):
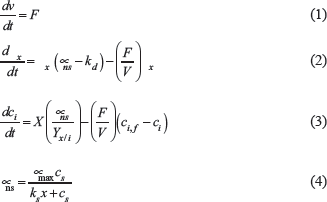 | [Click here to view] |
where F is the feed rate (when applicable, during fed-batch), V is the volume, Ci is the instantaneous concentration of species i at time t, Ci,f is the concentration of species i in the feeding, γx is the dry cell weight (DCW) biomass concentration, μns is the growth rate in the nonstructured model, μmax is the maximum experimentally observed growth rate, kd is the death constant, ks is the Monod constant for the substrate, and Cs is the substrate concentration.
The instantaneous FBA problem was solved by accounting for sub-optimal growth by means of a weighted objective function of maximum enzymatic efficiency and growth maximization. The rate of biomass production was conciliated by minimizing the error between the specific growth rates (μ) estimated by both, the unstructured dynamic model and the steady-state genome-scale model. The formulation of the optimization problem is shown as follows (Equation 5):
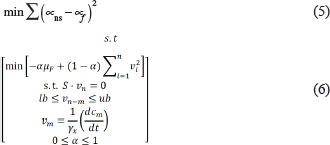 | [Click here to view] |
where μf is the growth rate estimated in the instantaneous optimization for solving the FBA problem, and vi is a vector of instantaneous intracellular fluxes. Similarly, vm is the constrained set of uptake fluxes calculated by solving the unstructured dynamic model system, and vn is the set of fluxes calculated by solving the FBA problem. The dFBA was implemented in MATLAB R2022b with Gurobi 10.0 optimization API.
Simulations were performed according to the concentration of the substrates, operation volume, feeding rate, and cultivation time reported in the literature for each set of conditions. The carbon utilization to synthesize the product of interest was assessed in terms of the ratio of the total carbon used in CA biosynthesis to the total carbon influx in batch and feed media. In addition, time profiles of reaction fluxes toward key pathways and enzymes impacting CA biosynthesis were analyzed to adjust further fed-batch operation for the media under evaluation. Finally, a medium modification for enhanced fed-batch production of CA was proposed based on experimental data and in silico fluxes prediction.
Strain and media compositions
The literature search was carried out in ScienceDirect, Scopus, and PubMed databases using predetermined terms such as “S. clavuligerus fermentation” and “CA production,” exclusively focusing on research articles that used a wild-type strain. The scenarios yielding the best carbon utilization for its biosynthesis in silico were tested after the initial screening of fed-batch and batch cultivations available in the literature. Fed-batch operation was adjusted according to the observed in silico distribution of metabolic fluxes to improve the carbon utilization in CA biosynthesis. According to this rationale, a complex medium and two synthetic media were selected to promote the highest CA production.
Streptomyces clavuligerus DSM 41826, cryo-preserved, was activated in seed medium (50 ml) and pre-cultured in 250 ml as indicated in previous works [28]. All cultivations were carried out in a 5.0 l stirred-tank bioreactor (Labfors 5, Infors AG, Bottmingen, Switzerland). Batch cultivations were performed in a volume of 2.0 l. Fed-batch cultivations were operated up to a final volume of 4.5 l. Inoculum of 10% v/v from the preculture was used. The pH was maintained at 6.8 by using 4 M solutions of either hydrochloric acid or sodium hydroxide. Air was supplied at 0.6 vvm and temperature was controlled at 28°C. Agitation was controlled in the range of 300–600 rpm, assuring dissolved oxygen ranging between 40% and 75%. Sampling was performed as described elsewhere [28].
After the screening, the media for experimental testing were selected. The complex medium, referred to here as isolated soy protein (ISP), was used and prepared as specified previously [21,29,30]. Similarly, a synthetic medium named here as glycerol-sucrose-proline-glutamate (GSPG), optimized by Saudagar and Singhal was prepared [31,32]. Likewise, the so-called restricted medium modified with ornithine (RMO), previously used by Gómez-Ríos [23], was also evaluated. All reactants were from Merck-Sigma-Aldrich (Darmstadt, Germany).
Preliminary simulations were performed to set up the feeding start and flow rate to avoid substrate accumulation during the culture (data not shown). All the fed-batch cultures were operated at 120 hours with the same basal media as that used for batch operation. The carbon and nitrogen feeding composition corresponded to those reported by the authors who previously used those media in CA production with remarkable results in wild-type cultivations [21,25,31]. The fed-batch cultivation with the ISP medium had the following composition in g L−1: glycerol, 180.04; L-ornithine HCl, 4.72; K2HPO4, 2.0; and (NH4)2SO4, 8.0; feed rate 0.01 L−1, feeding time 19 hours. For the GSPG medium, the fed-batch composition was the following in g L−1: L-arginine HCl, 21.07; L-threonine, 119.12; K2HPO4, 2.0; and (NH4)2SO4, 8.0; feed rate 0.02 L−1, feeding time 18 hours. In the case of the RMO cultivation, the feed was prepared as follows (concentration in g L−1): glycerol, 27.6; K2HPO4, 2.0; (NH4)2SO4, 8.0 and mono-sodium glutamate, 25.4; feed rate 0.035 L−1, feeding time 25 hours [28].
Experimental testing of the proposed scenario of improved CA yield was performed at the same bioreactor scale, using a modified GSPG medium prepared as follows (concentrations in g L−1): sucrose, 12.09; glycerol, 9.3; NaCl, 5.0; L-proline, 11.36; monosodium glutamate, 9.8; K2HPO4, 2.0; CaCl2, 0.4; MnCl2·4H2O, 0.1; MgSO4·7H2O, 1.0; ZnCl2, 0.05; FeCl3·6H2O, 0.1; and trace elements solution (1.0 ml). The feeding conditions were the same as those used for the RMO fed-batch culture, delaying the feeding time up to 34 hours to promote a precise substrate addition and optimal carbon utilization according to the in silico study of the production scenario.
RESULTS AND DISCUSSION
The impact of several complex nitrogen sources on CA production has been studied in batch and fed-batch operations. Rich nitrogen sources such as peptone, isolate soybean protein, yeast extract, and soybean meal, are commonly used for CA production, usually leading to high concentrations in bioreactor scale (over 600 mg/l) with wild-type strains [17,29,33]. The highest CA concentrations in the bioreactor scale for both batch and fed-batch operations have been reported for glycerol and isolate soy protein-based media [21] while the highest concentration for a synthetic media was reported by Saudagar and Singhal for a medium based on glycerol, sucrose, proline, and glutamate [31]. In this study, we examined the effect of fed-batch operation with amino acid supplementation, from a dynamic-fluxomic standpoint as an approach for tuning of fed-batch S. clavuligerus cultures.
