INTRODUCTION
Among all diseases, cancer poses the highest economic, social, and clinical burden in terms of cause-specific disability-adjusted life years [1]. With a 20.2% lifetime risk of developing cancer and a mortality rate that is second worldwide after ischemic heart disease, the need to address cancer is paramount. In the Philippines, the latest statistics show that cancer is responsible for more than 60,000 deaths in the year 2017. Over the years, patients with cancer are treated with chemotherapy, radiotherapy, and surgery. However, it is common for cancer cells to develop resistance to treatment [2]. Furthermore, patients with late-stage diagnosis are often ineligible for surgery. Hence, there is a clear and urgent need for effective drugs for the treatment of patients with cancer. Recent advances in cancer treatment include the application of targeted therapies which have been successful in reducing mortality and minimizing toxicity [3]. Targeted therapy is an approach using designed drugs that specifically bind to aberrant proteins, such as those implicated in malignant transformation. The design of drugs used in targeted therapy is continuously influenced and inspired by natural products [4]. These natural products are unrivaled sources of anticancer compounds in this modern era of drug discovery due to applicability, accessibility, and diminished cytotoxicity. They act by modulating the cancer microenvironment and various signaling pathways [5].
Chronic myeloid leukemia (CML) is one of the many types of cancer. It is a clonal malignancy with a global incidence of around 2 per 100,000 persons. In the Philippines, CML along with other leukemias ranked fifth leading cause of cancer-related mortalities [6]. With this, the occurrence of leukemia and its changing trends were highly varied, and more preventive strategies must be adapted to each country [7]. A large majority of this disease is caused by chromosomal translocation between chromosomes 9 and 22, which results in the formation of the hybrid gene BCR::ABL1. The gene product BCR::ABL kinase is responsible for the impaired cellular signaling, thereby dysregulating normal cell proliferation, invasion, migration, and apoptosis which are hallmarks of cancer pathophysiology [8,9]. Thus, anti-CML agents have been continuously discovered and developed to target this hybrid protein [10,11]. The chemotherapeutics imatinib, nilotinib, and ponatinib being BCR-ABL-targetting tyrosine kinase inhibitors (TKIs) mitigate strongly CML progression. In the case of imatinib, which is an inhibitor of tyrosine kinase, it allowed CML patients to experience a life expectancy that is near-normal [12]. Due to concerns related to drug resistance brought by BCR::ABL mutations and overexpression, more chemical-based CML therapeutics are still warranted.
In search for new anticancer agents, natural products have long been tapped due to their purported efficacy and safety. Antiproliferative natural products comprised of alkaloids, flavonoids, glycosides, lignans, polyketides, and oxidized terpenoids inhibit CML proliferation via induction of apoptosis. Apart from differentiating CMLs into monocyte/erythroid cell types, natural products also prevent the occurrence of multidrug resistance in CML cells. The Philippines being a megadiverse archipelagic country hosts native and endemic plants. Among them are medicinal plants reported for their anticancer activities [13–16]. The Philippines’ Department of Health enlisted the “Ten scientifically validated” Philippine medicinal plants in 1992 (R.A. No. 8423—Philippine Institute of Traditional and Alternative Health Care), in which Momordica charantia (ampalaya) and Quisqualis indica (niyug-niyogan) were identified to have anticancer properties [17]. In the Northern Philippines, locals and natives from provinces such as Apayao and Cagayan utilize Syzygium lineatum (Roxb.) (DC.) Merr & Perry (locally known as “Malubeg” and “Alebadu”), a traditional medicine used to treat cancer and as a souring agent for cooking. Syzygium lineatum is a fruiting tree that grows 4–5 m in height and belongs to the family Myrtaceae [18,19]. Different species of the genus Syzygium have been reported to exhibit anticancer activities [20–22]. Syzygium natural products such as dimethyl cardamonins, phenolics, oleanolic, and betulinic acids have been reported to inhibit cell proliferation and induce apoptosis [23]. In this paper, we report the antiproliferative and cytotoxic activities of S. lineatum extracts, sub-extracts, and its alkenylated phenolic compounds 5-(8Z-pentadecenyl)resorcinol (gingkol, 1) and 5-(8Z-pentadecenyl)resorcinol (bilobol, 2) as well as β-sitosterol (3) and fatty acids 4–6 against HUVEC, K-562, and HeLa cell lines (Fig. 1). To understand the mechanism of action of the antiproliferative compounds against CML, in silico binding characteristics were interrogated against clinically approved target kinases in CML such as BCR::ABL kinase, c-Src kinase, and protein kinase B.
