INTRODUCTION
Breast cancer (BC) is the most prevalent malignancy in women worldwide. In 2020, the World Health Organization (WHO) projects that 2.3 million new cases of BC will be diagnosed. The incidence rates in various regions and countries vary considerably. Certain mutations in BRCA1/2 have been found to be more prevalent in Asian populations, despite the fact that mutations can vary between populations. For instance, the BRCA1 c.3331_3334delCAAG mutation has been observed more frequently in Asians. The clinical consequences of germline mutations in BRCA1/2 among BC patients of Asian descent are of considerable importance [1]. According to existing literature, these mutations have been found to be correlated with a heightened susceptibility to breast and ovarian malignancies, just like in other demographic groups [2–4].
The proteins BRCA1 and BRCA2 play crucial roles in the homologous recombination (HR) pathway, which is responsible for the repair of DNA double-strand breaks. HR repair is a meticulous and accurate mechanism employed to mend damaged DNA. This process relies on an intact sister chromatid or a homologous chromosome, which serves as a template for the repair of DNA breaks. BRCA1 serves as a scaffold protein that coordinates and regulates DNA repair-related proteins. It interacts with a variety of proteins implicated in HR repair, such as BRCA2, RAD51, and PALB2. BRCA1 aids in the recruitment and stabilization of the RAD51 protein, which is essential for the strand invasion phase of HR repair. BRCA2 interacts directly with RAD51 and facilitates its transfer onto single-stranded DNA at the DNA break site. This formation of RAD51 filaments facilitates the search for and invasion of the homologous DNA template, resulting in the repair of the double-stranded break. Mutations in BRCA1 or BRCA2 that impede their function can impair HR repair and heighten susceptibility to DNA damage accumulation. These mutations can result in genomic instability, as cells become more susceptible to accumulating DNA alterations such as mutations, deletions, and translocations. This can contribute to the eventual development of cancer [5,6].
The availability of genetic mutation data pertaining to the susceptibility and prognostic implications of cancer can offer guidance for therapeutic interventions in cases of BC. At present, ongoing clinical trials are being undertaken to assess the potential of poly (ADP-ribose) polymerase (PARP) inhibitors in the management of BC, specifically in relation to their effectiveness in managing BC with BRCA mutations. Hence, the administration of systemic therapy for BC in individuals with BRCA mutations can be tailored depending on the unique state of the BRCA mutation, rather than relying just on the manifestation of disease symptoms [7].
A number of distinct molecular subtypes that differ considerably in prognosis and therapeutic targets present in cancer cells have been identified by gene expression studies. Microarray technology is a powerful instrument used in molecular biology and genomics to investigate gene expression, DNA sequencing, genotyping, and other genomic applications. It enables researchers to analyze the expression or presence of thousands to millions of genes or DNA sequences simultaneously in a single experiment [8]. Several prior studies have discussed the identification of differentially expressed genes (DEGs) in BC when compared to healthy individuals [9–11]. However, there is a lack of research that specifically examines the analysis of DEGs in BC patients with BRCA1/2 mutations in comparison to those without such mutations. There is a limited study as well that analyzes the DEGs of PARP inhibitor treatments in BC with BRCA1/2 mutations. Therefore, it seems important to understand more about those conditions that can be more developed as biomarkers as well.
In this study, BC gene expression profiles from three separate studies (two types of datasets) using microarrays were analyzed. The microarray expression profile datasets GSE25835, GSE40115, and GSE55399 were obtained from the Gene Expression Omnibus (GEO). The DEGs were retrieved based on parameter adjustment p-value of 0.05, log2FC > 1 (upregulated DEGs) and log2FC < −1 (downregulated DEGs). By using this approach, we aimed to identify the biomarkers from DEGs of BRCA1/2 mutation carriers and BRCA1/2 mutation carriers that received PARP inhibitor treatment. We hypothesized that there are some DEGs in patients with BRCA1/2 mutations and that the use of PARP inhibitor treatments will change the biological functions (BFs) as well.
MATERIAL AND METHODS
Identification of differentially expressed genes
The GSE25835 [12], GSE40115 [13], and GSE55399 [14] datasets were retrieved from the GEO database. Dataset GSE25835 uses the Affymetrix HT Human Genome U133A Array Platform (GPL3921), dataset GSE40115 uses Agilent-029949 Custom Sureprint G3 Human GE 8x60K Microarray (GPL15931), and dataset GSE25835 uses the Affymetrix HT Human Genome U133A Array (GPL3921) platform. In this study, we used two different categories of datasets. The first datasets were GSE25835 and GSE40115, which compared the DEGs of BC with BRCA1/2 mutations and wild-type. Whereas in the GSE55399 dataset, the DEGs were compared between PARP inhibitor treatment and no PARP inhibitor treatment.