Batch and fed-batch operation mode for CA production
Figure 2 displays the specific CA concentrations (bCA) and the biomass (DCW) in batch and fed-batch operations with the three media tested. Batch production is still the most used operation mode in the pharmaceutical industry [5,34]. In the case of S. clavuligerus, the low specific growth rate constitutes a challenge for continuous operation. In batch conditions, the complex ISP and synthetic GSPG showed higher biomass accumulation, 17.0 and 18.9 gDCW/l, respectively, than the RMO medium (10.8 gDCW/l). The GSPG medium contains a high amount of free amino acids that promoted more growth than the complex medium ISP did. As has been extensively reported, phosphate limitation is key in the activation of secondary metabolism and for achieving high CA secretion rates [24,31]; this condition is attained rapidly in the RMO medium, inducing CA biosynthesis since the first hours of cultivation. In the case of the rich media, CA biosynthesis was detectable from 10 hours onward. The exponential phase lasted between 35 and 45 hours in both the GSPG and the ISP medium with a μmax = 0.09/hours. More limited RMO medium led to a longer growth phase and lower biomass levels, with an up to the 55th hour with μmax = 0.07/hours. In terms of CA secretion, both synthetic media yielded higher mean specific production rates (0.016 mmol CA/gDCW.h and 0.012 mmol CA/gDCW.h for the GSPG and RMO media, respectively), which are higher than that obtained in the rich nitrogen medium ISP (0.010 mmol CA/gDCW.h). This synthetic medium was selected to promote high uptake of glycerol (C-3) and amino acids (C-5), as CA precursors and hence, for better carbon channeling via glycolysis and urea cycle toward the CA pathway. The RMO medium provides less carbon and total nitrogen than the ISP and GSPG media (Table 1), hence lower maximum biomass and specific growth rate were expected. The final specific CA concentration in RMO cultivations was lower than that reached in the GSPG cultures, but it was expected given the lower initial concentration of C-3 and C-5 precursors. Figure 2a and b shows the biomass and specific CA time courses for batch cultures of S. clavuligerus showing the absence of a lag phase, possibly as a consequence of the high volume of inoculum and the high concentration of cells in the seed culture.
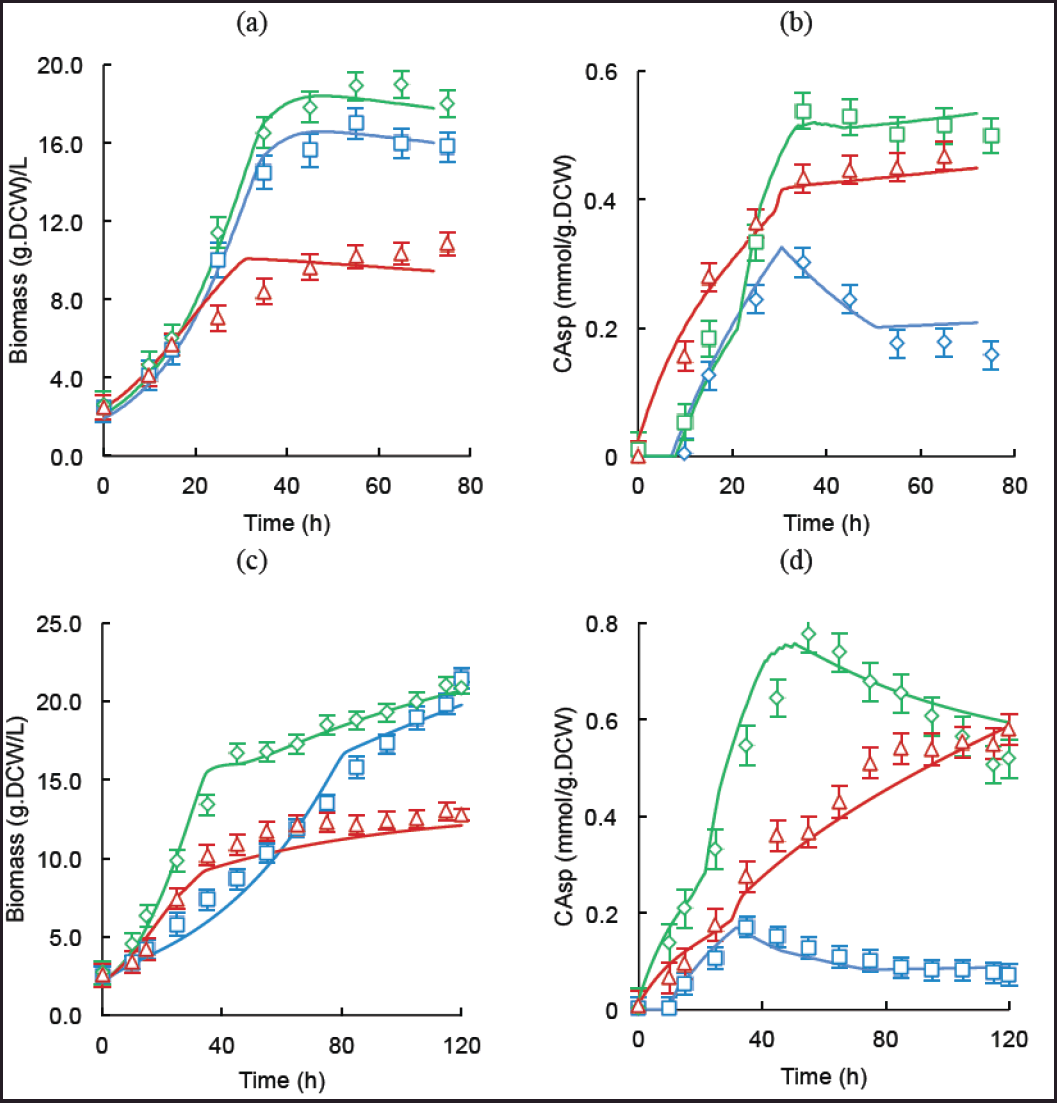 | Figure 2. Experimental and simulated time courses of batch and fed-batch cultivations of S. clavuligerus in ISP (blue), GSPG (green) and RMO (red) media. (a) Biomass concentration (γx) in batch cultures. (b) CA specific concentration (bCA) in batch cultures. (c) Biomass concentration (γx) in fed-batch cultures. (d) CA specific concentration (bCA) in fed-batch cultures. [Click here to view] |
Figure 2c and d displays the biomass and specific CA time courses for the fed-batch operation of S. clavuligerus cultures. In the case of fed-batch cultures, the results agreed with those of the batch operation, yielding higher biomass and CA concentrations, as a consequence of more extended growth and stationary phases with maintenance of the nutritional conditions that promoted continuity of the secondary metabolism and CA accumulation. The biomass concentrations reached 21.5, 20.9, and 13 g L−1 for the ISP, GSPG, and RMO media, respectively. An earlier feeding than the reported for those media allowed to avoid drops in biomass concentration and CA secretion, as well as a sustained growth considering the dilution rate due to feed addition. The addition of phosphate in small amounts in the feed media regulated growth while maintaining CA biosynthesis. The CA concentrations attained in the fed-batch cultures were 0.17, 0.77, and 0.58 mmol CA/gDCW of biomass for the ISP, GSPG, and RMO media, respectively. The high synthesis rate of biomass in the ISP medium did not reflect a high CA production, while the synthetic media supplying a high concentration of C-3 and C-5 precursors favored the CA accumulation concomitantly with biomass. The RMO medium promoted considerably lower biomass, indicating a strong limitation of nutrients, but this favored high synthesis rates of CA considering the low biomass concentrations.
