INTRODUCTION
A large number of people in developing countries are approaching herbal species for their medicinal needs because of their safety over synthetic drugs. There is an equal range of international markets for herbal drugs and pharmaceutical products [1]. India is a rich source of herbal plants. Most human and animal diseases can be treated and prevented with the help of herbal plants [2,3]. Approximately 80% of the world’s population still rely on medications from herbal formulas [4]. The adulteration of herbal medicinal materials is becoming a global concern, coinciding with the growth of the herbal market. Therefore, more research should be supported to demonstrate the efficacy and security of natural herbal products [5]. Anatomical characteristics can be used to identify a small portion of a significant herbal product that is sold commercially as well as the actual plant source. The characteristics may be helpful in positively identifying plants [6].
DNA barcoding is a technique for identifying species depending on the analysis of short, uniform, and universal DNA region(s) (also known as “barcode sequence(s)”). This technique is widely used to identify a species by comparing the sequence of an unknown item to barcodes in a sequence database of recognized species [7,8]. Because of their physical resemblance to other plants in the same known species that are challenging to distinguish can be authenticated using DNA barcoding [9]. By comparing new species’ sequences to assembled reference libraries of barcode sequences, this technique enables the identification of new species. Many DNA sections, including matK, rbcL, trnH-PsbA, and ITS2, have been investigated for potential use as DNA barcodes [10].
The process of identifying, describing, and assessing the quality and purity of natural goods and plant-based medicines is known as pharmacognostic evaluation. Justicia species are essential to many drug formulations used in Indian traditional medicine. “Vasa,” among other drug preparations, is a significant drug in Ayurveda. Justicia adhatoda and Justicia beddomei are reliable sources of “Vasa.” Particularly when dried, the morphological characteristics of Justicia species can be confusing. Intentionally or unintentionally, J. adhatoda and J. beddomei are replaced with species such as J. carnea, J. betonica, J. gendarussa, J. santapaui, and J. wynaadensis when making the Ayurvedic medication “Vasa.” To evaluate the botanical identification, authenticity, purity, and quality of medicinal plant resources and their products, a variety of scientific procedures and approaches are applied [11–14].
In Justicia species, 56 different varieties are recorded in continental India [15]. Of these, 30 species are distributed in the Western Ghats regions and 26 species are present in Kerala [16]. Justicia beddomei (C.B. Clarke) Bennet [syn. Adhatoda beddomei C.B. Clarke] is a member of the Acanthaceae family [14]. Treatment of colds, coughs, asthma, bronchitis, and tuberculosis has long been practiced in India using tropical plants of the genus Adhatoda (Acanthaceae), usually referred to as “Vasa” [17]. The plant has many morphological traits with J. adhatoda, except for a few minor differences [14]. The main difference is in floral characteristics and similarities are in leaf and stem parts. The plant is an erect perennial shrub, 1.5–3 m tall; branches glabrous, terete to subtetragonal. This species is solely restricted to India’s Southern Western Ghats [17]. Recent literature has mentioned J. beddomei from Kerala as Chittadalodakam [14,17]. Justicia beddomei has historically been used to treat a number of illnesses, including fever, cough, leprosy, heart problems, blood disorders, hemorrhage, and tuberculosis. Some articles describe the Malayalam name as Cheriya adalodakam/Chittadalodakam for A. beddomei [18,19] which is a variant of Justicia adathoda L and is not J. beddomei (C.B. Clarke) Bennet. The commonly found plant is Justicia adathoda L. J. beddomei is a different species with very similar macroscopic features to J. adathoda L. Therefore, the taxonomy experts found difficulty in authenticating this plant in the nonflowering season based on morphological characters [20]. The main identification features of the plant are the inflorescence and floral characteristics. The flowering season of the plant is between December and June. The current article proposes an alternate method to identify the plant in the nonflowering season by DNA barcoding technique and also updates the information regarding the main microscopic difference of various parts of J. beddomei with J. adathoda L [17].
