INTRODUCTION
Cephalosporin class antibiotics are widely used due to their broad activity against gram-positive and negative bacteria. In 2003, the sales of cephalosporin antibiotics were the highest in the world [1]. WHO Global Antimicrobial Resistance Surveillance System report in 2019 stated that cephalosporins (generations 3, 4, and 5) were one of the five antibiotics included in the Highest Priority Category [2]. A survey that collected antibiotic consumption data from 204 countries from 2018 to 2020 showed that there was a large increase in the consumption of third-generation cephalosporin antibiotics in North Africa, Central Asia, and South Asia [3]. As well as other beta-lactam antibiotics, resistance to cephalosporins has emerged. Nonetheless, researchers continue to work on modifying their side chains due to their promising potential activities, such as cefiderocol discovered in 2018 and then approved by the US Food and Drug Administration for complicated urinary infections and pyelonephritis in 2019 [4,5].
Cephalosporin class antibiotics are synthesized from a precursor named 7-aminocephalosporanic acid (7-ACA) obtained from the deacetylation of cephalosporin C, a natural antibiotic isolated from fungi Acremonium chrysogenum in 1945 by Giuseppe Brotzu [1,6]. Synthesis of 7-ACA can be carried out enzymatically from cephalosporin C through two stages involving D-amino acid oxidase (DAAO) and Glutaryl 7-ACA acylase (GA) enzymes. The two-stage synthesis process is costly so researchers are trying to find another method to simplify the process so 7-ACA can be directly synthesized from cephalosporin C [6].
Researchers found that there is a GA enzymes that naturally have the activity to directly convert cephalosporin C into 7-ACA. Some types of GA enzymes that are known to have cephalosporin C acylase (CA) activity are GA enzymes produced by Pseudomonas sp. SE83 (1987), V22 (1987), and N176 (1992). However, the activity of these GA enzymes toward cephalosporin C is very low, approximately about 4% compared to Glutaryl-7-ACA [6–9]. Therefore, increasing the strength of its bond with cephalosporin C is necessary so that its activity increases and becomes more suitable for industrial applications. To achieve this goal, several methods can be used, such as modification of enzyme structure and optimization of enzyme expression in certain host organisms, such as Escherichia coli. This article aims to discuss the importance of cephalosporin CA enzyme, obstacles faced in its expression in E. coli, and strategies for optimization.
CEPHALOSPORIN C
Cephalosporin C was the second beta-lactam class antibiotic discovered and used in clinical therapy after penicillin. Cephalosporin C was discovered in 1945 by a doctor named Giuseppe Brotzu. He found that a fungus from seawater near a sewage drain in the city of Cagliari (Italy) had antibacterial activity against Staphylococcus aureus, Vibrio cholera, and Bacillus anthracis. The fungus was initially identified as Cephalosporium acremonium but later was reclassified as A. chrysogenum. In 1953, Guy Newton and Edward Abraham from Oxford isolated and purified the antibacterial substance produced by the fungus which was initially named as Penicillin N. In 1962, Robert Morin from Lily Company discovered a process that could break the D-α-amino adipoyl group on the side chain of cephalosporin C and produce 7-ACA. This became an inspiration for the development of cephalosporin-class antibiotics with better activity. The advantage of cephalosporin antibiotics over penicillins in their development is that cephalosporin C can be modified by two sites, unlike penicillins which only have one site. The two sites are the 7-amino group and C3 atom in the acetoxy group (Fig. 1). However, modifying the side chain of cephalosporin C by chemical reaction is not easy. The reaction requires harsh conditions and produces toxic by-products that require complicated elimination processes [6,10].
ROLE OF CEPHALOSPORIN CA
The use of CA enzymes to directly convert cephalosporin C into 7-ACA provides many advantages, such as not resulting in toxic chemicals, i.e., trimethoxychlorosilane, phosphate pentachloride, dichloromethane, dimethylaniline, and others; reactions are selective, so that the use of temporary protective groups is not necessary; low energy consumption allows the reactions to occur at temperatures 20°C–30°C; the use of relatively simpler and cheaper tools because the reagents involved are safe for the environment; better quality because of specific results, hence contamination of the final product can be avoided [6].
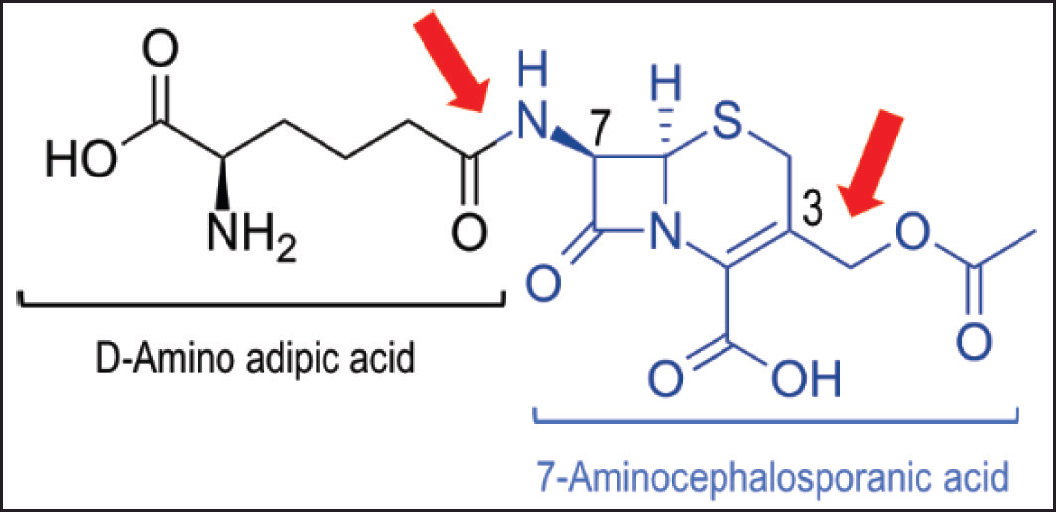 | Figure 1. Structure of cephalosporin C. Cephalosporin C consists of D-amino adipic acid 7-ACA groups. The 7-amino group and C3 on the acetoxy group which are key sites for modification of cephalosporin-class antibiotics are indicated by red arrows. Modified from Pollegioni et al. 2013 [6] . [Click here to view] |
It has been previously mentioned that at the beginning, bioconversion of cephalosporin C to 7-ACA involves two stages (Fig. 2). In the first stage, DAAO converts cephalosporin C into keto-adipoyl-7-ACA, hydrogen peroxide released automatically induces oxidative decarboxylase to form Glutaryl 7-ACA. In the second stage, 7-ACA is formed with the help of GA. The bioconversion of cephalosporin C to 7-ACA using a two-stage reaction is more efficient than using a chemical reaction. However, the presence of hydrogen peroxide resulting from the DAAO enzyme reaction also has a negative effect on affecting stability of the GA enzyme. Several ways have been found to overcome this problem, such as the use of catalase [11] and the replacement of DAAO with D-amino acid transaminase from Bacillus licheniformis ATCC 9945 [12].