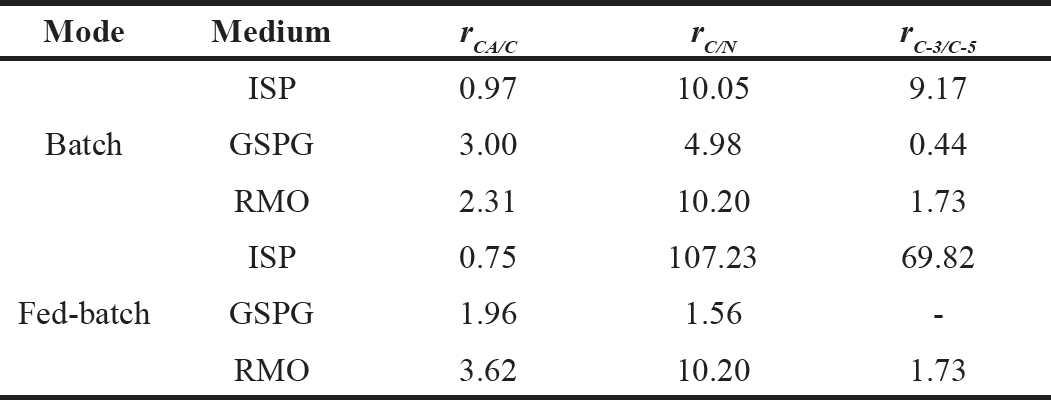 | Table 1. CA carbon utilization (rCA/C), carbon to nitrogen (rC/N) and C-3/C-5 precursors (rC-3/C-5) ratios in S. clavuligerus cultivations. [Click here to view] |
Table 1 presents the percentage of carbon dedicated to CA biosynthesis regarding the total carbon in the media, the carbon to organic nitrogen, and the C-3 to C-5 precursors ratio. The ISP medium has a high glycerol concentration, and it is expected that most of the carbon uptake proceeds via the glycolytic pathway. The medium is rich in biomass precursors but not only C-5 amino acids, which promote biomass synthesis but comparatively lower CA production, leading to a ~1.0% of carbon effectively dedicated to synthesizing CA: The GSPG medium has a more balanced C/N and C-3/C-5 composition, which presumably give a better cellular response to phosphate limitation, increasing the carbon usage for CA biosynthesis (3.0%). The RMO medium showed an intermediate carbon utilization in CA biosynthesis (2.3%) in batch operation. However, in fed-batch operation, the C/N and C-3/C-5 ratio was maintained in the feeding medium, yielding a closer carbon utilization to the batch culture, while the feeding composition for the ISP and GSPG media reduced the carbon diverted to secondary metabolism, possibly due to lack of one precursor, either C-3 or C-5. This suggests a longer and more stable production phase when providing both precursors in the feed.
Genome-scale dynamic flux-distribution of S. clavuligerus cultures in batch and fed-batch operation
Figures 3 and 4 summarize the dynamic distribution of metabolic fluxes for the main pathways related to CA biosynthesis in batch and fed-batch operation modes for the three media tested, i.e., ISP, GSPG, and RMO media. As mentioned, CA accumulation depends on reaching the phosphate limitation but having precursors available. Although the availability of the C-3 precursor sustains the CA biosynthesis even after a C-5 precursor starvation, the CA secretion rate is lower in this condition, as shown in previous experimental reports [28,33].
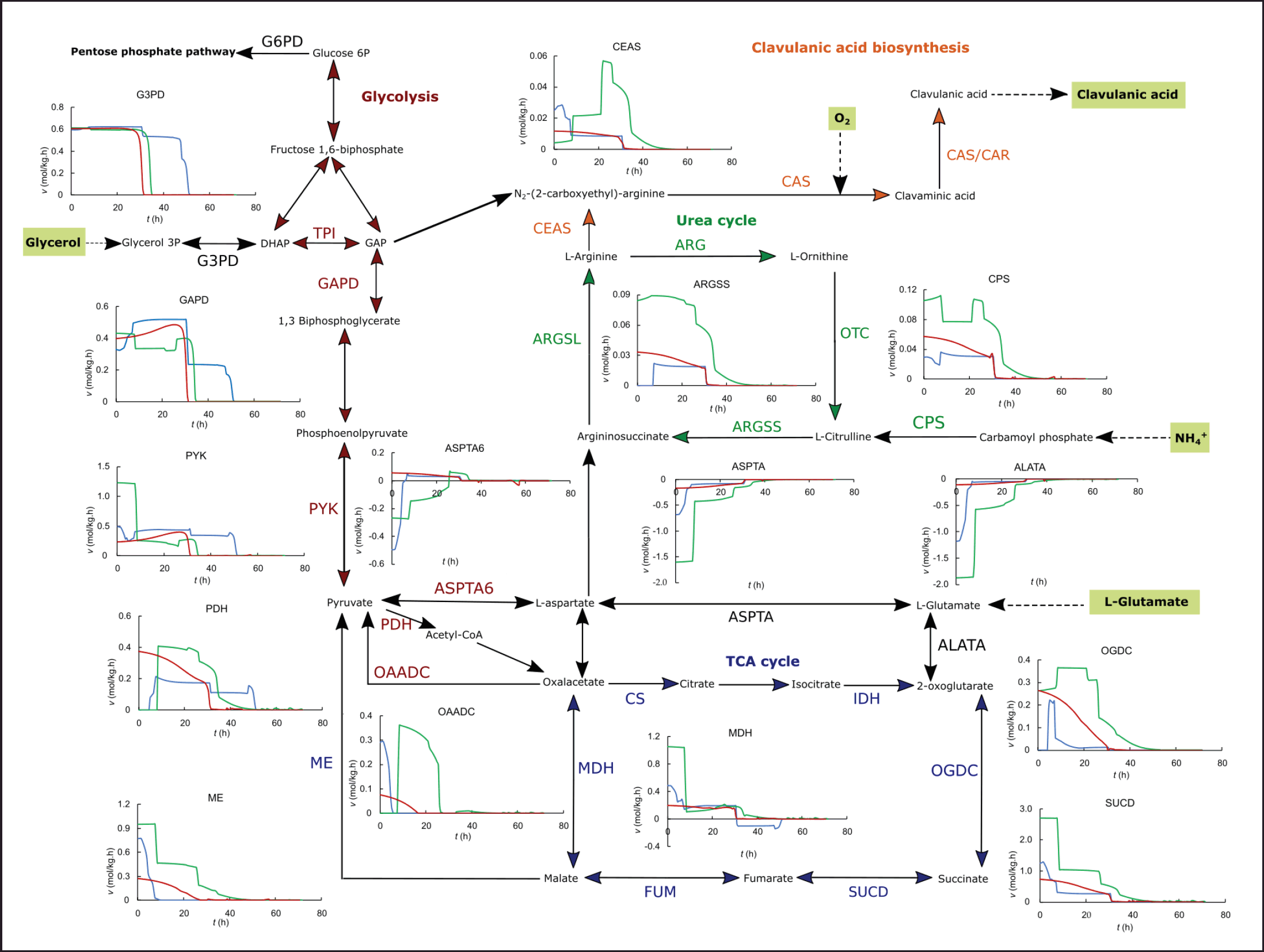 | Figure 3. In silico dynamic flux distribution in the central metabolism and CA biosynthesis for the ISP (blue), GSPG (green), and RMO (red) media, in batch operation. [Click here to view] |
Streptomyces clavuligerus has a deficient transportation of glucose, thus glycerol is often used as the carbon source for CA production [5,22]. The dynamic flux distribution shows that the flux through the glyceraldehyde-3-phosphate dehydrogenase (GAPD) is highly dependent on the extracellular glycerol concentration, and hence the glycerol uptake. In all cases, and more pronounced in the batch operation, glycerol depletion leads to a strong decrease in the glycolysis flux, also shown by the trend of pyruvate dehydrogenase (PDH). However, in the case of the GSPG cultivations, the C/N ratio is ~5, and the intracellular fluxes in silico show that the activity in central metabolism is higher in the direction of the urea cycle and upstream of the TCA cycle, as a consequence of high uptake of amino acids. Proline and glutamate are direct biomass precursors, the high availability of those free amino acids in the medium leads to a decrease in the demand for glycerol, as suggested by the lower fluxes through GAPD. The effect of proline and glutamate uptake during the cultivation also occurs in reactions that use 2-oxoglutarate as a reactant, as is the case of reactions catalyzed by 2-oxoglutarate dehydrogenase and succinyl-CoA synthetase. 2-oxoglutarate is produced by various amino acid degradation reactions, as well as pyruvate, which would stimulate the flux through PDH.