Despite the fact that J. beddomei (C.B. Clarke) Bennet (Fig. 1) contains a lot of documented phytochemical information and is listed in Critically Endangered Category [18,20]. There are only a few molecular studies on the Justicia species using ITS and matK genes [21]. There is no proof of its molecular identification using trnH-PsbA DNA barcode, detailed pharmacognostic (macroscopic and microscopic) features of various parts of this plant (leaf, stem, and flower), and power microscopic features of the whole plant of this plant. This study will facilitate the identification of this plant and improve the identification of adulterants in further studies of this plant.
 | Figure 1. Justicia beddomei (C.B. Clarke) Bennet. [Click here to view] |
MATERIALS AND METHODS
Plant material collection and authentication
The plant, J. beddomei (C.B. Clarke) Bennet was taken from the herb garden at the Arya Vaidya Sala in Kottakkal. The plant was identified by the Department of Forest Botany, Kerala Forest Research Institute (KSCSTE), Peechi, Thrissur, Kerala. The plant specimen’s herbarium is saved at Kerala Forest Research Institute (Herbarium), Peechi, Thrissur (Accession Number-19373).
Chemicals
All analytical grade chemicals used were commercially purchased from Chemind, Thrissur, Kerala.
Sample preparation for DNA extraction
Young, healthy leaves from the selected plant species were used to prepare the samples. After collection, the samples were produced using the sample identification number one. Leaves were professionally washed, dried, and stored at room temperature in sealed plastic packets [22]. Leaf samples were cut into tiny pieces after the midribs were removed. A mortar and pestle were used to grind 0.4 g of the sample into a powder using liquid nitrogen. The stored samples were later utilized to extract DNA in the molecular biology laboratory.
DNA extraction
DNA was isolated from a 0.4 g sample of leaf tissue using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions. Gel electrophoresis is used on a 0.8% agarose gel with 5 μl of sample standard DNA loaded (Sample M) per each lane, the quality of the isolated DNA was evaluated by a spectrophotometric approach using the Bio photometer plus (Eppendorf, Germany).
Polymerase chain reaction (PCR) amplification and purification
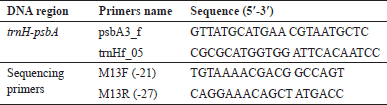
The PCR amplification was carried out using a 20 μl reaction mixture containing 5–20 ng of DNA: 1X PCR buffer (2 mM MgCl2), 200 μM each of dATP, dCTP, dGTP, and dTTP, 0.5 μM of each forward and reverse primer, 1U of Taq polymerase (Takara, Japan). The DNA amplification was carried out in a thermal cycler (Eppendorf, Germany). The list of primers and their nucleotide sequences are shown in Table 1. For 35 cycles of the PCR, the temperature was set at 92°C for 4 minutes, 94°C for 1 minute, 52°C for 1 minute, 64°C for 1 minute, and then 64°C for an additional 8 minutes. The 10 μl PCR reaction combination was used [23,24]. The tubes were taken out of the thermal cycler once the reaction was complete and kept at 4°C for storage. Gel-loading dye and PCR products were mixed, and the outcomes were separated using 1.8% agarose gel electrophoresis. The PCR products were purified after they had been separated on an agarose gel using the Machery-Nagel, Germany-based NucleoSpin® Gel, and PCR Clean-up kit. The purified PCR products were sequenced on an ABI 3730xl DNA sequencer at the AgriGenome laboratories facility in India for bidirectional sequencing (Sanger DNA sequencing) using M13Forward or M13Reverse primers, since all of our particular F/R primers were connected to M13F and M13R sequences, respectively.
Sequence editing, alignment, and analysis
The software, BioEdit sequence alignment editor version 7.2.6, was used to manually alter the unprocessed DNA sequences [25]. The CLUSTALW [26] algorithm in BioEdit was used to align the forward and reverse sequences, and the alignments were manually reviewed and corrected. The overlapping regions were joined together to obtain the contiguous DNA sequences [27]. The sequences were analyzed using the similarity-based method BLASTn available from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). NCBI BLASTn to find homologies between the fragments and identify species [28].
Pharmacognostical evaluation
Organoleptic and macroscopic evaluation
It is referred to as the evaluation of plant material employing elements such as color, odor, taste, form, and texture. Various parts leaf, stem, and flower of the plant were considered for the macroscopical study [29].