To analyze gene expression data, GEO2R software was used to calculate adjusted p-values and FDR by using T test and Benjamini and Hochberg (false discovery rate) [15]. To obtain a significant DEG from the dataset, we standardized the primary adjustment at a p-value of 0.05; the up-regulated DEG is considered if log2FC > 1 and the down-regulated DEG if log2FC < −1. The DEGs derived from the two data sets were searched for overlapping sections using Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/) and considered for further analysis [11].
Protein–protein interaction (PPI) networks
The search tool for the retrieval of interacting genes/proteins (STRING) database provides information on PPIs and functional associations. It seeks to facilitate the investigation of protein networks and their relationships with diverse biological processes [16]. By using Cytoscape software [17] (version 3.8.2), a confidence value of 0.9 was employed to achieve a robust interaction. In addition, the molecular complex detection (MCODE) algorithm was employed to acquire overlapping clusters from the resultant interaction networks. Multiple group determination parameters are employed, including a degree cutoff of 2, a k-score of 2, a maximum depth of 100, and a node score cutoff of 0.2 [8,18].
Gene ontology (GO) term and signaling pathway enrichment analysis
The DEGs that had been analyzed were enriched for their gene ontologies using Enrichr (http://maayanlab.cloud/Enrichr/). The ClueGO [19] (version 2.5.7), module of the Cytoscape software was used to investigate the gene annotation interrelationship analysis. The statistical analysis for ClueGO enrichment analysis involved the utilization of a two-tailed hypergeometric test with a significance level of p less than 0.05. In addition, the Benjamini–Hochberg correction, as well as the kappa score of 0.4 were applied to determine significant enrichment. The pathway analysis was conducted utilizing the REACTOME database, accessible at http://www.reactome.org [20].
Survival analysis
Gene expression profiling interactive analysis (GEPIA) [21] was utilized to do a statistical analysis of gene expression data to determine the correlation between gene expression levels and both time and % overall survival (OS). This analysis involved the calculation of Kaplan–Meier curves and performed on the BC dataset and using the Cox proportional hazards model. In addition, the hazard ratio was determined, along with its corresponding 95% confidence intervals. The analysis was conducted to predict the effect of DEG on the survival of BC patients. A significant difference being defined by the p-value score is less than 0.05 [22].
RESULT AND DISCUSSION
BC is the most commonly diagnosed cancer and the primary cause of cancer-related deaths worldwide among females. The prevalence of germline mutations in the BRCA1 or BRCA2 genes among BC patients in Asia is estimated to range from 2% to 3% [1]. Both BRCA1 and BRCA2 have significant involvement in the process of HR repair, hence contributing to the repair of DNA damage [5]. A dysfunction of BRCA1/2 results in heightened chromosomal instability, potentially contributing to the development of tumors. Significantly, the absence of BRCA1/2 also leads to increased cellular susceptibility to interstrand DNA cross-linking agents, including cisplatin, as well as PARP inhibitors, an auspicious new category of anticancer agents [23,24].
Cisplatin and its derivative, carboplatin, which are platinum compounds, exhibit efficacy as chemotherapeutic agents. Nonetheless, relapse does transpire in the majority of female patients who have BRCA1/2-mutated cancer which then develop resistance to platinum. Several mechanisms have been identified in scientific research as potential contributors to cisplatin resistance, such as increased glutathione expression and altered drug transport via altered copper transporter expression.
Due to the high sensitivity of BRCA1/2-mutant to PARP inhibitors, numerous ongoing clinical trials are evaluating the efficacy of PARP inhibitors in BC patients with BRCA1/2 mutation. Synthetic lethality is the consequence of inhibiting PARP in HR-deficient cells, such as BRCA1/2-mutant, and encouraging clinical trial outcomes in BRCA-associated carcinomas have been documented [25]. Selection of systemic therapy for BC can be done by looking at BRCA mutation status not only on disease characteristics [7].
Bioinformatics plays an important role in the study of cancer at the molecular level related to prognostics, diagnostics, and therapeutics. The comprehensive bioinformatics analysis and biological database are innovative techniques to bring future perspectives to the field of oncology [26]. In this regard, bioinformatic analysis by using computational gene profiling strategies for BC has been advocated. This study employs a methodology that utilizes two distinct types of datasets, a practice that remains infrequently employed in other research endeavors. Some previous studies applied only one type of dataset, as mentioned in Deng et al. [27], Fei et al. [28], and Zhou et al. [29].The objective is to obtain more extensive findings by identifying DEGs in individuals with BRCA1/2 mutations and understanding how these genes impact the effectiveness of PARP-inhibitor therapy.
Differentially expressed genes
In this work, we analyzed the gene expression of two types of microarray datasets, namely, GSE25835, GSE40115, and GSE55399, obtained from the GEO database (Table 1). The data set was analyzed using GEO2R to obtain DEG by setting the cutoff criteria, namely the p-value adjustment < 0.05; log2FC > 1 (upregulated DEGs) and log2FC < −1 (downregulated DEGs). From the GSE25835 and GSE40115A datasets, we found DEGs in BC patients with BRCA1/2 mutation carriers compared to the wild-type carriers. Meanwhile, from the GSE55399 dataset, we found DEGs in BC cell lines treated with PARP inhibitors compared to those without PARP inhibitors.