As research progressed, the two-stage bioconversion was replaced with a simpler and more efficient one-stage reaction. One-stage bioconversion can be performed by cephalosporin CA enzyme. As shown in Figure 1, CA breaks the amide bond connecting the cephem core to the acyl group [13]. CA and GA mechanisms of action are similar because they act on the same site (Fig. 3), but only some classes of GA that have CA activity can act directly on cephalosporin C substrates, namely GA classes I and III. GA enzymes that have CA activity are produced by several microorganisms including Pseudomonas sp. strain N176 [7,14], Pseudomonas sp. V22 [7], and Pseudomonas sp. strain SE83 [8,15]. However, GA activity toward cephalosporin C is naturally very low [6].
CEPHALOSPORIN CA AND ITS STRUCTURAL MODIFICATION
Cephalosporin CA belongs to N-terminal nucleophile aminohydrolase (Ntn hydrolase) superfamily based on the structural classification of proteins classification [16]. The open reading frame gene structure of this superfamily members consists of a signal peptide, followed by α subunit, spacer, and β subunit [17]. Members of this superfamily have several specific characteristics, such as:
a. Enzymes structure are composed of four layers consisting of α-helices and β-sheets forming heterodimer (αβ) or heterotetramer (αββα) structures [18].
b. Enzymes are expressed as inactive precursors and then undergo autoproteolytic reaction to become active enzymes after being translocated to the periplasm [18,19].
c. N-terminal residue of β-chain plays two catalytic functions for amide bond hydrolysis, as a nucleophile and proton donor. Ser170 residue plays a role in this process [18,19].
Relative activity of wild-type CA produced by Pseudomonas sp. strain N176, Pseudomonas sp. V22, Pseudomonas sp. strain SE83 (acyII) toward cephalosporin C when compared to Glutaryl-7-ACA substrate were 2.4%, 3.2%, and ~4% respectively [6,7]. To obtain higher CA activity, modification of enzyme structure is necessary, i.e., through mutation techniques by involving knowledge of the relationship between structure and function of GA presented in Table 1.
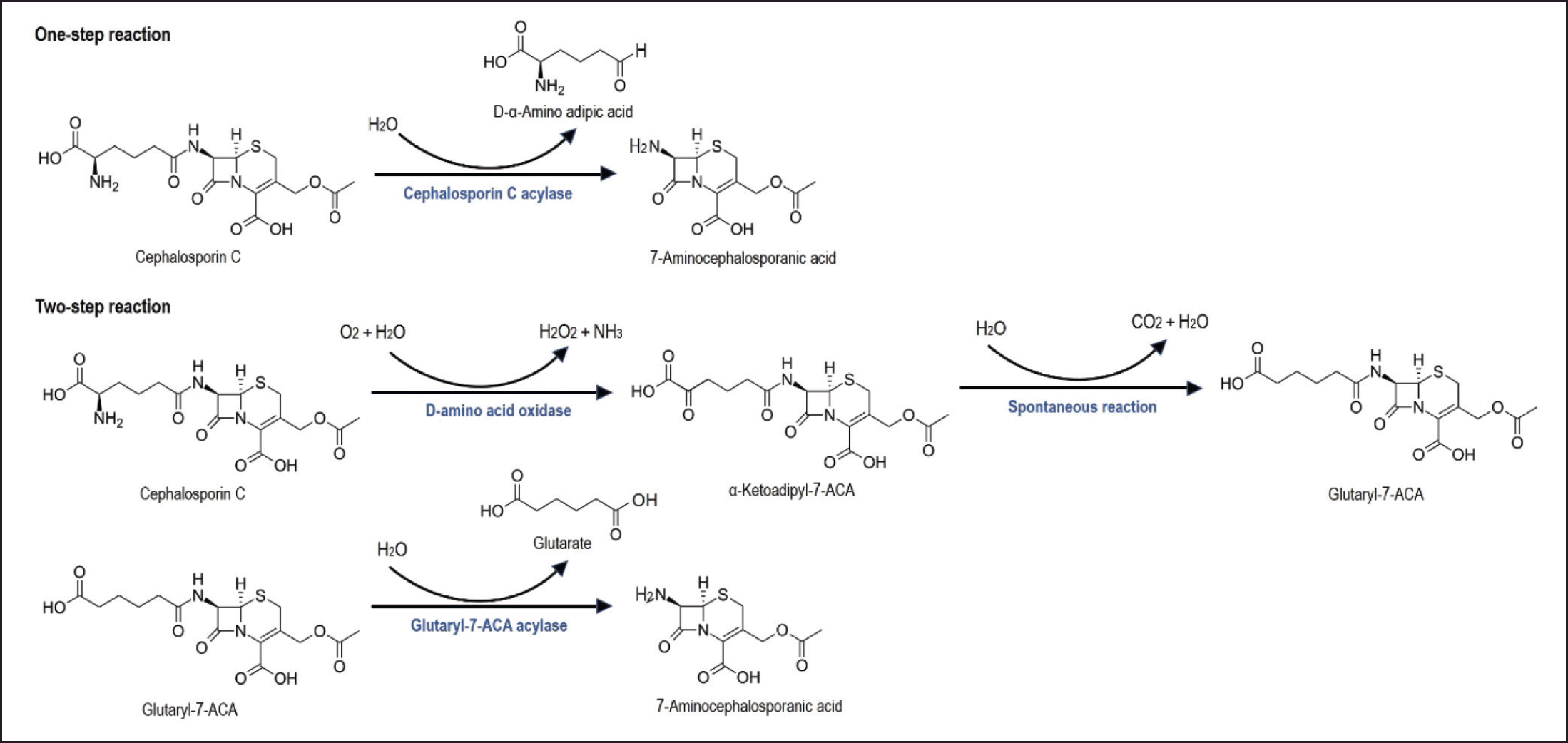 | Figure 2. Bioconversion of cephalosporin C to 7-ACA. One-stage bioconversion of cephalosporin C to 7-ACA (top) using cephalosporin C acylase (CA) enzyme. While, two-stage bioconversion (bottom) using DAAO) and GA enzyme. Modified from Pollegioni et al. 2013 [6]. [Click here to view] |
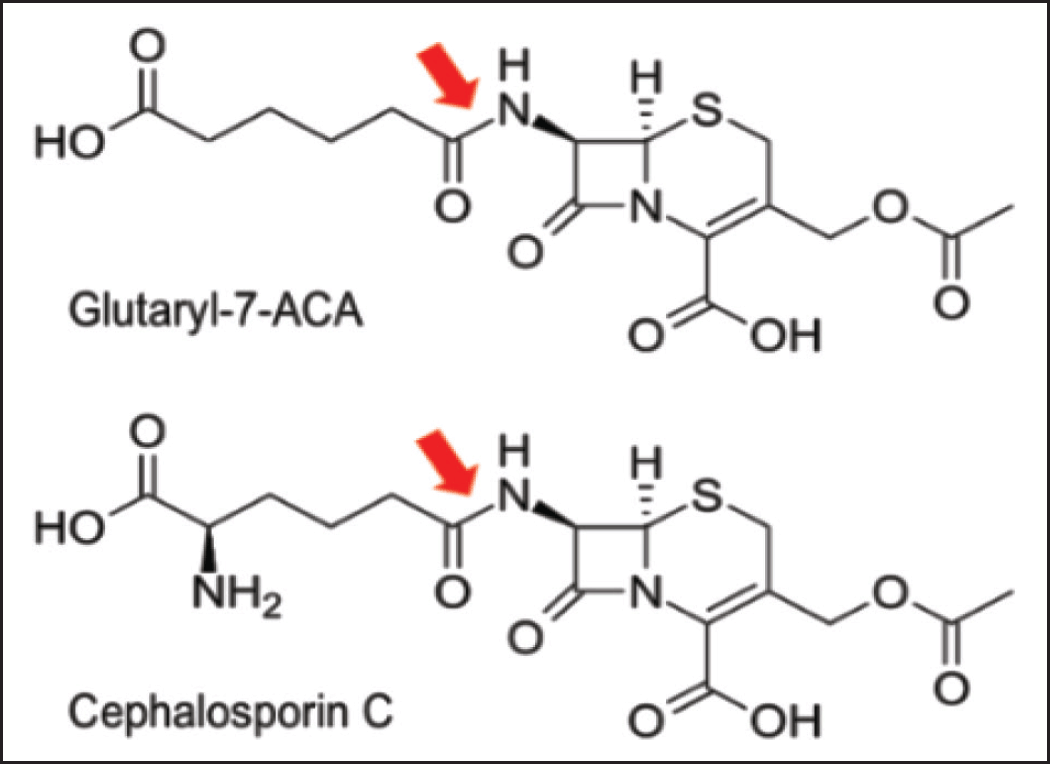 | Figure 3. GA and CA action sites in their substrate. GA with Glutaryl-7-ACA as substrate (top) and cephalosporin C-acylase (CA) with cephalosporin C as substrate (bottom) have the same site of action in their substrate, that is amide bond connecting the cephem nucleus to the acyl group indicated by the red arrow. Modified from Fritz-Wolf et al. 2002 [13]. [Click here to view] |
In 2007, Shin et al performed a six-point mutation of CA (acyII) encoding gene from Pseudomonas sp. SE83 resulting S12 variant (CAs12). The mutations are by replacing Val121α to Ala, Gly139α to Ser, Phe58β to Asn, Ile75β to Thr, Ile176β to Val, and Ser471β to Cys. CAs12 has specific activity toward cephalosporin C 5.8 units/mg protein, which was increased 8.5-fold compared to wild-type CA. In addition, there was also an increase in Ki value from 0.4 to 1.9 µM which indicates a reduction of 7-ACA production inhibition [20]. In 2012, Wang et al. published their findings that the replacement of alanine at position 675 with glycine was able to increase the specific activity of CA to 11.3 U/mg protein. The chemical structures of alanine and glycine are shown in Figure 4. The mutation is in the cavity associated with substrate transport in CA from Pseudomonas sp. SE83 [21]. The absence of a methyl group on the amino acid glycine as on alanine, probably widened the cavity so that cephalosporin C could more easily enter and bind to the active side of CA.
ESCHERICHIA COLI AS EXPRESSION HOST
Advantages of E. coli as expression host