Microscopic evaluation
Cross sections of live plant materials from the leaf, stem, and flower portions were cut, and each part separately under a microscope. Microscopic features of plant powder were analyzed [30,31]
 | Table 1. List of primers used for amplification/sequencing of trnH-ps34bA locus. [Click here to view] |
RESULT AND DISCUSSION
PCR amplification
Using the trnH-psbA region, the juvenile leaf of the J. beddomei genome was characterized. The trnH-psbA intergenic spacer region has been used in many DNA barcoding studies. This area of the plant was amplified by the primers psbA3_f and trnHf_05. Figure 2 shows that the nuclear trnH-psbA barcode sections of the sample were amplified using universal primers with 100% PCR performance after DNA extraction and nuclear trnH-psbA region PCR amplification in plant J. beddomei (C.B. Clarke) Bennet. A clear band of about 400 bp in length was visible in the sample (Fig. 2). All the primers amplified a specific target region of 400 bp.
Sample 1 amplified a ~400 bp product using the trnH-psbA primers. After sequencing the amplified products, the final DNA sequence contigs contain 319 bp. The obtained sequence was analyzed in the NCBI—BLASTn for species identification by comparing the nucleotide query sequence to a nucleotide sequence database. The top 10 results obtained were selected (Table 2). From the results, trnH–psbA barcode region of the selected plant was compared with the BLASTn and showed 100% similarity with the plant J. beddomei nucleotide NCBI database with Accession no: MK347214.1. The BLASTn result’s percentage of identity verified the taxonomic identity of the plant, indicating that the plant was successfully identified by the DNA barcoding method.
Pharmacognostical evaluation
Macroscopy
The leaves of the plant are light green in color, simple, opposite, and exstipulate with the short petiole (Fig. 3A). Leaf shape is oblong-lanceolate. Leaves are entire and glabrous. Leaves measure 7.2–14.1 cm in length and 0.8–3.5 cm in breadth. The inflorescence is a short peduncled, condensed spike, 4.5–5 cm in length. Spike is axillary and stout. The leaf of the morphologically similar plant J. adathoda is in dark green color and possesses a long petiole [14].
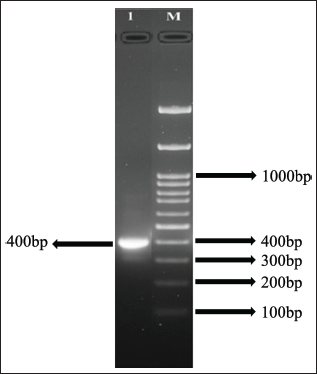 | Figure 2. Purification of PCR products: 100 bp DNA ladder (Lane M) J. beddomei (Lane 1). [Click here to view] |
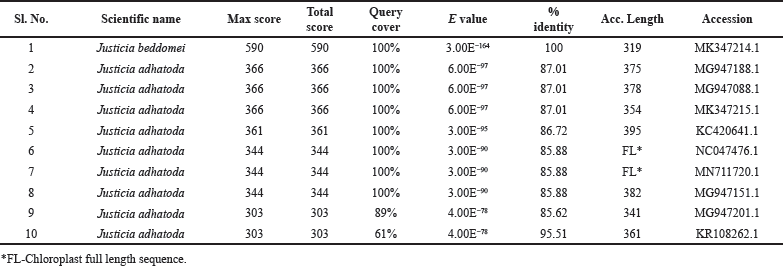 | Table 2. The top 10 BLASTn results from the NCBI database. [Click here to view] |
A small piece of stem is separated from a leafed twig (Fig. 3B). It measures 6.1 cm in length and 0.5 cm in width. The stem is green in color. It is unbranched, quadrangular, and thick. It is hard to touch, rough and woody texture. It has several brown patches. The odor of the plant is not characteristic and is bitter in taste.
The flower is the main identification part of the plant. Flowering season is from the month of December to June. Flowers are zygmomorphic, bisexual, sessile, and 1.5 cm long (Fig. 3C and D) and the inflorescence, number of flowers, is 4. The flower of J. adhatoda plant is 3–3.5 cm long and the inflorescence contains a minimum of 10 flowers [14].