In the GSE25835, GSE40115, and GSE55399 datasets, a total of 4,503 DEGs were identified (2,312 upregulated and 2,191 downregulated), 900 (495 upregulated and 405 downregulated), and 620 (441 upregulated and 179 downregulated). As shown in Figure 1D–E (Tables S1 and S2), 113 DEGs (60 upregulation and 53 downregulation) were discovered to overlap in the GSE25835 and GSE40115 datasets. Figure 1A–C depict the volcano graphs for DEGs of GSE25835, GSE40115, and GSE55399, respectively. The DEGs that overlapped between the GSE25835 and GSE40115 datasets were analyzed further.
 | Table 1. Sample information in GSE25835, GSE40115, GSE55399, and GSE173839 datasets. [Click here to view] |
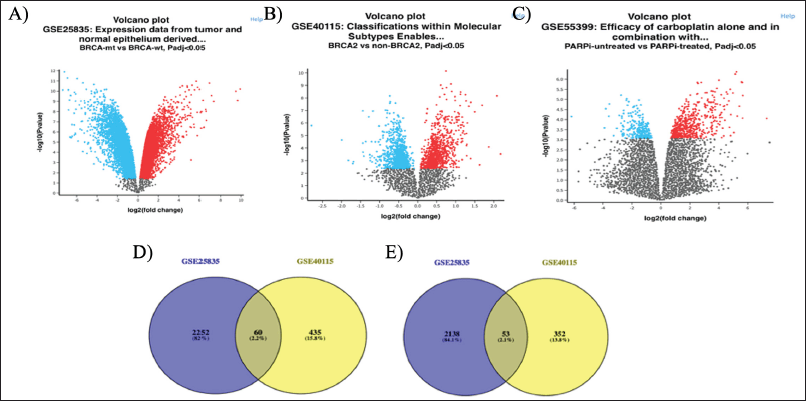 | Figure 1. Volcano plot of all the DEGs of A) GSE25835, B) GSE40115, C) GSE55399; Venn diagram of GSE25835 and GSE40115, D) upregulated overlapping DEGs, and E) downregulated overlapping DEGs. [Click here to view] |
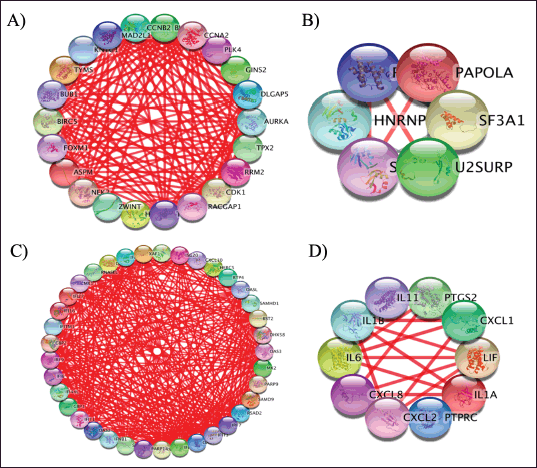 | Figure 2. PPI network of overlapping GSE25835 and GSE40115 DEGs: A) upregulated DEGs and B) downregulated DEGs; PPI network of GSE55399 DEGs: C) upregulated DEGs and D) downregulated DEGs. [Click here to view] |
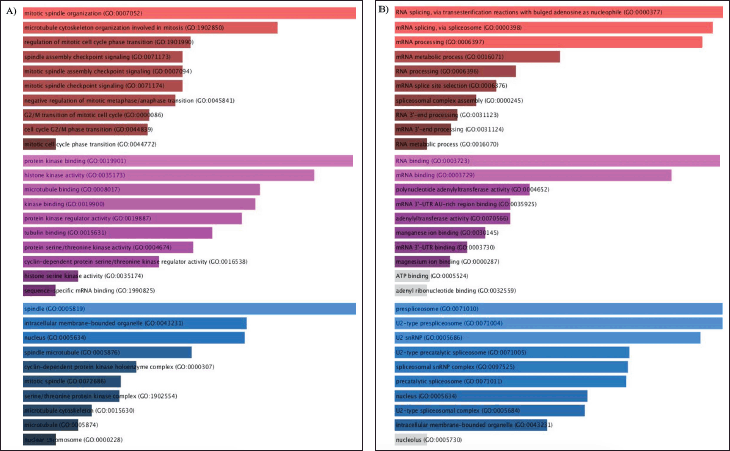 | Figure 3. A) GO enrichment analysis of overlapping upregulated GSE25835 and GSE40115 DEGs, B) GO enrichment analysis of overlapping downregulated GSE25835 and GSE40115 DEGs. The top 10 annotations based on their p-values are shown for biological processes (red), MFs (magenta), and CCs (blue). [Click here to view] |
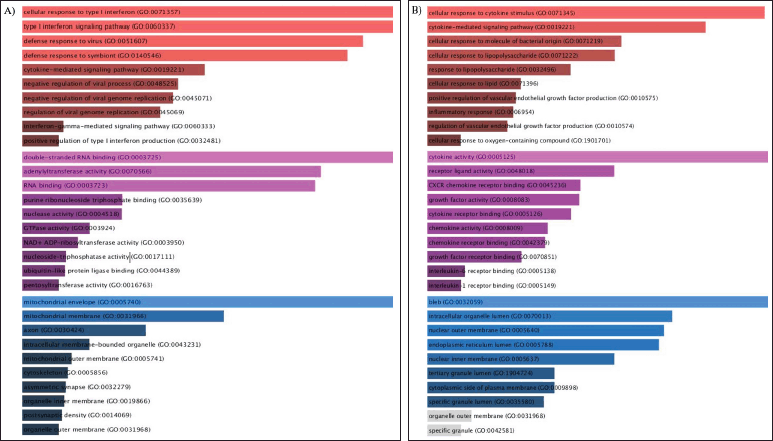 | Figure 4. A) GO enrichment analysis of the upregulated GSE55399 DEGs, B) GO enrichment analysis of the downregulated GSE55399 DEGs. The top 10 annotations based on their p-values are shown for biological processes (red), MFs (magenta), and CCs (blue). [Click here to view] |
Prediction of hub genes through PPI network