In batch operation, the highest fluxes in the urea cycle and through carboxyethyl arginine synthase (CEAS) as the first reaction of the CA pathway occur for the GSPG medium since proline and glutamate incorporate directly via aspartate toward arginine and later, CA biosynthesis. Proline, the major component of the medium, is degraded via glutamate semialdehyde, which also yields glutamate. Thus, glutamate and proline increase substantially the fluxes of aspartate transaminase (ASPTA), argininesuccinate synthase (ARGSS) in the urea cycle, as well as CEAS. The mix of amino acids in the ISP medium provides free C-5 precursors to a lesser extent than those added directly to the GSPG medium, leading to lower fluxes in the arginine direction of the urea cycle and CA biosynthesis. The fluxes in the CA biosynthesis direction from the urea cycle were marginally higher for the RMO medium than for the ISP, given the higher concentration of glutamate, which is a direct C-5 precursor; while the major contribution of C-5 from ISP comes from arginine and proline in lower concentrations. In both synthetic media, the flux results confirm the importance of adding high concentrations of C-5 precursors in the glutamate-aspartate direction for promoting high CA biosynthetic fluxes in batch operation.
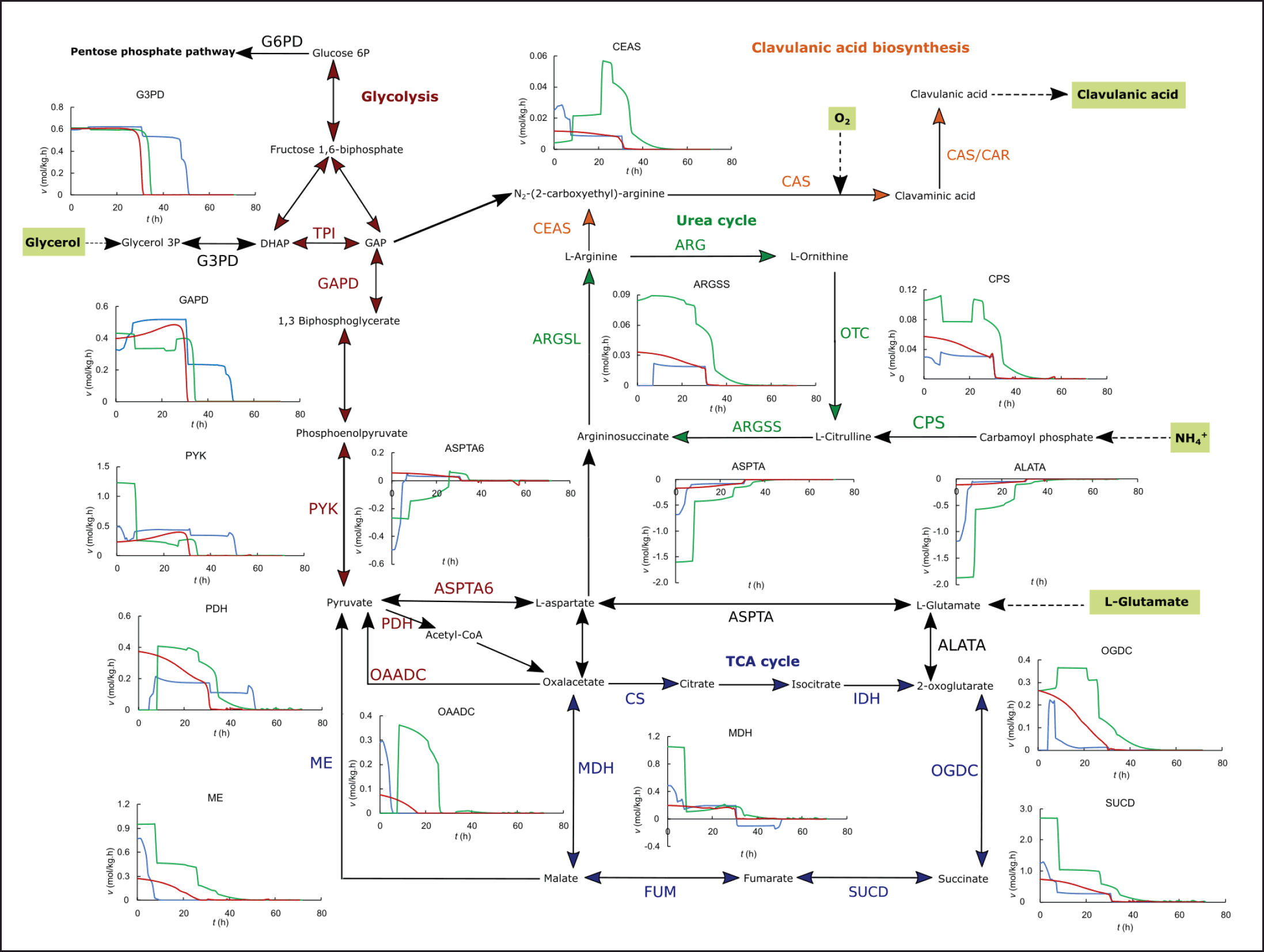 | Figure 4. In silico dynamic flux distribution in the central metabolism and CA biosynthesis for the ISP (blue), GSPG (green), RMO (red), and modified GSPG (yellow) media, in fed-batch operation. [Click here to view] |
The effects of amino acid supplementation on CA production are still under discussion since a wide range of conditions and supplements have been shown to increase CA secretion rate [5,22]. However, several experimental studies have concluded that the addition of urea cycle intermediates such as arginine and ornithine, although it has a positive effect, it has not been as high as expected in boosting CA production, considering that arginine is the precursor of the first product in the CA pathway [21,31,35]. In silico fluxes suggest that supplementation with C-5 precursors in the line of glutamate/aspartate degradation has a more significant effect on increasing CA secretion rates. This is likely due to the role of 2-oxoglutarate in CA biosynthesis and TCA cycle regulation. Previously, it was verified experimentally that 2-oxoglutarate supplementation may favor CA secretion, but not as much as C-5 amino acids [31,32]. This experimental test was supported by the fact of the requirement of 2-oxoglutarate for CA production since three molecules of 2-oxoglutarate are consumed per molecule of CA synthesized; specifically, in the oxidizing steps catalyzed by clavaminate synthase, which yields succinate [5]. The in silico fluxes suggest that the addition of either glutamate or glutamate precursors might increase the cataplerotic flux of 2-oxoglutarate from the glutamate and aspartate transaminations by ASPTA and alanine transaminase (ALATA) as observed in simulations of cultures with high organic nitrogen content (Fig. 3). This would increase the fluxes in the urea cycle toward arginine and thus, toward carboxyethyl arginine in the CA pathway (see CEAS in Fig. 3), under an adequate C-3 uptake via glyceraldehyde 3-phosphate. The increasing anaplerotic flux toward 2-oxoglutarate also stimulates the further oxidative steps of the TCA cycle, such as those catalyzed by 2-oxoglutarate dehydrogenase (OGDC) and succinyl-CoA synthetase, as shown in Figure 3, while increasing the cataplerotic disposal of malate and oxalacetate via the malic enzyme (ME) and oxalacetate decarboxylase (OAADC), both to pyruvate (see ME and OAADC in Fig. 3).