Microscopy
Leaf
Justicia beddomei’s transverse section is slightly depressed centrally, on the upper side and convex on the bottom side. Upper and lower surfaces have a single-layered epidermis which is square-to-rectangular, radially organized cells with a cuticle. A few simple, uniserate, long, straight or bent, thick-walled nonglandular covering trichomes were found. Sessile, four-celled glandular trichomes were also found. Hypodermis is collenchymatous, tightly packed followed by parenchymatous ground tissue. The midrib and laminar region contains calcium oxalate crystals. Vascular bundles are located at the center of the midrib composed of the xylem and phloem. Vascular bundles include 3–4 conjoint, collateral meristeles. Centrally located arc-shaped meristele is the largest among them. Smaller meristeles are located on both lateral sides (Fig. 4).
The Laminar region is composed of the upper epidermis, mesophyll layer, and lower epidermis. The mesophyll layer is composed of palisade and spongy parenchyma (Fig. 5). Palisade parenchyma is thin-walled, columnar cells, found below the upper epidermis followed by circular to oval-celled spongy parenchyma found in the lower epidermis. Oil globules are found in the mesophyll region. Crystals of calcium oxalate and elongated warty cystoliths are also present in the mesophyll region. Stomata is diacytic and is found more in the lower epidermis than in the upper epidermis (Table 3).
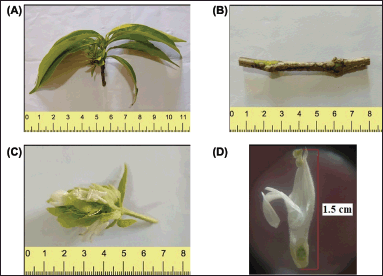 | Figure 3. (A) Leaf twig of J. beddomei; (B) Stem of J. beddomei; (C, D) Flower of J. beddomei. [Click here to view] |
The Transverse section of J. adhatoda leaf [32] is almost similar to J. beddomei. The main difference is in the cystolith region. The cystolith part extends from the epidermis to the lower cells of the palisade in J. adhatoda. Whereas, in J. beddomei cystolith part is in the mesophyll region. The stomatal index of J. beddomei is similar to J. adhatoda.
Stem
The transverse section of J. beddomei stems shows cork, followed by a parenchymatous cortex. Crystals of calcium oxalate are found in the collenchymatous hypodermis. A band of stone cells, mostly rectangular in shape with distinct pits are also seen in the cortical region. The xylem is composed of lignified vessels, fibers, parenchyma, and rays. Phloem is composed of phloem elements. Medullary rays are also found in the vascular region. Pith is composed of pitted parenchyma. A number of raphids are seen in the pith region (Figs. 6 and 7). The transverse section of the stem of J. adhatoda [33] is similar to J. beddomei.
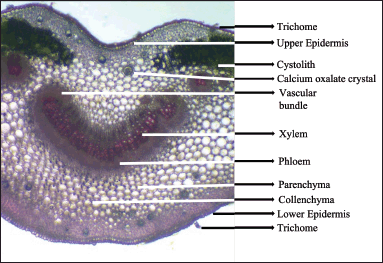 | Figure 4. Transverse section of midrib of J. beddomei. [Click here to view] |
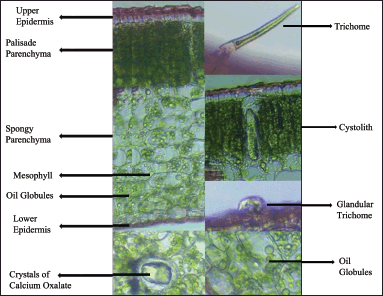 | Figure 5. Transverse section of leaf lamina of J. beddomei. [Click here to view] |
 | Table 3. Stomatal index of J. beddomi leaf. [Click here to view] |
Longitudinal section of flower
The longitudinal section of J. beddomei flower shows corolla, androecium, and gynoecium. Corolla is white, and imbricate. Corolla tube is pubescent inside, 2-lipped, the lower lip is 3-lobed, striations absent on the lip and the upper lip is bifid. There are two stamens, basifixed and light green. Filaments are dull white, and glabrous except for hirsute towards the base. Anther is 2-celled and cells are superposed. The ovary is superior, light green, glabrous, bicarpellate. Style is filiform, pubescent and Stigma is minutely bifid, dull-white, sparsely pubescent (Fig. 8).