Physical and functional associations between DEG proteins were evaluated using STRING. The confidence threshold for the minimum required interaction score was set at 0.90. Groups resulting from STRING analysis were visualized using Cytoscape. The nodes represent the number of proteins, while the edges represent their interactions. The MCODE plugin in Cytoscape was utilized to visually depict clusters of closely interconnected network interactions (Fig. 2). This analysis identified hub genes in 22 and 6 nodes for upregulated and downregulated overlapping GSE25835 and GSE40115 DEGs, respectively. These were 37 and 10 nodes for upregulated and downregulated GSE55399 DEGs, respectively. This hub genes were utilized for additional analysis. The DEGs were listed in Tables S1 and S2.
GO enrichment analysis
Based on the GO analysis of significant DEGs, the top 10 annotations were selected for biological process ontology (BP), molecular function (MF), and cellular component (CC). The DEGs that overlapped from GSE25835 and GSE40115, as depicted in Figure 3A, were significantly enriched in the mitotic spindle organization and mitotic cell cycle. Protein kinase binding, histone kinase activity, and cyclin-dependent regulator activity were observed to be upregulated in MF. For CC, significant enrichment was observed in the spindle, cyclin-dependent protein kinase holoenzyme complex, and serine/threonine protein kinase complex. On the other hand, in Figure 3B, RNA splicing was observed to be downregulated in BP. For CC, prespliceosome and spliceosome downregulation was observed. The downregulation of RNA binding and adenyltransferase activity were observed in MF.
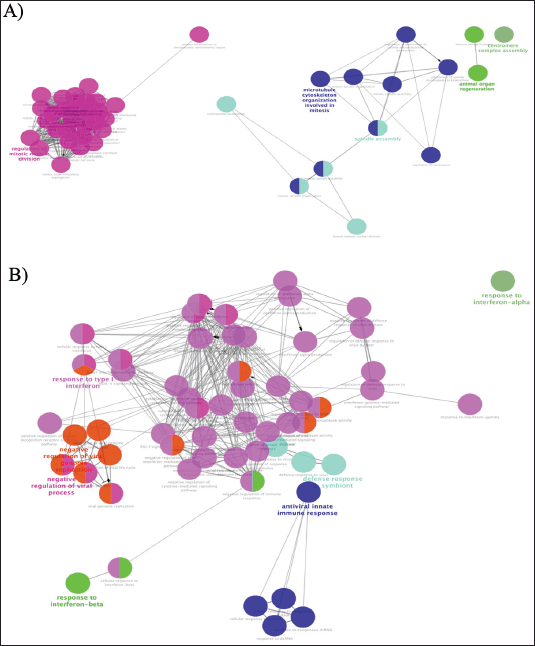 | Figure 5. The GO enrichments were visualized using the ClueGO plugin from Cytoscape. A) biological processes (BP) interactions of overlapping upregulated GSE25835 and GSE40115 DEGs; B) biological processes (BP) interactions of upregulated GSE55399 DEGs. The bold fonts indicate the most important functional GO terms. [Click here to view] |
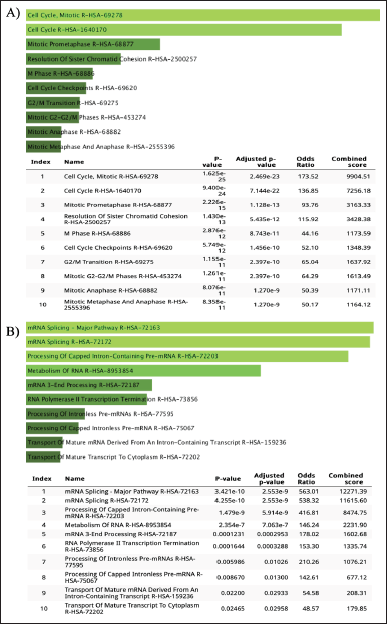 | Figure 6. Pathway enrichment analysis for overlapping GSE25835 and GSE40115 DEGs: A) upregulated DEGs and B) downregulated DEGs by using REACTOME. [Click here to view] |
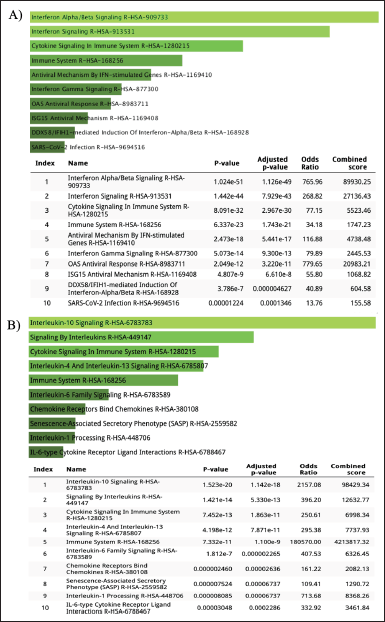 | Figure 7. Pathway enrichment analysis for GSE55399 DEGs: A) upregulated DEGs and B) downregulated DEGs by using REACTOME. [Click here to view] |