The selection of the host organism is the first thing to be determined in the stages of the recombinant protein expression process. Host organisms will affect the overall technology used. Some host microorganisms that can be used for recombinant protein expression include bacteria, yeast, filamentous fungi, and unicellular algae. Those host microorganisms have their advantages and disadvantages. One of the most popular host microorganisms used in recombinant protein production is E. coli [22–24]. The advantages of E. coli are well characterized; fast growth kinetics (on optimal growth media has a doubling time of 20 minutes), easy-to-obtain cultures with large cell densities (theoretically on liquid media E. coli can reach a cell density of 200 g dry cell weight/L or 1 × 1013 live bacterial cells/ml; in batch cultures on LB media at 37°C the growth limit is lower, which is less than 1 × 1010 cells/ml); and DNA transformation can be performed quickly and easily [25–28].
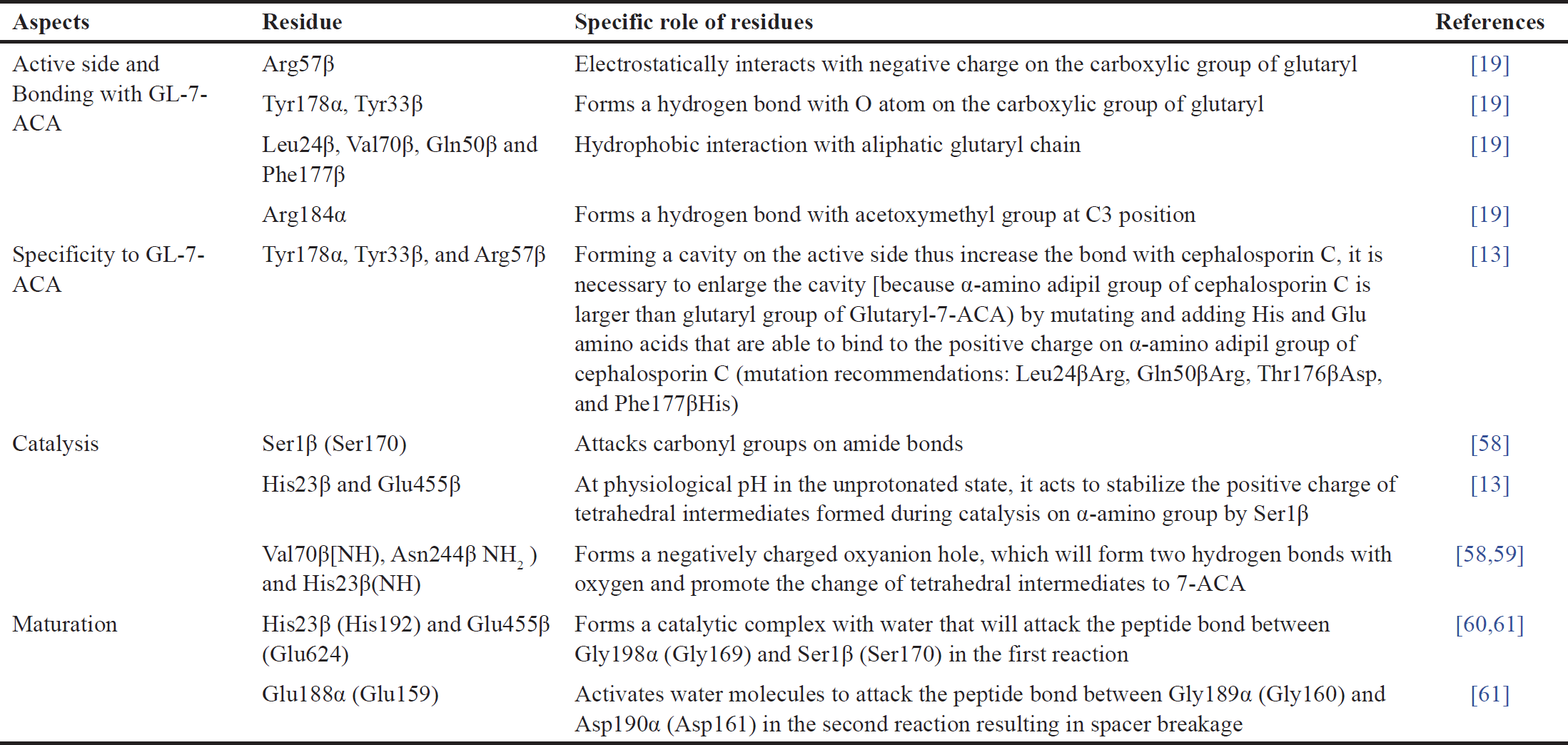 | Table 1. Structure and function relationships of Gas. Adapted from Pollegioni et al. [6]. [Click here to view] |
 | Figure 4. Chemical structure of alanine and glycine. The only difference in the chemical structure of alanine and glycine is the presence of a methyl group. [Click here to view] |
Various E. coli strains have specific characteristics according to their intended use. For expression purposes, there are two commonly used E. coli strains, namely E. coli BL21 (DE3) and K-12 derivatives. Escherichia coli BL21 (DE3) is characterized by lacking Lon protease, an enzyme that can degrade proteins, which is advantageous for stabilizing recombinant proteins [29]; missing the gene encoding Escherichia coli outer membrane protease (OmpT) protease that degrades extracellular proteins. The presence of OmpT disrupts the stability of proteins isolated by cell lysis [30]; mutations in hsdSB are beneficial for maintaining plasmid stability from DNA methylation and degradation; and λDE3 prophage inserted in chromosome BL21 contains a gene encoding T7 RNA polymerase enabling certain genes placed under T7 promoter on the plasmid being expressed [31].
Disadvantages of E. coli as expression hosts
Besides its various advantages, the use of E. coli in general and E. coli BL21 (DE3) in particular for recombinant protein production also has several obstacles (Table 2) that need to be overcome so that the expression of recombinant proteins in E. coli can be maximized [31,32].
The first obstacle is low production of recombinant proteins or no production at all. This is caused by two things, either the expression of toxic proteins or bias codons. To determine the expression of toxic proteins, we can monitor the growth of cells containing empty plasmids and plasmids containing the gene to be expressed. If there is a slowdown of growth in cells with plasmids containing the gene to be expressed, it can be confirmed that there is toxic protein production. Toxic proteins can be produced before and after induction [31]. Meanwhile, codon bias occurs when the frequency of foreign codons is high, resulting in a lack of tRNA. Consequently, the expression and activity of the recombinant protein become lower [33]. In general, this first obstacle has been overcome by the use of an appropriate promoter system in the expression host used, host modification, and codon optimization [31].
The second and third obstacles, namely inclusion body formation and inactive protein production, are more difficult to overcome thus being the main disadvantages of using E. coli as a host for heterologous protein expression. There were many solution options that required optimization to find the one that best suits the expected expression conditions [31,32,34]. In the next section, we will discuss how to minimize the formation of inclusion bodies and increase the activity of the proteins produced using chaperones and osmolytes as well as optimization of fermentation conditions and transcriptional control.
PRODUCTION STRATEGY OF CEPHALOSPORIN CA IN E. COLI
Due to the widespread use of E. coli as a host expression system, several strategies need to be implemented to increase efficiency in the production of recombinant proteins, as described in this chapter.