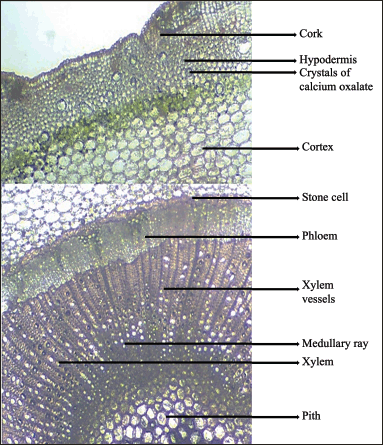 | Figure 6. Enlarged portion of J. beddomei stem. [Click here to view] |
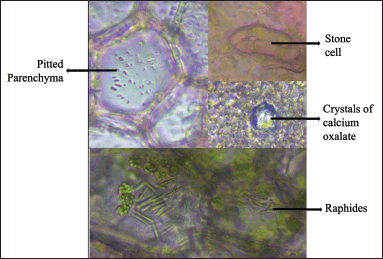 | Figure 7. Some characters of J. beddomei stem. [Click here to view] |
Microscopy transverse section
The transverse section (T.S.) of gynoecium is almost oval in outline. It shows a bicarpellary ovary with two locules. Each locule contains one ovule in each chamber. The ovary is hypogynous (Fig. 9). The main difference between J. adhatoda and J. beddomei is in the T.S of gynoecium, i.e., the presence of abundant unicellular to multicellular trichomes in the outer epidermis of J. adhatoda [31] which is absent in J. beddomei. The major difference in macroscopy and microscopic features of J. beddomei and J. adhatoda is illustrated in a tabular form (Table 4).
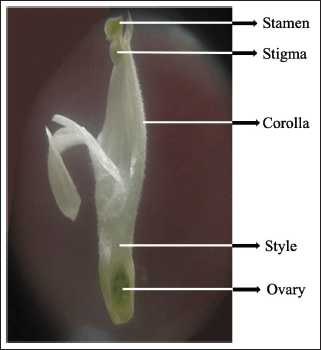 | Figure 8. Longitudinal section of J. beddomei flower. [Click here to view] |
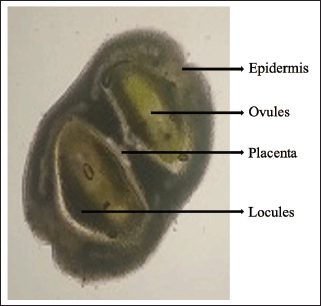 | Figure 9. Transverse section of J. beddomei flower gynoecium. [Click here to view] |
Powder microscopy
Powder microscopy of the whole plant of J. beddomei powder shows spiral vessels, vessels with reticulated thickening, pitted vessels, a fragment of lignified and nonlignified fiber, a large number of diacytic stomata, and few crystals. Uniseriate, covering, straight trichomes and four-celled glandular trichomes are also found along with fragments of the mesophyll layer. Colored contents, fragments of pitted parenchyma, sclerids, and stone cells are also found (Fig. 10).
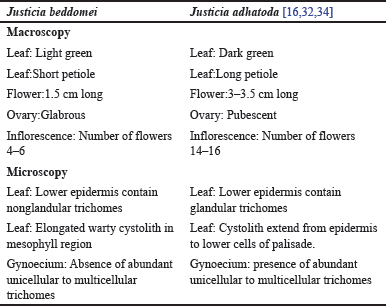 | Table 4. Difference between macroscopy and microscopic features of J. beddomei and J. adhatoda. [Click here to view] |
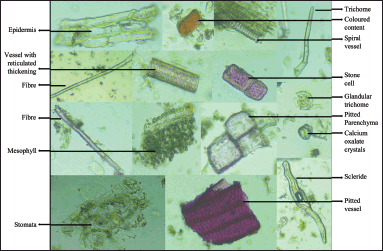 | Figure 10. Powder microscopy of J. beddomei whole plant. [Click here to view] |
Powder microscopy of J. beddomei and J. adhatoda showed prismatic calcium oxalate crystals [17]. Different epidermal trichomes exist in both plants [34]. In J. beddomei, only glandular trichomes are observed, whereas, in J. adathoda, both glandular and nonglandular trichomes are visible. Justicia beddomei is the sole species with the colored content.