The GO analysis of upregulated GSE55399 DEGs revealed significant interferon signaling enrichment (Fig. 4A). RNA binding, adenyltransferase activity, and nuclease activity were observed to be upregulated in MF. For CC, a notable mitochondrial membrane enrichment was observed. Significant enrichment of the cytokine-mediated signaling pathway was observed for the GSE55399 DEGs for BP (Fig. 4B). We observed a decrease in cytokine signaling and growth factor activity in MF. For CC, a noteworthy enrichment in blebs and the nuclear membrane’s outermost layer was observed.
ClueGO enrichment analysis
The Cytoscape ClueGO plugin allows the performance of functional enrichment analysis on genes that have been identified as differentially expressed. ClueGo enables the visualization of gene annotation clusters within PPI networks. Figure 5 depicts the GO for MFs and BF. The statistical method employed for the ClueGO enrichment analysis was the utilization of a two-tailed hypergeometric test with a significance threshold set at 0.05. In addition, the Benjamini–Hochberg correction was applied, and a kappa score of 0.4 was used as the key criterion. The enrichment results obtained from ClueGO suggest that mutations in BRCA1/2 genes have the potential to alter the regulation of mitosis. In addition, the administration of PARP inhibitors to individuals with BRCA1/2 mutations is expected to enhance the interferon response and reduce cytokine signaling. To validate our bioinformatics findings, it is necessary to conduct functional validations.
Enrichment of pathways
REACTOME, one of the largest metabolic pathway information databases, was used to analyze the DEG pathway. As a result of mutations in the BRCA1/2 gene (dataset GSE25835 and GSE40115), DEGs that are upregulated are involved in the cell cycle pathway, while DEGs that are downregulated are involved in the mRNA splicing pathway (Fig. 6). On the other hand, in PARP inhibitory therapy (dataset GSE55399), DEGs that were upregulated were involved in interferon signaling, whereas DEGs that were downregulated were involved in cytokine signaling (Fig. 7).