MATERIALS AND METHODS
Plant material
The leaves of S. lineatum were collected at Adams, Ilocos Norte, Northern Philippines (18.4602680°N; 120.9038960°E), in December 2016 and were identified by Mr. Michael A. Calaramo of Northwestern University Ecotourism Park and Botanic Gardens (NUEBG) and Dr. Cecilia Banag-Moran of University of Santo Tomas Herbarium (USTH). Voucher specimens were submitted to USTH (USTH015667) and the Herbarium of NUEBG (HNUL14950).
Extraction and isolation
The air-dried and ground leaves of S. lineatum (2 kg) were percolated with 1:1 dichloromethane-methanol (16.0 l). The combined extracts were evaporated until dry under reduced pressure to afford a green syrup (313.0 g). A suspension of the crude extract in water was sequentially extracted with petroleum ether (4.7 l), followed by dichloromethane (6.9 l), and finally with n-butanol (2.0 l) yielding after evaporation 98 g of the petroleum ether sub-extract (SlP), 58.9 g of dichloromethane sub-extract (SlD), and 32.4 g of the n-butanol sub-extract (SlB). The petroleum ether sub-extract was subjected to silica gel vacuum liquid chromatography with increasing amounts of ethyl acetate (EtOAc) in petroleum ether as eluent (10%). Fractions were pooled according to their thin layer chromatography (TLC) profile. Iterative separation using silica gel column chromatography of the SlP fraction 2 (20.8 g) with 20:1 petroleum ether: EtOAc resulted in the isolation and purification of 3-(8Z-pentadecenyl)phenol or gingkol (1) along with the known compounds β-sitosterol (3) and a mixture of fatty acids (4–6). Meanwhile, repetitive separation of SlP fraction 4 (SlPE4) yielded compound 2 (bilobol) [24].
Antiproliferative and cytotoxicity assay
To measure the antiproliferative and cytotoxic activities of S. lineatum extracts, sub-extracts, and compounds, inhibitory and cytotoxic concentrations against CML (K-562), human umbilical vein endothelial cord (HUVEC), and HeLa cell lines were measured using the CellTiter-Blue® viability assay based on the method developed by Krauth et al. [25]. The GI50 and CC50 values were determined as the value at the intersection of the dose curve with the 50% line, compared to the untreated control. Imatinib and doxorubicin, which are standard anticancer drugs, were used as positive controls [26].
Molecular docking studies
Ligand and protein preparation
To elucidate potential binding propensities and modalities of the compounds, they were formatted as ligands and were prepared and optimized in Avogadro (1.20). Target proteins were prepared and minimized in UCSF Chimera (1.16) based on previously reported methodologies using the Gasteiger charge method [27]. SMILES formats of compounds were fetched from PubChem while proteins were obtained from the Protein Data Bank (PDB) through their PDB IDs. (PDB ID: 2GQG), inactive (PDB ID: 2HYY), and T334I_D382N imatinib-resistant (PDB ID: 5MO4) BCR::ABL kinase, c-Src kinase (2SRC), and protein kinase B (4EJN) were selected as potential therapeutic targets due to their established roles in CML pathophysiology [28–30].
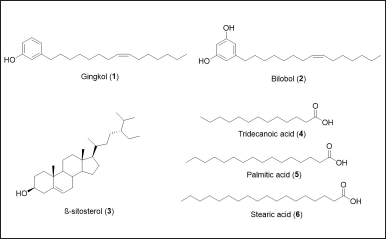 | Figure 1. Structures of 5-(8Z-pentadecenyl)phenol (gingkol, 1), 5-(8Z-pentadecenyl)resorcinol (bilobol, 2), β-sitosterol (3), and fatty acids 4–6 from S. lineatum. [Click here to view] |
Docking simulation
Molecular docking, an in silico tool to assess binding affinities of compounds, especially natural products, to target receptors or proteins, was performed using UCSF Chimera (1.16) [31]. A grid-based “flexible ligand into flexible active site” protocol was followed in the entire simulation [32]. To visualize and enumerate the amino acid residues involved in the binding as well as evaluate the kind of intermolecular interactions, BIOVIA Discovery Studio (4.1) was utilized.
ADME properties and BOILED-Egg prediction
To predict the drug-likeness of the compounds, their absorption, distribution, metabolism, and excretion (ADME) properties were assessed based on the number of violations in Lipinski’s rule of five (LRo5). Meanwhile, pharmacokinetic behaviors [gastrointestinal (GI) absorption and brain penetration] were predicted using the Brain Or IntestinaL EstimateD permeation (BOILED)-Egg methodological model [33,34]. The SMILES notations of the ligands were inputted in SWISSADME (ttp://www.swissadme.ch/index.php) and parameters such as molecular weight, lipophilicity, and number of H-acceptors and donors were recorded [35].