In all cases, the initial high fluxes in the anaplerotic reactions of amino acid synthesis/degradation catalyzed by L-alanine-alpha-keto acid aminotransferase (ASPTA6 and ALATA, in Fig. 3) decrease as the concentration of C-5 precursors in the media depletes. The higher the concentration of C-5 precursors in the basal media, the longer those fluxes remain during the exponential growth as displayed in Figure 3 for the batch operation. Given the lower initial concentration of the glutamate family precursors in the ISP medium, the fluxes in the anaplerotic metabolism of amino acids drop in the first 10 hours of cultivation, turning into the lowest among the three media tested. The time course of the fluxes during the batch culture suggests that most of the carbon proceeds via glycolysis-TCA toward the urea cycle, given the high C-3/C-5 ratio (9.17). Although this would stimulate fluxes in the TCA, it would do so to a lesser extent in the urea cycle and CA biosynthesis, compared to those media with a narrower C-3/C-5 balance. The CA biosynthetic flux for the ISP medium approaches that estimated for the RMO medium after 10 hours, despite the considerably lower C-3 and amino acid concentrations of the latter. In batch operation, the simulations show that the high concentration of proline and glutamate in the GSPG medium manages to maintain high fluxes through reactions related to CA biosynthesis, such as ALATA, ASPTA, ARGSS, and CEAS, for much longer; even in the face of the high demand for C-5 precursors. This is not so in the ISP medium, in which the uptake of C-5 precursors leads to a rapid depletion that negatively impacts fluxes in CA synthesis.
Interestingly, pyruvate shows a relationship with CA synthesis despite not being a direct precursor as previously hypothesized [36]. The fluxes through PDH estimated for the ISP medium are approximately half of those estimated for GAPD, considering the various bifurcations of the glycolytic pathway and that the assimilable carbon source is glycerol, given its high initial concentration in the medium. Conversely, in the case of both synthetic media, the fluxes through PDH are almost the same or even higher than those in GAPD, which is probably connected to the high uptake of C-5 precursors downstream of the TCA cycle and the conversion of oxalacetate and malate into pyruvate. This behavior was observed in silico only for the scenarios of high concentration of C-5 precursors in the basal media, i.e., for GSPG and, to a lesser extent, for RMO media. The high flux through PDH drops once the C-5 is depleted in the culture and the flux in the anaplerotic reactions of glutamate and aspartate decreases (see ASPTA6 and ALATA in Fig. 3). This scenario would cause a small accumulation of pyruvate, especially at the end of the culture, since the demand for pyruvate decreases substantially in the stationary phase. Recently, Fu et al. reported metabolomic and transcriptomic data comparing rich organic nitrogen media [16]. Media enhancing CA accumulation upregulated intracellular glutamate, pyruvate, and succinate, concomitantly with decreasing pools of 2-oxoglutarate, ornithine, citrulline, aspartate, argininosuccinate, and arginine, compared to a complex medium producing a lower CA secretion in batch operation [16].
It has been widely reported that the highest CA production in bioreactors for wild-type strains of S. clavuligerus is achieved by fed-batch operation. Different feeding media have been reported with good results in enhancing CA secretion rates and increasing maximum biomass and exponential growth phase. Threonine and arginine were added in fed-batch operation to the GSPG medium, based on the experimental results of Saudagar and Singhal [31]. However, several studies have highlighted the importance of adding a C-3 source, typically glycerol. The feeding medium consisting of threonine and arginine modulates the fluxes through glycolysis to maintain the flux of C-3 toward CA biosynthesis. The in silico flux distribution suggests that threonine degradation to glycine and further to serine contribute to supply pyruvate and, in consequence, a C-3 precursor in the absence of glycerol (see glycerol 3-phosphate dehydrogenase (G3PD), GAPD, PDH, and CEAS in Fig. 4). As a biomass precursor, glycine may be synthesized from serine, and this occurs during the batch operation. On the contrary, when threonine feeding starts, the conversion of glycine to serine by hydroxymethyl transferase increases the flux in the direction of serine, which is further metabolized into pyruvate. Nevertheless, most of the relevant reports about fed-batch production of CA point to a glycerol-based feeding medium to support biomass synthesis and energetic metabolism as well as to provide a direct C-3 precursor of CA.
The purpose of the fed-batch operation is to extend the productive phase of the cultivation; the dynamic simulation suggests that this goal is better achieved by maintaining more stable fluxes along the pathways related to CA [25]. As observed in the simulation of batch cultures, the intracellular fluxes tend to drop drastically as the substrates deplete, and cells must adapt to the scarcity by deaccelerating growth and secreting metabolites that help energetic metabolism regulation [24,37–39]. As observed in the dynamic simulations, the fed-batch operation extends a stable flux toward central metabolism beyond 50 hours of cultivation, i.e., during the stationary phase, even in the secondary metabolism favoring CA continued secretion under phosphate limitation. The fed-batch operation for the ISP medium, consisting of glycerol and a small amount of ornithine, would maintain the fluxes along glycolysis and the TCA cycle, but is less effective in promoting fluxes in the urea cycle and CA biosynthesis (Fig. 4). In the case of the GSPG medium with the feeding of threonine and arginine, it is plausible that the effect on maintaining the fluxes in the urea cycle and CA biosynthesis during the stationary phase but with low fluxes through glycolysis owing to the earlier depletion of C-3 and C-6 as the most assimilable carbon sources. Despite the lack of a direct C-3 precursor, the fluxes through CEAS are slightly higher than those in the RMO medium. However, the RMO medium has a lower content of total carbon, C-3 and C-5 precursors in batch and fed-batch (Table 1). This leads to more efficient carbon usage, considering the product of interest. The GSPG medium promotes both high biomass and high CA production in batch, but a feeding consisting mainly of highly concentrated threonine does not lead to better carbon utilization than the feeding medium consisting of glutamate and glycerol used for the RMO fed-batch cultivation.