 | Table 2. Obstacles to recombinant protein production in E. coli and solutions. Adapted from Francis and Page [32] and Rosano and Ceccarelli [31]. [Click here to view] |
Minimizing inclusion body formation: chaperones and osmolytes
Inclusion bodies are protein aggregates that form when foreign genes are expressed in E. coli. Inclusion bodies can arise as a result of E. coli losing spatiotemporal control. Recombinant proteins are expressed in a microenvironment that is different from the source organism, in terms of pH, osmolarity, redox potential, cofactor availability, and folding mechanism. In addition, high expression levels lead to high hydrophobic stretching that causes interactions at similar parts between proteins [35,36]. Inclusion bodies can also form due to errors in disulfide bond formation. In E. coli, the majority of disulfide bonds occur in the periplasm because cysteine oxidation occurs in the periplasm. Cysteine residues are part of the catalytic site of many enzymes. Enzymes that undergo maturation in the periplasm and fail to translocate will remain in the cytosol and consequently fail to form disulfide bonds. This occurs because cytosol has more negative redox potential that is controlled by thioredoxin-thioredoxin reductase (trxB) and glutaredoxin-glutaredoxin reductase (gor) systems, leading to reduction reactions instead of oxidation [37].
Chaperones are key in protein quality control that helps proteins achieve their final structure. Examples of chaperones include GroEL/GroES, DnaK/DnaJ, and ClpB [34]. High protein expression in the cytosol as occurs in the production of recombinant proteins causes the protein quality control system in the cell to become saturated resulting in misfolding [35,38]. To overcome this, chaperone co-expression can be performed. DnaK/DnaJ binds to the hydrophobic part of the polypeptide thus preventing aggregation and maintaining solubility. GroEL/GroES binds to the polypeptide and helps its folding [39]. ClpB helps in the prevention of aggregation and also resolubilization of aggregated proteins in bacteria This chaperone is naturally produced by Mycobacterium tuberculosis to encounter several stresses within the host [40]. Utilization of chemical chaperones, also called osmolytes, can be carried out in vitro by adding osmolytes to the protein isolate or mimicking in vivo conditions by adding osmolytes to the culture medium. Examples of osmolytes are proline, glycine betaine, and trehalose [41,42].
Proline (Fig. 5) plays an important role in bacteria. It can be used as an osmoprotectant, a stabilizer of protein, and help cells to survive a variety of stresses. Escherichia coli can naturally synthesize proline from glutamate which is tightly controlled via the feedback inhibition mechanism of γ-glutaryl kinase reaction. Moreover, accumulation of proline also can be achieved by active transport across plasma membrane via three transporter systems, that is putP, proP, and proW [43].
 | Figure 5. Chemical chaperones structure. Chemical structure of proline, glycine-betaine, and trehalose, some chemical chaperones used for osmoprotectants in bacteria [42, 46]. [Click here to view] |
Glycine-betaine (Fig. 5) is an important osmolyte for bacteria. Glycine-betaine is a potent osmoprotectant widely distributed in nature and its intracellular accumulation is achieved through uptake from the environment or synthesis from choline precursors [44]. Glycine-betaine protects bacterial cells from suddenly increased environmental osmolarity that can lead to rapid expulsion of water from the cell and result in decreased turgor that spurs rapid uptake of K+ ions [45]. K+ ions that enter the cell can affect cell physiology which may also affect the maturation of certain enzymes in the cell. In addition, osmolytes can also act as chemical chaperones by increasing protein stability and aiding the folding of unfolded polypeptides. Glycine-betaine has been reported capable of preventing protein aggregation in E. coli under heat stress [42,46]. However, the use of glycine-betaine in soil bacteria Ralstonia eutropha H16 is reported to have a partial inhibitory effect when the concentration is excessive [47]; therefore, its use in E. coli needs to be optimized for concentration.