RESULTS
In vitro antiproliferative and cytotoxic activities of S. lineatum leaf extracts and fractions, and compounds 1–6
Antiproliferative and cytotoxicity assessments of the crude extract (SlC), petroleum ether (SlPE), dichloromethane (SlD), and butanol sub-extracts (SlB) were carried out using a CellTiter-Blue assay. The antiproliferative activity versus normal HUVEC and human leukemia cells (K-562) is reported as GI50, while cytotoxicity versus cervical cancer cells (HeLa) as CC50. The petroleum ether sub-extract (SlPE) showed the strongest anti-proliferative and cytotoxic activity among the three sub-extracts against the three cell lines (Table 1). In addition, fraction 4 of the petroleum sub-extract (SlPE4) exhibited antiproliferative activity against K-562 at 12.6 (±1.1) ug/ml and cytotoxicity against HeLa at 22.2 (±0.9) ug/ml. Chromatographic purifications of fractions SlPE4 and SlPE2 (the next antiproliferative and cytotoxic fraction) afforded six compounds and were spectroscopically identified as 3-(8Z-pentadecenyl)phenol or gingkol (1), 3-(8Z-pentadecenyl)resorcinol or bilobol (2), β-sitosterol (3), and mixture of fatty acids (4–6). Compounds 1–6 were subjected to CellTiter-Blue assay wherein 1 and 2 exhibited potent cytotoxic activity against K-562 cells. Bilobol (2), an alkenylated resorcinol, showed better selective antiproliferative activity compared to gingkol (1), a phenolic congener (Table 2). For comparison, imatinib was used as a positive drug control.
In silico binding propensities and interactions between test compounds 1–2 and target CML-associated proteins
To further elucidate the potential mechanism of action of compounds 1 and 2 against CML (K-562), these were docked onto the adenosine triphosphate (ATP)-binding site of an active, inactive, and T334I_D382N imatinib-resistant BCR::ABL kinase; c-Src kinase; and protein kinase B (Table 3; Fig. 2). These proteins are well-established promising pharmaceutical targets of small molecules [28–30]. Interestingly, the findings from the in silico docking simulations corroborated with the results from the in vitro antiproliferative assessment. Both gingkol (1) and bilobol (2) which exhibited antiproliferative activity versus K-562 showed moderate to favorable antagonistic binding activities onto the active sites of selected CML-associated protein targets (Table 3; Fig. 2). Gingkol (1) showed its best binding propensity to imatinib-resistant BCR::ABL kinase (BE = −8.1 kcal/mol) with H-bonds formed with M337 and E335; pi-sigma interactions with L267, L389, and Y272; pi-pi bonding with F336; and pi-alkyl bonds with F401, I334, K290, V318, A399, M309, and V275 (Fig. 2a).
Meanwhile, bilobol (2) showed the best binding energy (BE) against the active and imatinib-resistant BCR::ABL kinases with BE of −8.0 kcal/mol. Congruent to the result of in vitro assay versus K-562, bilobol (2) showed a slightly better BE of −8.0 kcal/mol against active BCR::ABL kinase compared to that of gingkol (1) (BE = −7.9 kcal/mol) (Table 3). For active BCR::ABL kinase, compound 2 established pi-sigma interactions with Y253; pi-pi bonding with Y253; and pi-alkyl or alkyl bond with several other residues (V299, A380, K271, V256, A269, L370, and L248) (Fig. 2b). For the resistant structure, 2 exhibited pi-sigma interactions with L267, Y272, and L389; pi-alkyl or alkyl bridges with V318, M309, A399, K290, I334, V275, I332, F401, and A288; and pi-pi bond with F336 (Fig. 2c). Interestingly, both compounds 1 and 2 showed either higher or similar binding propensity to the imatinib-resistant BCR::ABL kinase while imatinib showed decreased binding affinity to the imatinib-resistant structure (BE = −9.7 kcal/mol) compared to its binding activities to the nonresistant targets. Residues M337, I332, F336, Y272, A288, V275, L267, and L389 were common targets in the imatinib-resistant structure of the two test compounds and the positive control. Finally, although the positive control imatinib showed the highest binding propensity to BCR::ABL kinase (active conformation) ATP-binding site (BE = −9.8 kcal/mol), notable residues targeted by imatinib have also been bound to either compounds 1 or 2, or both such as Y253, A269, V256, L370, and L248.