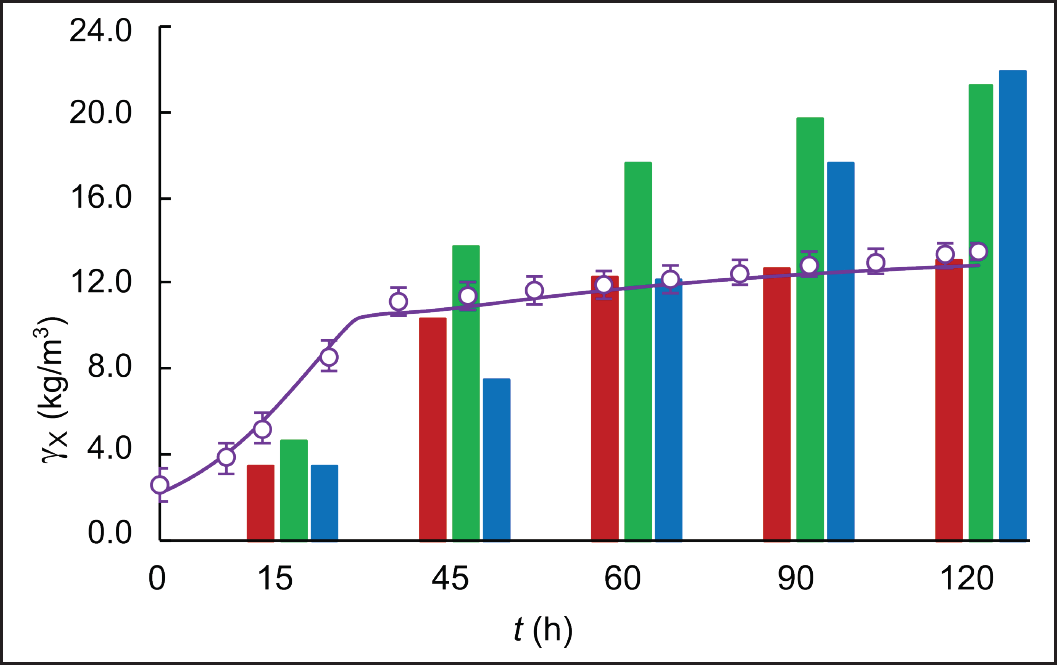 | Figure 5. Experimental (circles) and simulated (solid line) time course of biomass concentration in fed-batch cultivations of S. clavuligerus in modified GSPG medium (purple). Comparison of experimental points of biomass concentration (bars) in cultures with RMO (red), GSPG (green), and ISP (blue) media. [Click here to view] |
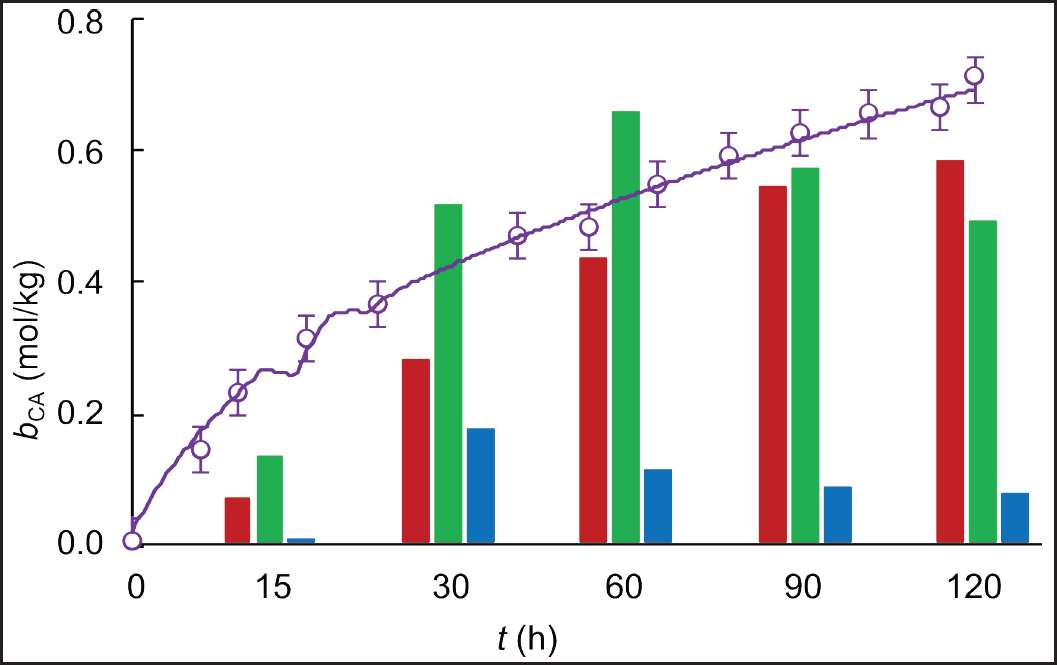 | Figure 6. Experimental (circles) and simulated (solid line) time course of CA-specific concentration in fed-batch cultivations of S. clavuligerus in modified GSPG medium (purple). Comparison of experimental points of CA-specific concentration (bars) in cultures with RMO (red), GSPG (green), and ISP (blue) media. [Click here to view] |
A fed-batch cultivation scenario for enhancing CA production
Dynamic simulations at the genome scale have provided insights into the stoichiometric effect of amino acid supplementation on S. clavuligerus batch and fed-batch cultivations. According to those observations and contrasting with the experimental results at the bioreactor scale, a fed-batch production scenario was proposed by modifying the GSPG basal medium and the respective feeding for better carbon utilization in terms of CA biosynthesis. Dynamic simulation of CA production using the proposed medium modification was done to confirm the potential CA enhancement, adjustment of the feeding time according to the carbon sources uptake rate, and effect on intracellular fluxes through key enzymes related to CA. The reaction fluxes for this proposed fed-batch production scenario were included in Figure 4 for the sake of comparison with the other conditions previously tested. The composition of the GSPG medium followed the same C/N and C-3/C-5 ratio which led to a carbon usage of 3.0% toward CA. The feeding medium was the one used in the RMO medium, which gave a better carbon utilization during the fed-batch operation owing to the provision of both precursors, glycerol, and glutamate. The feeding time was delayed to 34 hours considering the higher amount of carbon sources in the medium, thus preventing substrate accumulation and overflow metabolism [24,28].
Moreover, the dynamic FBA applied to batch and fed-batch cultivations with the ISP, GSPG, and RMO media has shown the importance of 2-oxoglutarate and pyruvate in modulating CA biosynthesis in connection with C-5 amino acids metabolism. The experiments and in silico flux distribution suggest a fed-batch operation at a low dilution rate considering the specific growth rate of S. clavuligerus under phosphate limitation. Previously, it was shown that starting the fed-batch operation before phosphate and carbon limitation prevents sharp declines in carbon fluxes. As observed, a feed composed of C-3 and C-5 direct precursors might maintain higher and more stable fluxes through the urea cycle and CA biosynthesis, than when using glycerol or amino acids as the only assimilable substrate. The feed medium formulation for S. clavuligerus cultures for CA production should provide sufficient glycerol as a C-3 precursor and a C-5 precursor such as glutamate or proline, as well as ammonia and phosphate in narrow concentrations to maintain the limitation that releases the antibiotic synthesis, while maintaining the activity along central metabolism and avoiding substrate accumulation or strong starvation potentially hampering the antibiotic secretion.
A higher specific CA concentration was obtained in the fed-batch scenario with modified GSPG medium, 0.72 mmol CA/gDCW compared to 0.69 mmol CA/gDCW reached with the GSPG medium in the first fed-batch scenario (Fig. 6). Although closer, as observed in Figures 5 and 6, the highest CA-specific concentration was attained at a maximum biomass concentration of 13.5 g L−1, which is 55% lower than that obtained in the standard GSPG medium (see Fig. 5). This suggests a better carbon utilization by the secondary metabolism, since less biomass is synthesizing more CA. Indeed, the ratio of total carbon used in CA biosynthesis was higher for the fed-batch cultivation with the modified GSPG medium (3.34%), suggesting a higher selectivity toward the product of interest. Although CA is not strictly a growth-coupled product, in a defined medium, the more carbon is channeled to a particular pathway, the less carbon is available for biomass synthesis; since the antibiotic’s biosynthesis recruits precursors directly from the central carbon metabolism. By feeding carbon in a precise concentration and flow rate, overflow metabolism and deviations toward pathways synthesizing side products are avoided. Both the nutritional restriction and the cultivation conditions favored CA biosynthesis, allowing sustained production with a quasi-stationary biomass production due to the dilution and cell death, as occurred in chemostat cultivations. Moreover, during bioreactor operation, it is desirable to have high CA-specific production rates at low biomass concentrations, since the S. clavuligerus mycelium confers to the broth a non-Newtonian behavior, increasing density and viscosity as biomass increases [28]. The dense and dispersed mycelia make it difficult to maintain high dissolved oxygen tension at moderate agitation rates, facilitating bioprocess control.