Trehalose (Fig. 5) is a non-reducing sugar found in bacteria and yeast. These sugars can act as osmoprotectants that help maintain celuller integrity under various stress conditions by affecting the antioxidant defense system. The special feature of trehalose is its ability to induce the accumulation of several other osmolytes, such as proline and glycine-betaine. Therefore, the presence of trehalose helps cells to be able to adapt to more varied stress conditions [48].
Optimization of fermentation conditions
Fermentation conditions are an important factor in protein production, including the CA enzyme in this discussion. It is known that variations in the concentration of isopropyl β-D-1-thiogalactopyranoside (IPTG) inducer and the length of fermentation time affect the expressed CA activity [49]. Temperature during fermentation also affects CA activity through its effect on inclusion body formation. Lower fermentation temperature leads to a reduction in inclusion bodies formation, thus increasing the activity of CA [50]. Fermentation media composition also needs to be optimized to obtain high CA activity, such as media type, carbon source, and nitrogen source.
Optimization of fermentation media for CA production in Pseudomonas sp. host has been reported using various types of media, carbon sources, and nitrogen sources (organic and inorganic). The types of media used were nutrient broth, minimal broth, casein hydrolysate broth, soybean casein digest broth, and Luria Bertani (LB) broth where the highest CA activity was obtained in fermentation using soybean casein digest broth. Variations of carbon sources used were galactose, dextrose, sucrose, fructose, and maltose where the highest CA activity was obtained in fermentation using galactose carbon source. Variations of organic nitrogen sources used were peptone, yeast extract, casein enzyme hydrolysate, soybean lysate, and meat extract where the highest CA activity was obtained in fermentation with peptone organic nitrogen source. While the variation of inorganic nitrogen sources used were ammonium sulfate, ammonium chloride, ammonium phosphate, and ammonium nitrate where the highest CA activity was obtained in fermentation using ammonium sulfate inorganic nitrogen source [51].
Optimization of fermentation conditions for CA production also has been reported in Achromobacter xylosooxidans. The parameters optimized were temperature, pH, carbon source, inorganic/organic nitrogen source, and inoculum level. The maximum CA activity in LB broth was achieved at a temperature of 30oC after 72 hours of fermentation. At a temperature of 30oC with a changing initial pH of 5–10, the highest enzyme activity was measured to be 28.9 U/ml at pH 8.0. As a carbon source, 4 g/l galactose had the highest CA activity than other carbon sources used (dextrose, sucrose, maltose, fructose). Ammonium sulfate is the best inorganic nitrogen source at a concentration of 2.5 g/l, while yeast extract is the best organic nitrogen source at a concentration of 3 g/l. The optimum inoculum level used was 4% v/v although overall it did not give significantly different results compared to other parameters [52].
In 2012, Wang et al. optimized the fermentation conditions for the production of mutant CA in E. coli JM109 (DE3) host. The highest mutant CA activity (5349 U/L) was obtained in batch fermentation using 3 g/l lactose as an inducer in the semi-defined medium. Composition of semi-defined medium used was 50 g/l corn steep liquor; 10 g/l yeast extract; 2.5 g/l NH4 Cl; 2.3 g/l KH2 PO4; 16.4 g/l K2 HPO4; 5 g/l glycerol. While the fermentation condition was using pH 7.5; fermentation at 28oC for 24 hours, shaker rotation speed of 200 rpm. The inoculum concentration used was 2.5%. Inoculum was made using LB media containing 50 μg/ml kanamycin, incubation was carried out at 37oC overnight, shaker rotation speed of 200 rpm [21].
Transcriptional control
Reduction of GC contents in spacer coding sequences can increase transcription strength. Higher GC contents in the sequence can lead to the formation of more stable secondary structures and inhibit DNA to form a complex with RNA polymerase. Conversely, higher AT contents in the spacer coding sequence can lead to transcription strength increases [53].
 | Figure 6. Chemical Structure of IPTG, lactose, and arabinose. IPTG, lactose, and arabinose are inducers commonly used in protein expression using E. coli as organism host [56, 58]. [Click here to view] |
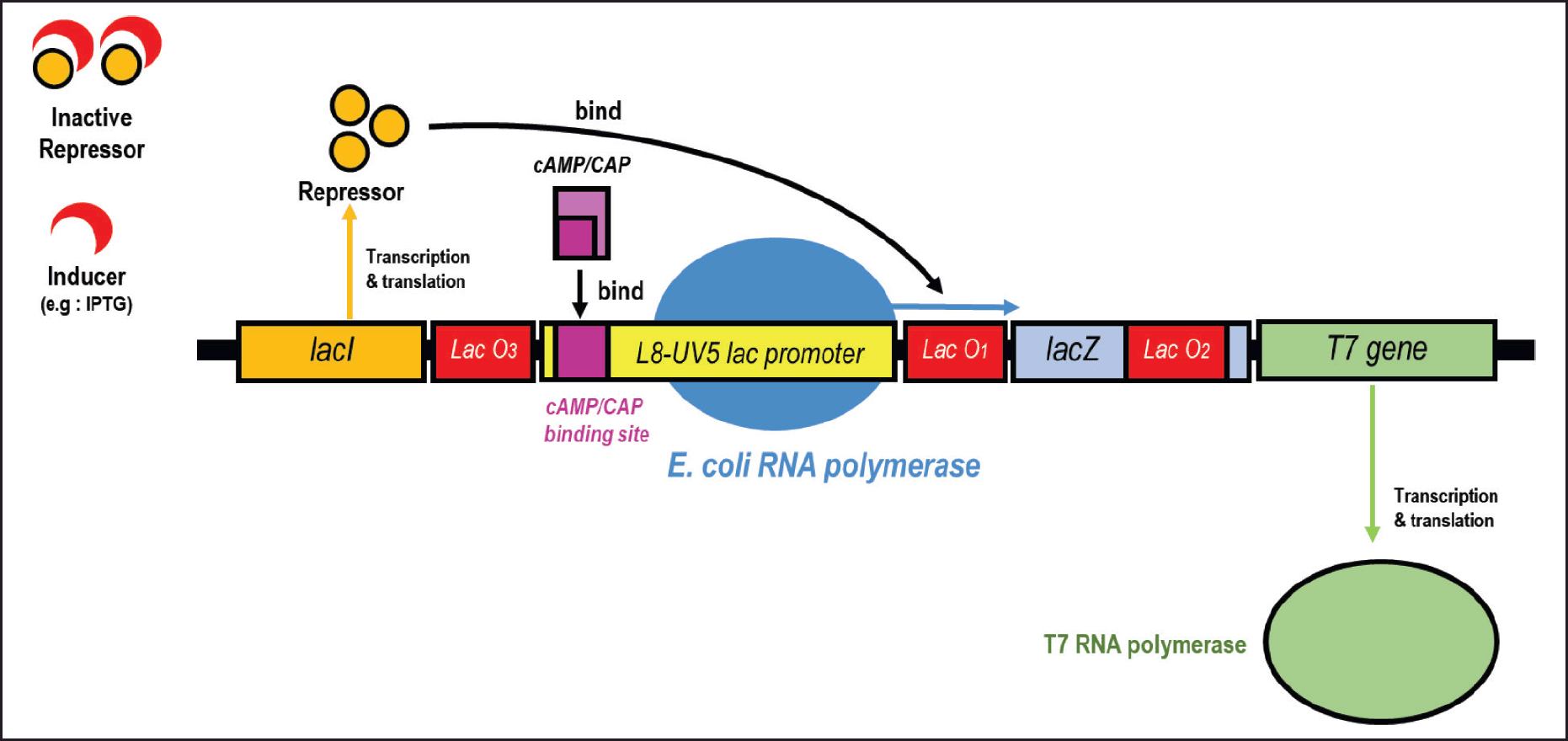 | Figure 7. Transcriptional control of T7 gene in λDE3. Transcription of T7 gene is controlled by L8-UV5 lac promoter and has undergone three mutations compared to the wild-type lac promoter. Lac repressor (encoded by lacI) binds to lacO1 and interacts with pseudo-operators (lacO2 and lacO3) to prevent transcription. IPTG inducer binds to the repressor thus decreasing its affinity with lacO1 and transcription can occur. When cAMP levels are sufficiently high, the CAP/cAMP complex binds immediately to the upstream position of the promoter and leads to transcription stimulation. If the CAP/cAMP complex is not formed, the transcription level will decrease, which is called catabolite repression. Modified from Novy and Morris [54]. [Click here to view] |