CONCLUSION
This study revealed the identification concerns of the plant J. beddomei (C.B. Clarke) Bennet with the morphologically similar plant J. Adathoda. The sequence analysis and NCBI Gen-Bank BLASTn database results after the amplification of the trnH–psbA barcode region of the plant facilitated the identification of this species. The distinguished morphological and microscopical features of J. beddomei (C.B. Clarke) Bennet are the four number of flowers in inflorescence, 1.5 cm flower length, and presence of cystolith in the mesophyll region of leaf, absence of trichomes in gynoecium, presence of glandular trichomes only in power microscopy, respectively. These features will help to find the adulteration in the selected plant studies and ensure the purity of the selected drug. The powder microscopic analysis helps to minimize the adulteration of drugs in powdered form. This article will be helpful for the preparation of J. beddomei (C.B. Clarke) Bennet monograph and pharmacopoeial standards. The distinguished features in microscopy of leaf, stem, and flower gave relevant information for the identification of the plant from J. Adathoda L. According to our knowledge, this is the first thorough analysis of molecular identification using trnH-psbA DNA barcoding and thorough macroscopic, microscopic, and powder microscopic features of J. beddomei (C.B. Clarke) Bennet parts.
ACKNOWLEDGMENT
The authors would like to thank CMPR, Arya Vaidya Sala, Kottakkal, Kerala, India, for providing the necessary facilities to carry out the DNA barcoding analysis.
AUTHOR CONTRIBUTIONS
SP and NR conceptualized and designed the research work, NR significantly contributed to the practical part and initial drafting of the manuscript; SP revised the article examining it carefully for significant intellectual content; consenting to submit to the current journal; SP gave the version’s final approval. All the authors are eligible to be authors as per the International Committee of Medical Journal Editors (ICMJEs) requirements/guidelines
FINANCIAL SUPPORT
There is no funding to report.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ETHICAL APPROVAL
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Khan S, Mirza KJ, Zainulabdin M. Authentication of the medicinal plant Sennaangustifolia by RAPD profiling. Saudi J Biol Sci. 2011;18(3):287–92. CrossRef
2. Rastogi S, Pandey MK, Prakash J, Sharma A, Singh GN. Veterinary herbal medicines in India. Pharmacogn Rev. 2015;9(18):155–63. CrossRef
3. Grover JK, Yadav S, Vats V. Medicinal plants of India with anti-diabetic poential. J Ethnopharmacol. 2002;81(1):081–100. CrossRef
4. Bahuguna Y, Zaidi S, Kumar N, Rawat K. Standardization of polyherbal marketed formulation Triphala Churna. Research and reviews. J Pharmacogn Phytochem. 2014;2(3):028–35.
5. Santhosh Kumar JU, Krishna V, Seethapathy GS, Ganesan R, Ravikanth G, Shaanker RU. Assessment of adulteration in raw herbal trade of important medicinal plants of India using DNA barcoding. 3 Biotech. 2018 Mar;8(3):135. CrossRef
6. Tilney PM, Wyk VBE. The value of antomy in pharmacognosy and forensic studies. South Afr J Bot. 2010;76(2):404. CrossRef
7. Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270(1512):313–21. CrossRef
8. De Mattia F, Bruni I, Galimberti A, Cattaneo F, Casiraghi M, Labra M. A comparative study of different DNA barcoding markers for the identification of some members of Lamiacaea. Food Res Int. 2011;44(3):693–702. CrossRef
9. Amin S, Ghosh S, Biswas B, Arifuzzaman M, Azad M, Siddiki A. Molecular identification of four medicinal plants using DNA barcoding approach from Chittagong, Bangladesh. J Adv Biotechnol Exp Ther. 2020;3(3):268. CrossRef
10. Hou D, Song J, Shi L, Ma X, Xin T, Han J, et al. Stability and accuracy assessment of identification of traditional Chinese materia medica using DNA barcoding: a case study on Flos Lonicerae Japonicae. Biomed Res Int. 2013;2013:549037. CrossRef
11. Handa SS. Indian efforts for quality control and standardization of herbal drugs/products. Proceedings of the 1st joint workshop on quality control and standardization of traditional medicine—Indo-China experience. Lucknow, India: National Botanical Research Institute; 2004. vol. 8, pp 008–010.