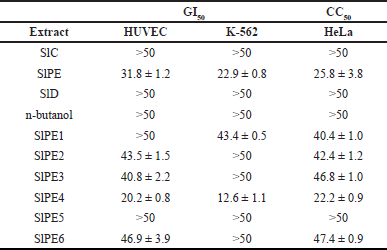 | Table 1. Growth inhibitory and cytotoxic concentration at 50% (μg/ml) of S. lineatum leaf extracts and fractions against HUVEC, K-562, and HeLa cells based on triplicates. [Click here to view] |
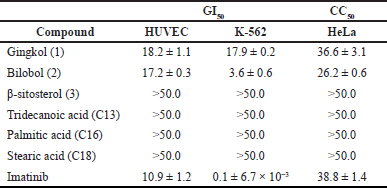 | Table 2. Growth inhibitory and cytotoxic concentration at 50% (μg/ml) of gingkol, bilobol, and the reference anti-CML drug imatinib against HUVEC, K-562, and HeLa cells based on triplicates. [Click here to view] |
ADME properties and BOILED-Egg predictions on compounds 1 and 2
Due to the favorable in vitro and in silico antiproliferative activities of gingkol (1) and bilobol (2) against CML cell line (K-562) and target kinases (BCR::ABL, c-Src, and protein kinase B), the drug-likeness and pharmacokinetic behaviors of both compounds were predicted using database-driven analysis in SWISSADME (Table 4). Compounds 1 and 2 showed only one LRo5 violation and thus are considered highly druggable pharmacophores. However, only compound 2 was predicted to be passively absorbed by the GI tract based on BOILED-Egg.
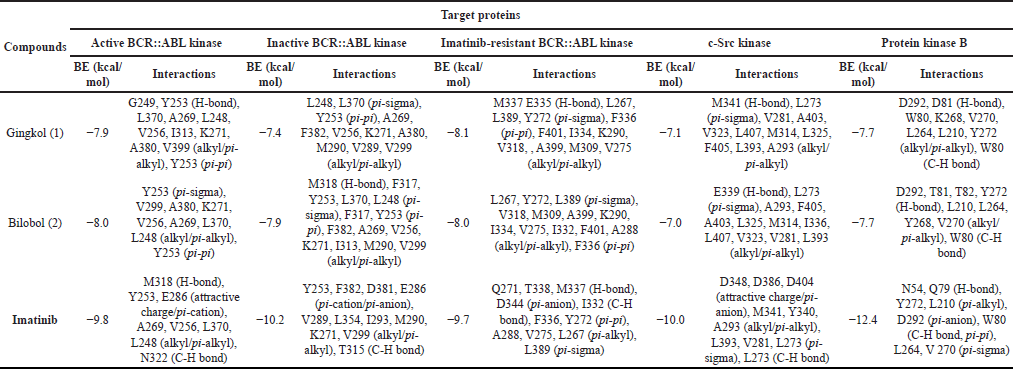 | Table 3. Binding energies (BEs) and interactions between test compounds 1–2 and target proteins. Imatinib served as positive control. [Click here to view] |
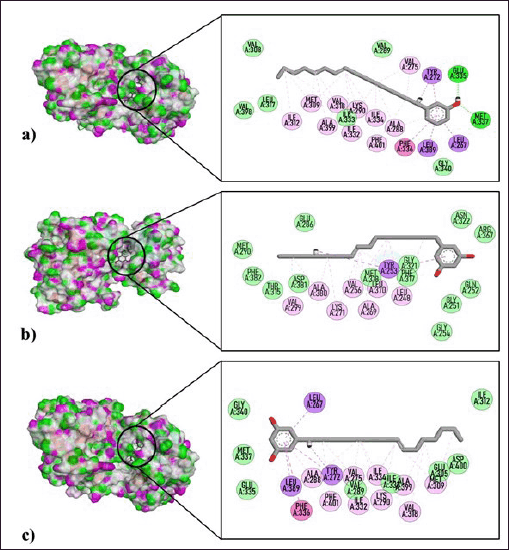 | Figure 2. Dock poses and interacting residues of (a) gingkol (1) versus imatinib-resistant BCR::ABL kinase, (b) bilobol (2) versus active BCR::ABL1 kinase structures, and (c) bilobol (2) versus imatinib-resistant BCR::ABL kinase. [Click here to view] |
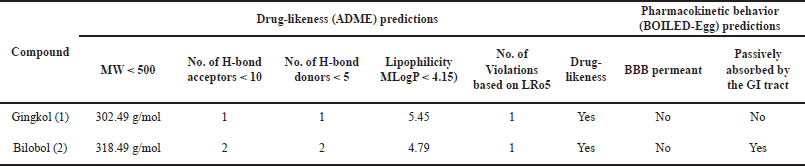 | Table 4. In silico predictions of ADME properties and pharmacokinetic behaviors of compounds 1 and 2. [Click here to view] |
DISCUSSION
In this study, extracts, sub-extracts, and alkenylated phenolic constituents gingkol (1) and bilobol (2) from the leaves of S. lineatum exhibited promising anticancer activities against HUVEC, K-562, and HeLa cell lines in vitro. In general, extracts from several members of the genus Syzygium have been reported to exhibit anticancer properties based on in vitro cell viability biological assay evidence. For example, S. aromaticum extracts showed cytotoxic activity versus breast cancer cell lines MCF-7 and MDA-MB-231 in a dose-dependent manner by inducing cell cycle arrest at the S phase and promoting apoptotic activities [36,37]. Interestingly, S. aromaticum conferred anticancer properties against colon, cervical, liver, ovarian, and pancreatic cancer cell lines. Water and ethanolic extracts of S. aqueum also conferred anticancer properties against MCF-7 cells. In addition, other Syzygium species such as S. samarangense and S. jambo extracts exhibited cytotoxic activities versus colon cancer and liver cancer cells, respectively [38–40]. In the Philippines, S. lineatum is among the traditional medicinal plants used by Ilocano natives and locals against cancer and other diseases [18,19]. This study contributes to the growing interest in Syzygium species as a bioresource of novel potential anticancer agents. More significantly, our results confirm the anticancer potentials and use of S. lineatum.