In 2018, Ammar et al. discovered a mechanism of catabolite repression in E. coli toward non-glucose sugars, where the most favored sugar would repress the expression of genes involved in the metabolism of other sugars. The metabolism of these sugars is hierarchically organized, with lactose as the most preferred sugar, followed by L-arabinose, then D-xylose. The mechanism of catabolite repression is closely related to the level of cyclic adenosine 3′,5′-monophosphate (cAMP) [46]. In the pET expression system, there is a component called lac operon that functions to regulate RNA transcription, both stimulating (promoter) and decreasing (repressor). Stimulation of transcription is not only influenced by inducers (Fig. 6) but also cAMP levels (Fig. 7). These cAMP levels are influenced by the carbon source used or sugar. CAMP levels will increase if the carbon source used is more unfavorable, for example, cAMP levels when using a carbon source in the form of glucose are lower than when using glycerol. Inducers stimulate transcription by binding to the lac repressor so that its binding to the lac operator decreases. Meanwhile, an increase in cAMP level stimulates transcription through binding of the CAP/cAMP complex with the CAP/cAMP binding site on the lac promoter. Thus, full transcriptional activation can only be achieved if there is an inducer and there is an increase in cAMP levels. Escherichia coli BL21 (DE3) carries the L8-UV5 promoter, which contains a three-point mutation of the wild-type lac promoter (Fig. 7). This mutation reduces sensitivity to catabolite repression while not eliminating it [54].
On the other hand, the optimization of the inducer itself has been widely studied in E. coli, especially to minimize inclusion body formation and maximize recombinant protein activity in the pET expression system. The use of IPTG is toxic to cells, increases stress conditions and stimulates the formation of inclusion bodies which reduce the activity of bioactive proteins. The use of lactose and arabinose is reported to be an alternative inducer of IPTG that is less toxic to cells and induces weaker expression, thus increasing the ability of cells to fold bioactive proteins [48,49]. Specifically, arabinose was shown to increase the expression of the active enzyme penicillin G-acylase where optimal induction occurred at a concentration of 15 g/l [55,56]. The chemical structures of IPTG, lactose, and arabinose are shown in Figure 6.
Besides mutations in the lac promoter gene, a new way to disable the catabolite repression effect was recently discovered by the crp gene mutation. This gene encodes the cyclic AMP receptor protein in several microbes, including E. coli. Experiments on E. coli BL21 (DE3) with a mutation in the crp gene showed the ability to consume glucose and xylose simultaneously, even though they are different types of sugar, namely pentose and hexose. Further proteomic analysis showed that glucose, which is a hexose/C6 sugar, can be metabolized via the C5 route to support de novo nucleotide synthesis and energy production in the crp mutant strain. However, the metabolic rate still needs to be monitored because overflow metabolism may inhibit bacterial cell growth [57].
CONCLUSION
Cephalosporin-class antibiotics have great potential and good prospects for further development. However, their development continues to face challenges, such as enzyme structural aspects related to strengthening enzyme bonds with cephalosporin C and enzyme production aspects in certain hosts. Especially in E. coli hosts, the main obstacles that occur are the formation of inclusion bodies and the production of inactive proteins. Several strategies are offered in this article based on the summary of previous studies. However, further proof in the laboratory is needed to find a suitable strategy formula that can fulfill industrial requirements. As a suggestion for further research in this field, further exploration is needed to direct the expression of CA enzyme in E. coli to cytoplasm instead of the periplasm because it has a wider space and thus reduces the spatio-temporal bottleneck that stimulates inclusion body formation. Finally, further research is also needed to explore possibilities for better host replacement without neglecting the criteria required by industry, especially economic aspects.
ACKNOWLEDGMENTS
The authors would like to convey many thanks to LPDP (Indonesia Endowment Fund for Education Agency) for funding the study.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this review article.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Barber MS, Giesecke U, Reichert A, Minas W. Industrial enzymatic production of cephalosporin-based beta-lactams. Adv Biochem Eng Biotechnol. 2004;88:179–215.
2. World Health Organization. Critically important antimicrobials for human medicine [Internet]. 6th rev. Geneva, Switzerland: World Health Organization; 2019 [cited 2023 Feb 17]. p. 45. Available from: https://apps.who.int/iris/handle/10665/312266
3. Browne AJ, Chipeta MG, Haines-Woodhouse G, Kumaran EPA, Hamadani BHK, Zaraa S, et al. Global antibiotic consumption and usage in humans, 2000–18: a spatial modelling study. Lancet Planetary Health [Internet]. 2021 Dec [cited 2024 Feb 15];5(12):e893–904. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2542519621002801
4. Aoki T, Yoshizawa H, Yamawaki K, Yokoo K, Sato J, Hisakawa S, et al. Cefiderocol (S-649266), a new siderophore cephalosporin exhibiting potent activities against Pseudomonas aeruginosa and other gram-negative pathogens including multi-drug resistant bacteria: Structure activity relationship. Eur J Med Chem. 2018 Jul 15;155:847–68.