12. Goverment of India Ministry of Ayush. Pharmacopoeia commission for Indian medicine and homeopathy. Ghaziabad, India: Goverment of India Ministry of Ayush; 2004.
13. Warrier PK, Nambiar VP, Ramankutty C. Indian medicinal plants. Chennai, India: Orient Longman Ltd.; 1993. vol. 268, 272–3 pp. CrossRef
14. Prabhukumar KM, Beegam F, George S, Balachandran I. Revisiting the taxonomy of Justicia beddomei (Acanthaceae). Phytotaxa. 2018;350(1):71. CrossRef
15. Karthikayan S, Murthy S, Sanjappa SM. Flowering plants of India, dicotyledons (Acanthaceae-Avicenniaceae) 1. Kolkata, India: Botanical Survey of India; 2009. 166–4 pp.
16. Prabhukumar KM, Robi AJ, Hareesh VS, Balachandran I. Justicia gambleana (Acanthaceae): a new species from Kerala, India. Kew Bull [Internet]. 2016;71(3):39. CrossRef
17. Nandhini S, Ilango K. Comparative study on pharmacognostical, phytochemical investigations and quantification of vasicine content in the extracts of Adhatoda vasica Nees and Adhatoda beddomei CB Clarke. Pharmacogn J. 2020;12(4):884–96. CrossRef
18. Karthikayan S, Murthy S, Sanjappa SM. Flowering plants of India, Dicotyledons (Acanthaceae-Avicenniaceae). Kolkata, India: Botanical Survey of India; 2009. 366 p.
19. Sasidharan N. Biodiversity documentation for Kerala 6. Flowering plants. Thrissur, India: Kerala Forest Research Institute; 2004.
20. SunilKumar KN, Ramya A. A review on critically endangered species of Acanthacea: Justicia beddomei (Clarke) Bennet. An immune booster. J Med Plants. 2020;8:94–8. CrossRef
21. Vijayakumar AS, Jeyaraj M, Kolanchinathan P, Selvakumar M, Praveenkumar D. Molecular identification of the medicinal plant Justicia gendarussa (burm) f. Using ITS gene. Life Sci Inform Publ. 2019;5:663–74. CrossRef
22. Sang T, Crawford D, Stuessy T. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am J Bot. 1997;84(8):1120. CrossRef
23. Tate JA, Simpson BB. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst Bot. 2003;28:723–37. CrossRef
24. Neerakkal I, Rajagopal R, Vijayakumar KR. Influence of inanimate shade on growth of rooted Adalodakam (Adhatoda beddomei C.B.Clarke) cuttings. J Trop Agric. 2005;43:91–3.
25. Haleshi C, Sringeswara AN, Danapur V. Pharmacognostic study of Justicia beddomei (C. B. Clarke) Bennet. Eur J Med Plants. 2020;31(9):83–8. CrossRef
26. Pathak MR, Mohamed AA, Farooq M. DNA barcoding and identification of medicinal plants in the Kingdom of Bahrain. Am J Plant Sci. 2018;9(13):2757–74. CrossRef
27. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8. CrossRef
28. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. CrossRef
29. Ross HA, Murugan S, Li WL. Testing the reliability of genetic methods of species identification via simulation. Syst Biol. 2008;57(2):216–30. CrossRef
30. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. CrossRef
31. Government of India 2, Ministry of Health and Family Welfare. New Delhi, India: Controller of Publications; 1996.
32. Oshi A. Pharmacognostical study of Justicia adhatoda Linn. Int J Herb Mede. 2014;1:14. Available from: https://api.semanticscholar.org/CorpusID:42510039
33. Reddy MP, Shantha TR, Rao VR, Venkateshwarlu G. Pharmacognostical and physicochemical evaluation on the flowers of Justicia adhatoda L. Res J Pharmacogn Phytochem. 2015;7(2):73. CrossRef
34. Datta Kalme RK. Pharmacognostic characterization of stem and root of Adhatoda zeylanica Medicus. J Pharm Sci Res. 2012;3(11):4264–9. CrossRef