With the high morbidity associated with CML in the present and in the next 20 years which is expected to increase by 60% by then, as well as the continuous occurrence of chemotherapeutic resistance in CML treatment, the discovery and development of new-generation drugs are warranted, thus, ushering the potentials of compounds 1 and 2 as anticancer agents and/or structural drug scaffolds [41,42]. Herein, the study reports the in vitro and in silico antiproliferative potentials of these two fatty phenolics, especially against K-562 (chronic myelogenous leukemic cell lines), of S. lineatum for the first time. We also report for the first time the isolation of alkenylated phenolics gingkol (1) and bilobol (2), and 3–6 from S. lineatum. Interestingly, both phenolic compounds also exhibited promising inhibitory activity and selectivity to CML. Comparing the activities of 1 and 2, the alkenylated resorcinol (bilobol, 2) conferred better cytotoxicity against the three cell lines (HUVEC, HeLa, and K-562). This could be attributed to the increase of hydroxylic functionalization in 2. In general, hydroxylation of compounds including alkenylated phenolic natural products has been widely reported to increase chemical stability, bioactivity including inhibitory effects on key enzymes in disease pathogenesis, and binding interactions to specific molecular disease targets [43–47].
In addition, three pharmaceutically important kinases (BCR::ABL, c-Src, and protein kinase B) have been targeted by compounds 1 and 2 as shown by the results of the docking simulations. BCR::ABL1 is a well-established target as it is a chimeric constitutively active tyrosine kinase that is associated with CML cell differentiation and survival. It is the protein product of the chromosomal translocation between chromosomes 9 and 22 [48]. Many anticancer drugs like imatinib have been developed as tyrosine kinase inhibitors [49]. In this paper, three BCR:ABL tyrosine kinases were targeted—active, inactive, and imatinib-resistant. Although BCR::ABL possesses both ATP-binding and allosteric sites, the earlier was chosen as a target as this is the usual mechanism of action of clinically approved TKIs. Competitive inhibition in the ATP-binding domain yields impaired protein phosphorylation, thereby decreasing intracellularly mediated cell proliferation, growth, and survival necessary in CML cellular pathophysiology [50,51]. Comparing the results of the docking studies to the three ABL1 kinases, the alkenylated phenolics 1 and 2 from S. lineatum conferred the best binding affinity to the imatinib-resistant structure that harbors T315I mutation. Meanwhile, the drug control imatinib showed a weaker propensity to this structure, although this is given due to the associated resistance. Furthermore, several residues such as M337, I332, F336, Y272, A288, V275, L267, and L389 were identified as common targets between imatinib, and the two test compounds and these could serve as initial target hotspots of the compounds. Thus, both compounds 1 and 2 may serve as prodrugs or templates in developing new-generation TKIs against cancer cells harboring chemotherapeutic resistance.
Other than the three ABL1 kinases, c-Src, and protein kinase B also served as CML targets. c-Src is associated with cellular invasion and metastasis brought about by decreased cell-to-cell adhesion when c-Src is overexpressed [26,28,52]. Meanwhile, protein kinase B (or AKT) is a downstream target in the PI3K/AKT signaling pathway which is involved in cell proliferation, apoptosis, and development of cancer chemotherapeutic resistance. Thus, inhibition of AKT negatively affects the PI3K/AKT pathway leading to apoptosis and inhibition of cell growth in many types of cancer [29,53,54]. In our results, both compounds 1 and 2 showed moderate binding to both c-Src and AKT kinases, thereby highlighting their multitargeting potentials as anticancer agents against cell proliferation and drug resistance while promoting apoptosis and cell growth arrest.