5. Fong IW. New Cephalosporins: fifth and sixth generations. In: new antimicrobials: for the present and the future [Internet]. Cham, Germany: Springer International Publishing; 2023 [cited 2024 Feb 14]. p. 25–38. (Emerging Infectious Diseases of the 21st Century). Available from: https://link.springer.com/10.1007/978-3-031-26078-0_2
6. Pollegioni L, Rosini E, Molla G. Cephalosporin C acylase: dream and(/or) reality. Appl Microbiol Biotechnol. 2013 Mar;97(6):2341–55.
7. Aramori I, Fukagawa M, Tsumura M, Iwami M, Yokota Y, Kojo H, et al. Isolation of soil strains producing new cephalosporin acylases. J Ferment Bioeng [Internet]. 1991 [cited 2023 May 24];72(4):227–31. Available from: https://www.academia.edu/30812477/Isolation_of_soil_strains_producing_new_cephalosporin_acylases
8. Matsuda A, Toma K, Komatsu K. Nucleotide sequences of the genes for two distinct cephalosporin acylases from a Pseudomonas strain. J Bacteriol [Internet]. 1987 Dec [cited 2023 May 24];169(12):5821–6. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC214163/
9. Tan Q, Qiu J, Luo X, Zhang Y, Liu Y, Chen Y, et al. Progress in one-pot bioconversion of cephalosporin c to 7-aminocephalosporanic acid. Curr Pharm Biotechnol. 2018;19(1):30–42.
10. Morin RB, Jackson BG, Flynn EH, Roeske RW, Andrews SL. Chemistry of cephalosporin antibiotics. XIV. The reaction of cephalosporin C with nitrosyl chloride. J Am Chem Soc. 1969 Mar 12;91(6):1396–400.
11. Tan Q, Zhang Y, Song Q, Wei D. Single-pot conversion of cephalosporin C to 7-aminocephalosporanic acid in the absence of hydrogen peroxide. World J Microbiol Biotechnol [Internet]. 2010 Jan 1 [cited 2023 May 24];26(1):145–52. Available from: https://doi.org/10.1007/s11274-009-0153-9
12. Aretz WDK, Sauber KDS. New D-amino Acid Transaminase and Their Use [Internet]. DE3447023A1, 1986 [cited 2023 May 24]. Available from: https://patents.google.com/patent/DE3447023A1/en
13. Fritz-Wolf K, Koller KP, Lange G, Liesum A, Sauber K, Schreuder H, et al. Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C. Protein Sci. 2002 Jan 1;11(1):92–103.
14. Aramori I, Fukagawa M, Tsumura M, Iwami M, Isogai T, Ono H, et al. Cloning and nucleotide sequencing of new glutaryl 7-ACA and cephalosporin C acylase genes from Pseudomonas strains. J Ferment Bioeng [Internet]. 1991 Jan 1 [cited 2023 May 24];72(4):232–43. Available from: https://www.sciencedirect.com/science/article/pii/0922338X9190155A
15. Matsuda A, Matsuyama K, Yamamoto K, Ichikawa S, Komatsu K. Cloning and characterization of the genes for two distinct cephalosporin acylases from a Pseudomonas strain. J Bacteriol [Internet]. 1987 Dec [cited 2023 May 24];169(12):5815–20. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC214159/
16. Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995 Apr 7;247(4):536–40.
17. Pollegioni L, Lorenzi S, Rosini E, Marcone GL, Molla G, Verga R, et al. Evolution of an acylase active on cephalosporin C. Protein Sci [Internet]. 2005 Dec [cited 2023 May 25];14(12):3064–76. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2253238/
18. Cho K, Kim KH. Cephalosporin acylase precursor, glutaryl-7-aminocephalosporanic acid acylase precursor. Handbook of Proteolytic Enzymes. Cambridge, MA: Academic Press; 2013 Dec 31. Vol. 3, pp 3659–63.
19. Kim Y, Kim S, Earnest TN, Hol WGJ. Precursor structure of cephalosporin acylase. Insights into autoproteolytic activation in a new N-terminal hydrolase family. J Biol Chem. 2002 Jan 25;277(4):2823–9.
20. Shin YC, Jeon JYJ, Jung KH, Park MR, Kim Y. Cephalosporin C acylase mutant and method for preparing 7-ACA using same [Internet]. US7592168B2, 2009 [cited 2023 Mar 18]. Available from: https://patents.google.com/patent/US7592168B2/pt
21. Wang Y, Yu H, Song W, An M, Zhang J, Luo H, et al. Overexpression of synthesized cephalosporin C acylase containing mutations in the substrate transport tunnel. J Biosci Bioeng. 2012 Jan;113(1):36–41.
22. Adrio JL, Demain AL. Recombinant organisms for production of industrial products. Bioeng Bugs [Internet]. 2010 [cited 2023 Jul 1];1(2):116–31. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3026452/
23. Demain AL, Vaishnav P. Production of recombinant proteins by microbes and higher organisms. Biotechnol Adv. 2009;27(3):297–306.
24. Sahdev S, Khattar SK, Saini KS. Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies. Mol Cell Biochem. 2008 Jan;307(1–2):249–64.
25. Lee SY. High cell-density culture of Escherichia coli. Trends Biotechnol. 1996 Mar;14(3):98–105.
26. Pope B, Kent HM. High efficiency 5 min transformation of Escherichia coli. nucleic acids res [Internet]. 1996 Feb 1 [cited 2023 Jul 3];24(3):536–7. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC145660/
27. Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli physiology in luria-bertani broth. J Bacteriol [Internet]. 2007 Dec [cited 2023 Jul 3];189(23):8746–9. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2168924/
28. Shiloach J, Fass R. Growing E. coli to high cell density--a historical perspective on method development. Biotechnol Adv. 2005 Jul;23(5):345–57.
29. Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506.
30. Grodberg J, Dunn JJ. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988 Mar;170(3):1245–53.
31. Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol [Internet]. 2014 [cited 2023 May 29];5. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2014.00172
32. Francis DM, Page R. Strategies to optimize protein expression in E. coli. Curr Protoc Protein Sci [Internet]. 2010 Aug [cited 2023 Jul 4];61(1):5–24. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7162232/
33. Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004 Jul;22(7):346–53.
34. Bhatwa A, Wang W, Hassan YI, Abraham N, Li XZ, Zhou T. Challenges associated with the formation of recombinant protein inclusion bodies in Escherichia coli and strategies to address them for industrial applications. Front Bioeng Biotechnol [Internet]. 2021 [cited 2023 Mar 18];9. Available from: https://www.frontiersin.org/articles/10.3389/fbioe.2021.630551
35. Carrió MM, Villaverde A. Construction and deconstruction of bacterial inclusion bodies. J Biotechnol. 2002 Jun 13;96(1):3–12.
36. Hartley DL, Kane JF. Properties of inclusion bodies from recombinant Escherichia coli. Biochem Soc Trans. 1988 Apr;16(2):101–2.