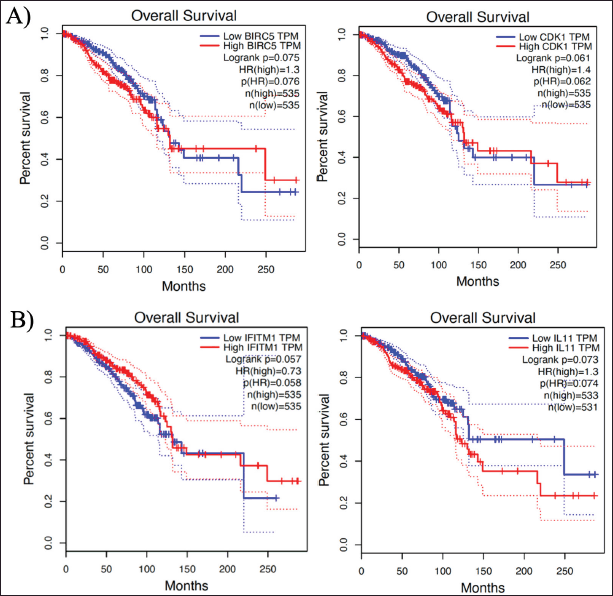 | Figure 8. Kaplan–Meir plots that showed OS analysis: A) poor OS due to the upregulated expression BIRC5 and CDK1 and B) poor OS due to the downregulated expression of IFITM1 and upregulated IL11 expression. [Click here to view] |
Survival analysis
The GEPIA results display an analysis of the hub genes’ OS. Figure 8 depicts the identification criteria: HR score > 1 (upregulated gene); HR < 1 (downregulated gene) and log-rank p < 0.05. This result suggested that the upregulated expression of BIRC5 (logrank p = 0.075; HR = 1.3) and CDK1 (logrank p = 0.061; HR = 1.4) were associated with an impaired OS, as were the downregulated expression of interferon-induced transmembrane 1 (IFITM1) (logrank p = 0.057; HR = 0.73) and the upregulated expression of Interleukin 11 (IL11) (logrank p = 0.073; HR = 1.3). Unfortunately, those results were not statistically significant because the log-rank p-score was above 0.05. However, the increased expression of BIRC5 and CDK1 in BRCA1/2 mutation carriers may be associated with a low OS. In contrast, treatment with a PARP inhibitor may improve OS by upregulating IFITM1 and downregulating IL11 expression.
This result is consistent with previous research indicating that BC patients with an upregulated BIRC5 gene, which codes for Survivin, have a lower OS rate [30,31]. Survivin is one of the apoptosis inhibitors implicated in cell division regulation and apoptosis inhibition [32]. In vitro research revealed that endogenously defective or mutant BRCA1 (MDA-MB-436) cells express a higher level of survivin than do normal cells [33]. Other research revealed that BRCA1 mutant mouse mammary tumors have low levels of SIRT1 and high levels of Survivin [34].
In mammalian cells, the cyclin-dependent kinases (CDKs), which include CDK2, CDK4, and CDK6, and also CDK1, are crucial regulators of cell cycle progression [35]. Several malignancies, including BC, have been found to exhibit CDK dysregulation that results in increased cell proliferation [36]. Our findings are consistent with previous studies, indicating that BC patients with high CDK1 expression had a worse 5-year recurrence-free survival compared to those with low CDK1 expression [37]. Therefore, the upregulation of BIRC5 and CDK1 expression in BRCA1/2 mutant carriers who may have a low OS rate is a promising marker.
The upregulation of IFITM1 gene expression was observed in individuals with BRCA1/2 mutations who underwent treatment with PARP inhibitors. IFITM1 is a member of the immune-related IFITM protein family that is expressed on the cell surface [37]. The lack of BRCA2 leads to the activation of cell-intrinsic immunological signaling. In addition, the administration of PARP inhibitors elicits the interferon response in cells and tumors that are defective in BRCA2, which aligns with the findings of our works [38]. IFITMs have been the subject of intense research due to their role in viral inhibition, and a considerable body of evidence has established a correlation between IFITMs and cancer growth. The increased expression of IFITM1 has an impact on the expression of HLA-B, potentially affecting the presentation and detection of cancer-causing antigens on the surface of cells by cytotoxic T cells. Consequently, this may hinder the complete elimination of tumor cells [37].
In our study, PARP inhibitor interventions for BRCA1/2 mutation carriers resulted in decreased IL11 expression. Analysis of OS revealed that downregulation of IL11 expression improved survival [39]. Based on our research, it has been observed in various studies that decreased expression of IL-11 is associated with enhanced survival rates among BC patients. IL-11 belongs to the IL-6 cytokine family, characterized by the utilization of the GP130 signal transducing receptor subunit. The cytokine GP130 facilitates the enhancement of the “cancer-specific capabilities” of the malignant epithelium within the tumor’s inflammatory microenvironment, while concurrently suppressing the immunological reaction of innate and adaptive immune cells toward the tumor. IL-11, aside from its documented role as a hemopoietic growth factor, is increasingly becoming recognized for its involvement in the biology of epithelial cancer [40]. Several studies showed as well that PARP inhibitors may improve the treatment of genomically unstable cancers by activation/modulation of the immune system and attenuating the inflammatory response [41–43]. Therefore, the upregulation of IFITM1 and the downregulation of IL11 can serve as a prospective evaluation marker for PARP inhibitor treatment in BRCA1/2 mutant carriers.
Nevertheless, our investigation has encountered some limitations. The DEGs discovered were derived from an online database. To validate our research, additional tests conducted using patient samples are required. Furthermore, we utilized a dataset sourced from a non-Asian population as a result of the scarcity of datasets available in the database. Further investigation is required to specifically examine the subsequent processes triggered by BRCA1/2 mutations in BC and the application of PARP inhibitor treatment.