CONCLUSION
Our study validated the traditional medicinal use of S. lineatum against cancer as we report the antiproliferative and cytotoxic activities of its extracts, sub-extracts, and alkenylated phenolics gingkol (1) and bilobol (2) against HUVEC, HeLa, and K-562 cell lines for the first time. In vitro results further suggested the sensitivity of K-562 (CML) cells to gingkol (1) and bilobol (2) with the latter showing better selectivity to K-562 cells. Molecular docking experiments supported putative multitargeting binding mechanisms of 1 and 2 in the active pockets of associated target kinases BCR::ABL1, c-Src, and protein kinase. In-depth studies on 1 and 2 are, therefore, warranted to explore further their in vitro and in vivo mechanisms of action against CML.
ACKNOWLEDGMENT
One of the authors acknowledges the support provided by the Commission on Higher Education (CHED) under the CHED K to 12 Transition Program (UNID 2016b-010405) for his graduate studies. This work was supported in part by the Alexander von Humboldt Foundation Equipment Grant.
AUTHOR CONTRIBUTIONS
FVI, VNODL, JAHM: conceptualization, investigation, data collection and analysis, manuscript preparation, and manuscript editing. ALC, APGM: conceptualization, design, manuscript preparation, editing, and review. All authors have read and approved the present version of the manuscript.
CONFLICTS OF INTERESTS
The authors declare that they have no conflict of interest in the publication.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Mattiuzzi C, Lippi G. Current cancer epidemiology. J Epidemiol Glob Health. 2019;9(4):217. CrossRef
2. Wang X, Zhang H, Chen X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019;2(2):141. CrossRef
3. Seebacher NA, Stacy AE, Porter GM, Merlot AM. Clinical development of targeted and immune based anti-cancer therapies. J Exp ClinCancer Res. 2019;38(1):1–39. CrossRef
4. Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 2020;83(3):770–803. CrossRef
5. Hashem S, Ali TA, Akhtar S, Nisar S, Sageena G, Ali S, et al. Targeting cancer signaling pathways by natural products: exploring promising anti-cancer agents. Biomed Pharmacother. 2022;150:113054. CrossRef
6. Thielen N, Ossenkoppele GJ, Schuurhuis GJ, Janssen JJ. New insights into the pathogenesis of chronic myeloid leukaemia: towards a path to cure. Neth J Med. 2011;69(10):430–40.
7. Dong Y, Shi O, Zeng Q, Lu X, Wang W, Li Y, et al. Leukemia incidence trends at the global, regional, and national level between 1990 and 2017. Exp Hematol Oncol. 2020;9(1):1–11. CrossRef
8. Amir M, Javed S. A review on the therapeutic role of TKIs in case of CML in combination with epigenetic drugs. Front Genet. 2021;12:742802. CrossRef
9. Wolfe HR, Rein LAM. The evolving landscape of frontline therapy in chronic phase chronic myeloid leukemia (CML). Curr Hematol Malig Rep. 2021;16(5):448–54. CrossRef
10. Zhou C, Liu L, Zhuang J, Wei J, Zhang T, Gao C, et al. A systems biology-based approach to uncovering molecular mechanisms underlying effects of traditional Chinese medicine Qingdai in chronic myelogenous leukemia, involving integration of network pharmacology and molecular docking technology. Med Sci Monit. 2018;24:4305. CrossRef
11. Chai Y, Chen F, Li Z, Yang P, Zhou Q, Liu W, et al. Mechanism of salidroside in the treatment of chronic myeloid leukemia based on the network pharmacology and molecular docking. Clin Transl Oncol. 2023;25:384–95. CrossRef
12. Braun TP, Eide CA, Druker BJ. Response and resistance to BCR-ABL1-targeted therapies. Cancer Cell. 2020;37(4):530–42. CrossRef
13. Macabeo APG, Lopez ADA, Schmidt S, Heilmann J, Dahse HM, Alejandro GJD, et al. Antitubercular and cytotoxic constituents from Goniothalamus gitingensis. Res Nat Prod. 2014;8(1):41.