The dynamic flux distributions shown in Figures 4 and 4 suggest that metabolic fluxes in this scenario would be rather stable with this operation, avoiding the sharp fluctuations of the instantaneous fluxes that are estimated for the richer media used in this study, favoring the continued secretion of CA, without an apparent decline during all the fed-batch operation. As expected, the composition of the GSPG-modified medium allowed high and stable intracellular fluxes in the TCA and urea cycles (e.g., through ARGSS and ASPTA in Fig. 4), favoring the flux toward CA biosynthesis. Although the fluxes in central carbon metabolism were marginally lower than those estimated for the GSPG standard medium due to the reduced glycerol, glutamate, and proline concentrations, the higher carbon dedicated to CA production instead of biomass would result in lower operation costs for achieving comparable CA final concentrations. In addition, by producing CA in defined media, downstream processing might be simplified since fewer metabolites, biomass, and debris are present in the broth.
CONCLUSION
The dynamic flux distribution obtained from simulations with experimental constraints provided insights into the importance of the availability of C-5 precursors and highlighted the importance of 2-oxoglutarate and pyruvate in modulating CA biosynthesis in S. clavuligerus. The application of a dynamic modeling approach highlighted the importance of adding C-5 precursors in the line of glutamate and aspartate, in promoting high CA biosynthetic fluxes in both, fed-batch and batch operation. The addition of glutamate or glutamate precursors might increase the fluxes in the urea cycle toward arginine and thus, carboxyethyl arginine in the CA pathway, leading to enhanced CA production in wild-type S. clavuligerus. The use of a fed-batch cultivation scenario with a modified GSPG medium and adjusted feeding strategy enhanced CA production at lower biomass concentrations, and better carbon utilization in CA biosynthesis, which are desirable conditions for culture operation. The dynamic simulations suggest that metabolic fluxes in this scenario would be rather stable, favoring the continued secretion of CA. Overall, the proposed fed-batch cultivation scenario can contribute to the development of efficient CA production processes in S. clavuligerus.
LIST OF ABBREVIATIONS
ALATA: alanine transaminase; ARGSS: argininosuccinate synthase; ASPTA: aspartate dehydrogenase; ASPTA6: L-alanine-alpha-keto acid aminotransferase; CEAS: N2-(2-carboxyethyl)-arginine synthase; G3PD: glycerol 3-phosphate dehydrogenase; GAPD: glyceraldehyde 3-phosphate dehydrogenase; GSPG: glycerol-sucrose-proline-glutamate medium.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
Authors acknowledge the financial support of Universidad de Valle, grant CI 21258.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This article does not contain any studies with human participants or animals.
INFORMED CONSENT
For this type of study formal consent is not required.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Cuozzo S, de Moreno de LeBlanc A, LeBlanc JG, Hoffmann N, Tortella GR. Streptomyces genus as a source of probiotics and its potential for its use in health. Microbiol Res. 2023 Jan;266:127248.
2. Petkovic H, Lukežic T, Šuškovic J. Biosynthesis of oxytetracycline by streptomyces rimosus: past, present and future directions in the development of tetracycline antibiotics. Food Technol Biotechnol. 2017;55(1):3.
3. Sladic G, Urukalo M, Kirn M, Lešnik U, Magdevska V, Benicki N, et al. Identification of lipstatin-producing ability in Streptomyces virginiae CBS 314.55 using dereplication approach. Food Technol Biotechnol. 2014 Jul 1;52:276–84.
4. Ramirez-Malule H. Bibliometric analysis of global research on clavulanic acid. Antibiotics (Basel) [Internet]. 2018 Nov 26;7(4):102. Available from: https://pubmed.ncbi.nlm.nih.gov/30486255
5. López-Agudelo VA, Gómez-Ríos D, Ramirez-Malule H. Clavulanic acid production by Streptomyces clavuligerus: insights from systems biology, strain engineering, and downstream processing. Antibiotics (Basel) [Internet]. 2021 Jan 18;10(1):84. Available from: https://pubmed.ncbi.nlm.nih.gov/33477401
6. Paradkar A. Clavulanic acid production by Streptomyces clavuligerus: biogenesis, regulation and strain improvement. J Antibiot (Tokyo) [Internet]. 2013;66(7):411–20. Available from: http://dx.doi.org/10.1038/ja.2013.26
7. Elson SW, Oliver RS. Studies on the biosynthesis of clavulanic acid. I. Incorporation of 13C-labelled precursors. J Antibiot (Tokyo) [Internet]. 1978;31(6):586–92. Available from: http://dx.doi.org/10.7164/antibiotics.31.586
8. Elson SW, Oliver RS, Bycroft BW, Faruk EA. Studies on the biosynthesis of clavulanic acid. III. Incorporation of DL-(3,4-13C2) glutamic acid. J Antibiot (Tokyo) [Internet]. 1982;35(1):81–6. Available from: http://dx.doi.org/10.7164/antibiotics.35.81
9. Stirling I, Elson SW. Studies on the biosynthesis of clavulanic acid. II. Chemical degradations of 14C-labelled clavulanic acid. J Antibiot (Tokyo) [Internet]. 1979;32(11):1125–9. Available from: http://dx.doi.org/10.7164/antibiotics.32.1125
10. Elson SW, Baggaley KH, Davison M, Fulston M, Nicholson NH, Risbridger GD, et al. The identification of three new biosynthetic intermediates and one further biosynthetic enzyme in the clavulanic acid pathway. J Chem Soc Chem Commun [Internet]. 1993;(15):1212. Available from: http://dx.doi.org/10.1039/c39930001212
11. Khaleeli N, Li R, Townsend CA. Origin of the β-lactam carbons in clavulanic acid from an unusual thiamine pyrophosphate-mediated reaction. J Am Chem Soc [Internet]. 1999;121(39):9223–4. Available from: http://dx.doi.org/10.1021/ja9923134
12. Gómez-Ríos D, Ramírez-Malule H, López-Agudelo VA. Dynamic in silico assessment of potential gene targets for enhancing clavulanic acid production in Streptomyces clavuligerus submerged cultures. J Appl Pharm Sci [Internet]. 2022; Available from: http://dx.doi.org/10.7324/japs.2023.130207
13. Kurt-Kizildogan A, Vanli-Jaccard G, Mutlu A, Sertdemir I, Özcengiz G. Genetic engineering of an industrial strain of Streptomyces clavuligerus for further enhancement of clavulanic acid production. Turk J Biol [Internet]. 2017;41:342–53. Available from: http://dx.doi.org/10.3906/biy-1608-17
14. Gómez-Ríos D, Ramírez-Malule H, Neubauer P, Junne S, Ríos-Estepa R. Degradation kinetics of clavulanic acid in fermentation broths at low temperatures. Antibiotics (Basel) [Internet]. 2019 Jan 17;8(1):6. Available from: https://pubmed.ncbi.nlm.nih.gov/30658482
15. Marques DAV, Oliveira RPS, Perego P, Porto ALF, Pessoa A, Converti A. Kinetic and thermodynamic investigation on clavulanic acid formation and degradation during glycerol fermentation by Streptomyces DAUFPE 3060. Enzyme Microb Technol [Internet]. 2009;45(2):169–73. Available from: http://dx.doi.org/10.1016/j.enzmictec.2009.03.005