CONCLUSION
We identified hub genes of cell cycle response (CDK1 and BIRC5) that are strongly linked to BRCA1/2 mutation. PARP inhibitor treatments in BRCA1/2 mutant carriers are strongly related to the upregulation of IFITM1 (interferon signaling) and the downregulation of IL11 (cytokine signaling). Therefore, PARP inhibitors may improve the treatment by activation/modulation the immune system and attenuating the inflammatory response. However, the dataset used to analyze the DEGs of PARP inhibitor treatments in BRCA1/2 mutant carriers still used BC cell lines, forthcoming research may be able to use clinical patients as the subjects. Moreover, functional studies are further needed to validate this finding.
AUTHOR CONTRIBUTIONS
RIP: Formal analysis, write the manuscript; SIW, SSP, FF: Conceptualize and supervise the research.
FINANCIAL SUPPORT
Publication costs were funded by Universitas Indonesia’s PUTI Grand 2022 (grant number: NKB-1422/UN2.RST/HKP.05.00/2022).
CONFLICTS OF INTEREST
The authors declare that they have no competing interests.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
Datasets were retrieved from the GEO (Gene Expression Omnibus) database (https://www.ncbi.nlm.nih.gov/geo/): GSE25835, GSE40115, and GSE55399.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Park S, Lee E, Park S, Lee S, Nam SJ, Kim SW, et al. Clinical characteristics and exploratory genomic analyses of germline BRCA1 or BRCA2 mutations in breast cancer. Mol Cancer Res. 2020;18(9):1315–25. CrossRef
2. Couch FJ, Johnson MR, Rabe KG, Brune K, De Andrade M, Goggins M, et al. The prevalence of BRCA2 mutations in familial pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(2):342–6. CrossRef
3. Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443–53. CrossRef
4. Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. J Am Med Assoc. 2015;313(13):1347–61. CrossRef
5. Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. J Am Med Assoc. 2017;317(23):2402–16. CrossRef
6. Dziadkowiec KN, Gasiorowska E, Nowak-Markwitz E, Jankowska A. PARP inhibitors: review of mechanisms of action and BRCA1/2 mutation targeting. Prz Menopauzalny. 2016;15(4):215–9. CrossRef
7. Wang YA, Jian J-W, Hung C-F, Peng H-P, Yang C-F, Cheng H-CS, et al. Germline breast cancer susceptibility gene mutations and breast cancer outcomes. BMC Cancer. 2018;18(1):315. CrossRef
8. Udhaya Kumar S, Thirumal Kumar D, Bithia R, Sankar S, Magesh R, Sidenna M, et al. Analysis of differentially expressed genes and molecular pathways in familial hypercholesterolemia involved in atherosclerosis: a systematic and bioinformatics approach. Front Genet. 2020;11(July):1–16. CrossRef
9. Zhu C, Hu H, Li J, Wang J, Wang K, Sun J. Identification of key differentially expressed genes and gene mutations in breast ductal carcinoma in situ using RNA-seq analysis. World J Surg Oncol. 2020;18(1):52. CrossRef
10. Zhang S, Jiang H, Gao B, Yang W, Wang G. Identification of diagnostic markers for breast cancer based on differential gene expression and pathway network. Front Cell Dev Biol. 2022;9(January):811585. CrossRef
11. Li J, Huang G, Ren C, Wang N, Sui S, Zhao Z, et al. Identification of differentially expressed genes-related prognostic risk model for survival prediction in breast carcinoma patients. Aging (Albany NY). 2021;13(12):16577–99. CrossRef
12. Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell [Internet]. 2011 Feb;8(2):149–63. CrossRef
13. Larsen MJ, Kruse TA, Tan Q, Lænkholm AV, Bak M, Lykkesfeldt AE, et al. Classifications within molecular subtypes enables identification of BRCA1/BRCA2 mutation carriers by RNA tumor profiling. PLoS One. 2013;8(5):e64268. CrossRef
14. Karginova O, Siegel MB, Van Swearingen AED, Deal AM, Adamo B, Sambade MJ, et al. Efficacy of carboplatin alone and in combination with ABT888 in intracranial murine models of BRCA-mutated and BRCA-wild-type triple-negative breast cancer. Mol Cancer Ther. 2015;14(4):920–30. CrossRef
15. Sean D, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–7. CrossRef
16. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–8. CrossRef
17. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res [Internet]. 2003 Nov;13(11):2498–504. CrossRef
18. Li Q, Aishwarya S, Li JP, Pan DX, Shi JP. Gene expression profiling of glioblastoma to recognize potential biomarker candidates. Front Genet. 2022;13:832742. CrossRef
19. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–3. CrossRef
20. Gillespie M, Jassal B, Stephan R, Milacic M, Rothfels K, Senff-Ribeiro A, et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50(D1):D687–92. CrossRef
21. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. CrossRef
22. Albogami S. Comprehensive analysis of gene expression profiles to identify differential prognostic factors of primary and metastatic breast cancer. Saudi J Biol Sci [Internet]. 2022;29(7):103318. CrossRef
23. Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–7. CrossRef
24. McCarthy N. A positive defect. Nat Rev Cancer [Internet]. 2005 May 20;5(5):333–333. CrossRef
25. Dhillon KK, Swisher EM, Taniguchi T. Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci. 2011;102(4):663–9. CrossRef
26. Kaur G, Gupta S, Kaur G, Verma M, Kaur P. Bioinformatics: an important tool in oncology. In: biomedical data mining for information retrieval [Internet]. Hoboken, NJ: Wiley; 2021. p. 163–95. CrossRef
27. Deng JL, Xu YH, Wang G. Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Front Genet. 2019;10(JUL):695. CrossRef
28. Fei H, Chen S, Xu C. RNA-sequencing and microarray data mining revealing: the aberrantly expressed mRNAs were related with a poor outcome in the triple negative breast cancer patients. Ann Transl Med. 2020;8(6):363. CrossRef
29. Zhou Q, Liu X, Lv M, Sun E, Lu X, Lu C. Genes that predict poor prognosis in breast cancer via bioinformatical analysis. Biomed Res Int. 2021;2021:6649660. CrossRef
30. Oparina N, Erlandsson MC, Beding AF, Parris T, Helou K, Karlsson P, et al. Prognostic significance of birc5/survivin in breast cancer: results from three independent cohorts. Cancers (Basel). 2021;13(9):1–16. CrossRef
31. Dai JB, Zhu B, Lin WJ, Gao HY, Dai H, Zheng L, et al. Identification of prognostic significance of BIRC5 in breast cancer using integrative bioinformatics analysis. Biosci Rep. 2020;40(2):1–12. CrossRef
32. Mehraj U, Aisha S, Sofi S, Mir MA. Expression pattern and prognostic significance of baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5) in breast cancer: a comprehensive analysis. Adv Cancer Biol - Metastasis. 2022;4(March):100037. CrossRef
33. Promkan M, Liu G, Patmasiriwat P, Chakrabarty S. BRCA1 modulates malignant cell behavior, the expression of survivin and chemosensitivity in human breast cancer cells. Int J Cancer. 2009;125(12):2820–8. CrossRef
34. Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, et al. Interplay among BRCA1, SIRT1, and survivin during BRCA1-Associated Tumorigenesis. Mol Cell [Internet]. 2008;32(1):11–20. CrossRef
35. Izadi S, Nikkhoo A, Hojjat-Farsangi M, Namdar A, Azizi G, Mohammadi H, et al. CDK1 in breast cancer: implications for theranostic potential. Anticancer Agents Med Chem [Internet]. 2020 Jul 3;20(7):758–67. CrossRef
36. Xing Z, Wang X, Liu J, Zhang M, Feng K, Wang X. Expression and prognostic value of CDK1, CCNA2, and CCNB1 gene clusters in human breast cancer. J Int Med Res. 2021;49(4):300060520980647.
37. Gómez-Herranz M, Faktor J, Yébenes Mayordomo M, Pilch M, Nekulova M, Hernychova L, et al. Emergent Role of IFITM1/3 towards splicing factor (SRSF1) and antigen-presenting molecule (HLA-B) in Cervical Cancer. Biomolecules. 2022;12(8):1–21. CrossRef
38. Reisländer T, Lombardi EP, Groelly FJ, Miar A, Porru M, Di Vito S, et al. BRCA2 abrogation triggers innate immune responses potentiated by treatment with PARP inhibitors. Nat Commun [Internet]. 2019 Jul 17;10(1):3143. CrossRef
39. Ernst M, Putoczki TL. Molecular pathways: IL11 as a tumor-promoting cytokine-translational implications for cancers. Clin Cancer Res. 2014;20(22):5579–88. CrossRef
40. Johnstone CN, Chand A, Putoczki TL, Ernst M. Emerging roles for IL-11 signaling in cancer development and progression: focus on breast cancer. Cytokine Growth Factor Rev. 2015;26(5):489–98. CrossRef
41. Rom S, Zuluaga-Ramirez V, Reichenbach NL, Dykstra H, Gajghate S, Pacher P, et al. PARP inhibition in leukocytes diminishes inflammation via effects on integrins/cytoskeleton and protects the blood-brain barrier. J Neuroinflammation [Internet]. 2016;13(1):1–16. CrossRef
42. Liu Z, Wang H, Wang S, Gao J, Niu L. PARP-1 inhibition attenuates the inflammatory response in the cartilage of a rat model of osteoarthritis. Bone Joint Res. 2021;10(7):401–10. CrossRef
43. Lee EK, Konstantinopoulos PA. PARP inhibition and immune modulation: scientific rationale and perspectives for the treatment of gynecologic cancers. Ther Adv Med Oncol [Internet]. 2020 Jan 24;12(6):1758835920944116. CrossRef
SUPPLEMENTARY MATERIAL
The supplementary material can be accessed at the journal's website: Link Here [https://japsonline.com/admin/php/uploadss/4310_pdf.pdf].