14. Macabeo APG, Letada AG, Budde S, Faderl C, Dahse HM, Franzblau SG, et al. Antitubercular and cytotoxic chlorinated seco-cyclohexenes from Uvaria alba. J Nat Prod. 2017;80(12):3319–23. CrossRef
15. Macabeo APG, Flores AIG, Fernandez RAT, Budde S, Faderl C, Dahse HM, et al. Antitubercular and cytotoxic polyoxygenated cyclohexane derivatives from Uvaria grandiflora. Nat Prod Res. 2021;35(23):5229–32. CrossRef
16. Quimque, MT, Notarte KI, Letada A, Fernandez RA, Pilapil DY, Pueblos, KR, et al. Potential cancer-and Alzheimer’s disease-targeting phosphodiesterase inhibitors from Uvaria alba: insights from in vitro and consensus virtual screening. ACS Omega. 2021;6(12):8403–17. CrossRef
17. Boy HIA, Rutilla AJH, Santos KA, Ty AMT, Yu AI, Mahboob T, et al. Recommended medicinal plants as source of natural products: a review. Digit Chin Med. 2018;1(2):131–42. CrossRef
18. Whittaker RJ, Bush MB, Partomihardjo T, Asquith NM, Richards K. Ecological aspects of plant colonisation of the Krakatau Islands. Geo J. 1992;28:201–11. CrossRef
19. Zarate-Manicad MC. Phytochemical analysis of Lubeg (Syzygium lineatum (DC). Merr & LM Perry) species in Apayao. Int J Novel Res Life Sci. 2016;3(6):1–5.
20. Batiha GES, Alkazmi LM, Wasef LG, Beshbishy AM, Nadwa EH, Rashwan EK. Syzygium aromaticum L.(Myrtaceae): traditional uses, bioactive chemical constituents, pharmacological and toxicological activities. Biomolecules. 2020;10(2):1–16. CrossRef
21. Ismail A, Ahmad WANW. Syzygium polyanthum (Wight) Walp: a potential phytomedicine. Pharmacogn J. 2019;11(2):429–38. CrossRef
22. Maroyi, A. Syzygium cordatum Hochst. Ex Krauss: an overview of its ethnobotany, phytochemistry and pharmacological properties. Molecules. 2018;23(5):1084. CrossRef
23. Chua LK, Lim CL, Ling APK, Chye SM, Koh RY. Anticancer potential of Syzygium species: a review. Plant Foods Hum Nutr. 2019;74:18–27. CrossRef
24. Lee HJ, Song JH, Kim JH. Synthesis of resorcinol/formaldehyde gel particles by the sol-emulsion–gel technique. Mater Lett. 1998;37(4–5):197–200. CrossRef
25. Krauth F, Dahse HM, Rüttinger HH, Frohberg P. Synthesis and characterization of novel 1, 2, 4-triazine derivatives with antiproliferative activity. Bioorg Med Chem. 2010;18(5):1816–21. CrossRef
26. Malaluan IN, Manzano JAH, Muñoz JER, Bautista TJL, Dahse HM, Quimque MTJ, et al. Antituberculosis and antiproliferative activities of the extracts and tetrahydrobisbenzylisoquinoline alkaloids from Phaeanthus ophthalmicus: in vitro and in silico investigations. Philipp J Sci. 2022;151(1):371–81. CrossRef
27. Manzano JAH, Cruz CLM III, Quimque MTJ, Macabeo APG. In silico potentials of Alpinia galanga constituents against human placental aromatase vital in postmenopausal estrogen-dependent breast cancer pathogenesis. Philipp J Sci. 2022;151(6A):2101–15. CrossRef
28. Princiotto S, Musso L, Manetti F, Marcellini V, Maga G, Crespan E, et al. Synthesis and biological activity evaluation of 3-(hetero) arylideneindolin-2-ones as potential c-Src inhibitors. J Enzyme Inhib Med Chem. 2022;37(1):2382–94. CrossRef
29. Bibi S, Arslanhan MD, Langenfeld F, Jeanningros S, Cerny-Reiterer, Hadzijusufovic, E, et al. Co-operating STAT5 and AKT signaling pathways in chronic myeloid leukemia and mastocytosis: possible new targets of therapy. Haematologica. 2014;99(3):417–29. CrossRef
30. Daniel JP, Mesquita FP, Da Silva EL, de Souza PFN, Lima LB, de Oliveira LLB, et al. Anticancer potential of mebendazole against chronic myeloid leukemia: in silico and in vitro studies revealed new insights about the mechanism of action. Front Pharmacol. 2022;13:952250. CrossRef
31. Quimque MTJ, Notarte KIR, de Leon VNO, Manzano JAH, Muñoz JER, Pilapil DYH, et al. Computationally repurposed natural products targeting SARS-CoV-2 attachment and entry mechanisms. In: Frontiers of COVID-19: scientific and clinical aspects of the novel coronavirus. Cham, Swtizerland: Springer International Publishing; 2019. pp 505–37. CrossRef
32. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. CrossRef
33. Lipinski CA. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–41. CrossRef
34. Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1):42717. CrossRef
35. Fernandez RA, Quimque MT, Notarte KI, Manzano JA, Pilapil IV DY, de Leon VN, et al. Myxobacterial depsipeptide chondramides interrupt SARS-CoV-2 entry by targeting its broad, cell tropic spike protein. J Biomol Struct Dyn. 2021;40(22):12209–20. CrossRef
36. Aisha AFA, Abu-Salah KM, Alrokayan SA, Siddiqui MJ, Ismail Z, Majid AMSA. Syzygium aromaticum extracts as good source of betulinic acid and potential anti-breast cancer. Rev Bras. 2012;22:335–43. CrossRef
37. Oyedeji OO, Shode FO, Oyedeji AO, Songca SP, Gwebu ET, Hill GM, et al. Semi-synthesis of nitrogen derivatives of oleanolic acid and effect on breast carcinoma MCF-7 cells. Anticancer Res. 2014;34:4135–9.