16. Fu J, Xie X, Zhang S, Kang N, Zong G, Zhang P, et al. Rich organic nitrogen impacts clavulanic acid biosynthesis through the arginine metabolic pathway in Streptomyces clavuligerus F613-1. Microbiol Spectr [Internet]. 2022/12/14. 2023 Feb 14;11(1):e0201722–e0201722. Available from: https://pubmed.ncbi.nlm.nih.gov/36515504
17. Feng T, Zhao J, Chu J, Wang YH, Zhuang YP. Statistical optimizing of medium for clavulanic acid production by Streptomyces clavuligerus using response surface methodology. Appl Biochem Biotechnol [Internet]. 2021;193(12):3936–48. Available from: http://dx.doi.org/10.1007/s12010-021-03627-4
18. Ramirez-Malule H, Junne S, Nicolás Cruz-Bournazou M, Neubauer P, Ríos-Estepa R. Streptomyces clavuligerus shows a strong association between TCA cycle intermediate accumulation and clavulanic acid biosynthesis. Appl Microbiol Biotechnol [Internet]. 2018;102(9):4009–23. Available from: http://dx.doi.org/10.1007/s00253-018-8841-8
19. Ramirez-Malule H, López-Agudelo VA, Gómez-Ríos D, Ochoa S, Ríos-Estepa R, Junne S, et al. TCA cycle and its relationship with clavulanic acid production: a further interpretation by using a reduced genome-scale metabolic model of Streptomyces clavuligerus. Bioengineering (Basel) [Internet]. 2021 Jul 22;8(8):103. Available from: https://pubmed.ncbi.nlm.nih.gov/34436106
20. Bushell ME, Kirk S, Zhao HJ, Avignone-Rossa CA. Manipulation of the physiology of clavulanic acid biosynthesis with the aid of metabolic flux analysis. Enzyme Microb Technol [Internet]. 2006;39(1):149–57. Available from: http://dx.doi.org/10.1016/j.enzmictec.2006.01.017
21. Teodoro JC, Baptista-Neto A, Araujo MLGC, Hokka CO, Badino AC. Influence of glycerol and ornithine feeding on clavulanic acid production by Streptomyces clavuligerus. Braz J Chem Eng [Internet]. 2010;27(4):499–506. Available from: http://dx.doi.org/10.1590/s0104-66322010000400001
22. Ser HL, Law JWF, Chaiyakunapruk N, Jacob SA, Palanisamy UD, Chan KG, et al. Fermentation conditions that affect clavulanic acid production in Streptomyces clavuligerus: a systematic review. Front Microbiol [Internet]. 2016 Apr 22;7:522. Available from: https://pubmed.ncbi.nlm.nih.gov/27148211
23. Gómez-Rios D, Ramírez-Malule H, Ochoa S, Ríos-Estepa R. Rational selection of culture medium for clavulanic acid production by Streptomyces clavuligerus based on a metabolic modeling approach. Agric Nat Resour [Internet]. 2022;56(2):267–76. Available from: http://dx.doi.org/10.34044/j.anres.2022.56.2.05
24. Gómez-Ríos D, López-Agudelo VA, Ramírez-Malule H, Neubauer P, Junne S, Ochoa S, et al. A genome-scale insight into the effect of shear stress during the fed-batch production of clavulanic acid by Streptomyces clavuligerus. Microorganisms [Internet]. 2020 Aug 19;8(9):1255. Available from: https://pubmed.ncbi.nlm.nih.gov/32824882
25. Gómez-Ríos D, Ramírez-Malule H, Neubauer P, Junne S, Ríos-Estepa R, Ochoa S. Tuning of fed-batch cultivation of Streptomyces clavuligerus for enhanced clavulanic acid production based on genome-scale dynamic modeling. Biochem Eng J [Internet]. 2022;185:108534. Available from: http://dx.doi.org/10.1016/j.bej.2022.108534
26. Mahadevan R, Edwards JS, Doyle J. Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys J [Internet]. 2002 Sep;83(3):1331–40. Available from: https://pubmed.ncbi.nlm.nih.gov/12202358
27. da Veiga Moreira J, Jolicoeur M, Schwartz L, Peres S. Fine-tuning mitochondrial activity in Yarrowia lipolytica for citrate overproduction. Sci Rep [Internet]. 2021 Jan 13;11(1):878. Available from: https://pubmed.ncbi.nlm.nih.gov/33441687
28. Gómez-Ríos D, Junne S, Neubauer P, Ochoa S, Ríos-Estepa R, Ramírez-Malule H. Characterization of the metabolic response of Streptomyces clavuligerus to shear stress in stirred tanks and single-use 2D rocking motion bioreactors for clavulanic acid production. Antibiotics (Basel) [Internet]. 2019 Sep 27;8(4):168. Available from: https://pubmed.ncbi.nlm.nih.gov/31569725
29. Rodrigues KCS, Costa CLL, Badino AC, Pedrolli DB, Pereira JFB, Cerri MO. Application of acid and cold stresses to enhance the production of clavulanic acid by Streptomyces clavuligerus. Appl Biochem Biotechnol [Internet]. 2019;188(3):706–19. Available from: http://dx.doi.org/10.1007/s12010-019-02953-y
30. Ribeiro RMMGP, Esperança MN, Sousa APA, Neto ÁB, Cerri MO. Individual effect of shear rate and oxygen transfer on clavulanic acid production by Streptomyces clavuligerus. Bioprocess Biosyst Eng [Internet]. 2021;44(8):1721–32. Available from: http://dx.doi.org/10.1007/s00449-021-02555-1
31. Saudagar PS, Singhal RS. Optimization of nutritional requirements and feeding strategies for clavulanic acid production by Streptomyces clavuligerus. Bioresour Technol [Internet]. 2007;98(10):2010–7. Available from: http://dx.doi.org/10.1016/j.biortech.2006.08.003
32. Saudagar PS, Singhal RS. A statistical approach using L25 orthogonal array method to study fermentative production of clavulanic acid by Streptomyces clavuligerus MTCC 1142. Appl Biochem Biotechnol [Internet]. 2007;136(3):345–59. Available from: http://dx.doi.org/10.1007/s12010-007-9030-x
33. Bellão C, Antonio T, Araujo MLGC, Badino AC. Production of clavulanic acid and cephamycin C by Streptomyces clavuligerus under different fed-batch conditions. Braz J Chem Eng [Internet]. 2013;30(2):257–66. Available from: http://dx.doi.org/10.1590/s0104-66322013000200004
34. Kunath S, Kühn M, Völker M, Schmidt T, Rühl P, Heidel G. MILP performance improvement strategies for short-term batch production scheduling: a chemical industry use case. SN Appl Sci [Internet]. 2022;4(4):87. Available from: http://dx.doi.org/10.1007/s42452-022-04969-2
35. Rodrigues KC da S, Souza AT de, Badino AC, Pedrolli DB, Cerri MO. Screening of medium constituents for clavulanic acid production by Streptomyces clavuligerus. Braz J Microbiol [Internet]. 2018/03/15. 2018;49(4):832–9. Available from: https://pubmed.ncbi.nlm.nih.gov/29588197
36. Saudagar PS, Survase SA, Singhal RS. Clavulanic acid: a review. Biotechnol Adv [Internet]. 2008;26(4):335–51. Available from: http://dx.doi.org/10.1016/j.biotechadv.2008.03.002
37. Virolle MJ. A challenging view: antibiotics play a role in the regulation of the energetic metabolism of the producing bacteria. Antibiotics (Basel) [Internet]. 2020 Feb 13;9(2):83. Available from: https://pubmed.ncbi.nlm.nih.gov/32069930
38. Millan-Oropeza A, Henry C, Lejeune C, David M, Virolle MJ. Expression of genes of the Pho regulon is altered in Streptomyces coelicolor. Sci Rep [Internet]. 2020 May 22;10(1):8492. Available from: https://pubmed.ncbi.nlm.nih.gov/32444655
39. Esnault C, Dulermo T, Smirnov A, Askora A, David M, Deniset-Besseau A, et al. Strong antibiotic production is correlated with highly active oxidative metabolism in Streptomyces coelicolor M145. Sci Rep [Internet]. 2017 Mar 15;7(1):200. Available from: https://pubmed.ncbi.nlm.nih.gov/28298624