37. Stewart EJ, Aslund F, Beckwith J. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J [Internet]. 1998 Oct 1 [cited 2023 Jul 4];17(19):5543–50. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1170883/
38. Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002 Mar 8;295(5561):1852–8.
39. Chen Y, Song J, Sui SF, Wang DN. DnaK and DnaJ facilitated the folding process and reduced inclusion body formation of magnesium transporter CorA overexpressed in Escherichia coli. Protein Expr Purif. 2003 Dec;32(2):221–31.
40. Singh D, Tripathi P, Sharma R, Grover S, Batra JK. Role of a substrate binding pocket in the amino terminal domain of Mycobacterium tuberculosis caseinolytic protease B (ClpB) in its function. J Biomole Struct Dyn [Internet]. 2023 Jul 7 [cited 2024 Feb 14];1–11. Available from: https://www.tandfonline.com/doi/full/10.1080/07391102.2023.2232032
41. de Marco A, Vigh L, Diamant S, Goloubinoff P. Native folding of aggregation-prone recombinant proteins in Escherichia coli by osmolytes, plasmid- or benzyl alcohol-overexpressed molecular chaperones. Cell Stress Chaperones. 2005;10(4):329–39.
42. Diamant S, Rosenthal D, Azem A, Eliahu N, Ben-Zvi AP, Goloubinoff P. Dicarboxylic amino acids and glycine-betaine regulate chaperone-mediated protein-disaggregation under stress. Mol Microbiol. 2003 Jul;49(2):401–10.
43. Deutch CE. Growth of Escherichia coli K-12 on L-proline in the absence of known proline transporters. J Adv Microbiol Res [Internet]. 2023 Jan 1 [cited 2024 Feb 15];4(1):01–10. Available from: https://www.microbiojournal.com/archives/2023.v4.i1.A.57
44. Hernandez-Leon SG, Valenzuela-Soto EM. Glycine betaine is a phytohormone-like plant growth and development regulator under stress conditions. J Plant Growth Regul [Internet]. 2023 Aug [cited 2024 Feb 15];42(8):5029–40. Available from: https://link.springer.com/10.1007/s00344-022-10855-3
45. Lucht JM, Bremer E. Adaptation of Escherichia coli to high osmolarity environments: osmoregulation of the high-affinity glycine betaine transport system proU. FEMS Microbiol Rev. 1994 May;14(1):3–20.
46. Diamant S, Eliahu N, Rosenthal D, Goloubinoff P. Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. J Biol Chem. 2001 Oct 26;276(43):39586–91.
47. Iskandaryan MK. Role of glycine-betaine in the growth and hydrogenases activity of ralstonia eutropha H16. Proceedings of the YSU B: chemical and biological sciences [Internet]. 2023 Jul 7 [cited 2024 Feb 15];57(2 (261)):154–63. Available from: https://journals.ysu.am/index.php/proceedings-chem-biol/article/view/vol57_no2_2023_pp154-163
48. Hassan MU, Nawaz M, Shah AN, Raza A, Barbanti L, Skalicky M, et al. Trehalose: a key player in plant growth regulation and tolerance to abiotic stresses. J Plant Growth Regul [Internet]. 2023 Aug [cited 2024 Feb 15];42(8):4935–57. Available from: https://link.springer.com/10.1007/s00344-022-10851-7
49. Isdiyono BW, Hardianto D, Ivan FX. Production of recombinant cephalosporin acylase as biocatalyst for 7-aminocephalosporanic acid production. J Bioteknol Biosains Indonesia (JBBI) [Internet]. 2017 Jul 7 [cited 2023 Jun 2];4(1):28–35. Available from: https://ejurnal.bppt.go.id/index.php/JBBI/article/view/2059
50. Martius E, Wibisana A, Ardiyani Y. The optimization of soluble cephalosporin C acylase expression in E. coli. IJES 2018;7:29–34.
51. Jobanputra AH, Vasait RD. Cephalosporin C acylase from Pseudomonas species: production and enhancement of its activity by optimization of process parameters. Biocatal Agric Biotechnol [Internet]. 2015 Oct 1 [cited 2023 Jun 2];4(4):465–70. Available from: https://www.sciencedirect.com/science/article/pii/S187881811500081X
52. Vasait RD, Jobanputra AH. Bacterial conversion of cephalosporin C: optimization in Achromobacter xylosooxidans. Biocatal Agric Biotechnol [Internet]. 2023 Aug [cited 2024 Feb 13];51:102772. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1878818123001731
53. Sun H, Liu T, Luo H, Nie Z, Chang Y, Yu H, et al. Optimization of cephalosporin C acylase expression in Escherichia coli by high-throughput screening a constitutive promoter mutant library. Appl Biochem Biotechnol. 2021 Apr;193(4):1056–71.
54. Novy R, Morris B. Use of glucose to control basal expression in the pET System. Novagen Inc [Internet]. n.d.; Available from: http://wolfson.huji.ac.il/expression/procedures/bacterial/Glucose%20supression.pdf
55. Kopp J, Slouka C, Ulonska S, Kager J, Fricke J, Spadiut O, et al. Impact of glycerol as carbon source onto specific sugar and inducer uptake rates and inclusion body productivity in E. coli BL21(DE3). Bioengineering (Basel) [Internet]. 2017 Dec 21 [cited 2023 Aug 8];5(1):1. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5874867/
56. Narayanan N, Hsieh MY, Xu Y, Chou CP. Arabinose-induction of lac-derived promoter systems for penicillin acylase production in Escherichia coli. Biotechnol Prog. 2006;22(3):617–25.
57. Kaplan NA, Islam KN, Kanis FC, Verderber JR, Wang X, Jones JA, et al. Simultaneous glucose and xylose utilization by an Escherichia coli catabolite repression mutant. Buan NR, editor. Appl Environ Microbiol [Internet]. 2024 Jan 30 [cited 2024 Feb 15];e02169–23. Available from: https://journals.asm.org/doi/10.1128/aem.02169-23
58. Duggleby HJ, Tolley SP, Hill CP, Dodson EJ, Dodson G, Moody PC. Penicillin acylase has a single-amino-acid catalytic centre. Nature. 1995 Jan 19;373(6511):264–8.
59. Kim Y, Hol WG. Structure of cephalosporin acylase in complex with glutaryl-7-aminocephalosporanic acid and glutarate: insight into the basis of its substrate specificity. Chem Biol. 2001 Dec;8(12):1253–64.
60. Kim JK, Yang IS, Shin HJ, Cho KJ, Ryu EK, Kim SH, et al. Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries. Proc Natl Acad Sci USA. 2006 Feb 7;103(6):1732–7.
61. Kim JK, Yang IS, Rhee S, Dauter Z, Lee YS, Park SS, et al. Crystal structures of glutaryl 7-aminocephalosporanic acid acylase: insight into autoproteolytic activation. Biochemistry. 2003 Apr 15;42(14):4084–93.