38. Ling LT, Radhakrishnan AK, Subramaniam T, Cheng HM, Palanisamy UD. Assessment of antioxidant capacity and cytotoxicity of selected Malaysian plants. Molecules. 2010;15:2139–51. CrossRef
39. Rabeta MS, Chan S, Neda GD, Lam KL, Ong MT. Anticancer effect of underutilized fruits. Int Food Res J. 2013;20(2):551–6.
40. Liu H, Schmitz JC, Wei J, Cao S, Beumer JH, Strychor S, et al. Clove extract inhibits tumor growth and promotes cell cycle arrest and apoptosis. Oncol Res. 2014;21:247–59. CrossRef
41. Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2018 update on diagnosis, therapy and monitoring. Am J Hematol. 2018;93:442–59. CrossRef
42. Sung H, Ferlay J, Siegel R L, Laversanne, M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. CrossRef
43. Xiao J, Mao F, Yang F, Zhao Y, Zhang C, Yamamoto K. Interaction of dietary polyphenols with bovine milk proteins: molecular structure–affinity relationship and influencing bioactivity aspects. Mol Nutr Food Res. 2011;55(11):1637–45. CrossRef
44. Magpantay HD, Malaluan IN, Manzano JAH, Quimque MT, Pueblos KR, Moor N, et al. Antibacterial and COX-2 inhibitory tetrahydrobisbenzylisoquinoline alkaloids from the Philippine medicinal plant Phaeanthus ophthalmicus. Plants. 2021;10(3):462. CrossRef
45. Liu Y, Qian J, Li J, Xing M, Grierson D, Sun C, et al. Hydroxylation decoration patterns of flavonoids in horticultural crops: chemistry, bioactivity, and biosynthesis. Hortic Res. 2022;9:uhab068. CrossRef
46. Quimque MT, Notarte KI, Adviento XA, Cabunoc MH, de Leon VN, Lugtu EJ, et al. Polyphenolic natural products active in silico against SARS-CoV-2 spike receptor binding domains and non-structural proteins—a review. Comb Chem High Throughput Screen. 2023;26(3):459–88. CrossRef
47. Manzano JAH, LLames LCJ, Macabeo APG. Tetrahydobisbenzylisoquinoline alkaloids inhibit target enzymes associated with type 2 diabetes and obesity. J Appl Pharm Sci. 2024;14(01):230–7. CrossRef
48. Haidary AM, Ahmed ZA, Abdul-Ghafar J, Rahmani S, Noor S, Erfani F, et al. Philadelphia chromosome positive chronic myeloid leukemia with 5q deletion at diagnosis. Mol Cytogenet. 2021;14(16):1–4. CrossRef
49. Shanmuganathan N, Hiwase DK, Ross DM. Treatment of chronic myeloid leukemia: assessing risk, monitoring response, and optimizing outcome. Leuk Lymphoma. 2017;58:2799–810. CrossRef
50. Rosti G, Castagnetti F, Gugliotta G, Baccarani M. Tyrosine kinase inhibitors in chronic myeloid leukaemia: which, when, for whom? Nat Rev Clin Oncol. 2017;14:141–54. CrossRef
51. Jiao Q, Bi L, Ren Y, Song S, Wang Q, Wang Y. Advances in studies of tyrosine kinase inhibitors and their acquired resistance. Mol Cancer. 2018;17(1):1–12. CrossRef
52. McLachlan RW, Kraemer A, Helwani FM, Kovacs EM, Yap AS. E-cadherin adhesion activates c-Src signaling at cell–cell contacts. Mol Biol Cell. 2007;18:3214–23. CrossRef
53. Martelli AM, Evangelisti C, Chappell W, Abrams SL, Bäsecke J, Stivala F, et al. Targeting the translational apparatus to improve leukemia therapy: roles of the PI3K/PTEN/Akt/mTOR pathway. Leukemia. 2011;25(7):1064–79. CrossRef
54. Gowda R, Madhunapantula SV, Desai D, Amin S, Robertson GP. Simultaneous targeting of COX-2 and AKT using selenocoxib-1-GSH to inhibit melanoma. Mol Cancer Ther. 2013;12(1):3–15. CrossRef