INTRODUCTION
The increase in diseases brought on by bacteria resistant to antibiotics (AMR) is a significant public health concern in many parts of the world. According to the World Health Organization (WHO), Pseudomonas, different Enterobacteriaceae (including Klebsiella and Escherichia coli), and Acinetobacter are the most dangerous carbapenem-resistant bacteria for human health and the healthcare system [1]. Several epidemiological maps for these bacteria are available. Examples include (vancomycin-resistant) Enterococcus faecium, which is a serious problem in Western and Northern Europe [2], and Acinetobacter baumannii, which is common in the Mediterranean region and the Arabian Peninsula [3]. AMR has a significant detrimental effect on development, animal welfare, and food security, resulting in 1.27 million illnesses and 929,000 fatalities annually worldwide [4].
There has been a notable increase in AMR observed in Jordan [5]. In the course of the year 2019, Jordan documented a cumulative count of 625 fatalities that were explicitly linked to AMR. Furthermore, an additional 2,400 deaths were identified as having a discernible association with AMR [6]. These figures position AMR as the fourth leading cause of death when compared to other causes in Jordan. Infections with methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant coagulase-negative staphylococci are widespread, while the resistance rates in Clostridioides difficile and enterococcal infections are moderate [7]. Moreover, S. pneumoniae and E. coli in medical isolates have shown high rates of antibiotic resistance [8]. In accordance with the global recommendations provided by the WHO and the United Nations (UN), a Jordanian initiative was developed to tackle the problem of AMR [5]. The program serves as a manifestation of the political dedication at the national level in Jordan to combat the issue of AMR, a decrease in the use of antimicrobials, and a concurrent enhancement in the general population’s awareness of appropriate antimicrobial consumption. In keeping with the pledge of this plan, effective surveillance to bolster information and proof is crucial.
The profusion of AMR data in medical health records has generated a significant resource that supports policies and research centered on the gathering, analysis, and dissemination of data pertinent to trends in resistance microorganisms. While most current research has focused on one or a small number of disease types or bacterial causative agents, it has also ignored a number of intricate details related to the AMR issue, such as the existence of multidrug resistant profiles (MDR) or factors that influence AMR more than others. This implies that a lack of interest or barriers exist for the analysis of patient records. Most likely, the first justification is false in that studying AMR-varied datasets is becoming more and more important to achieve the goals of AMR combating plans. Without a doubt, reviewing medical records is a laborious and time-consuming endeavor. However, there has been a growing emphasis on the potential to produce reliable forecasts regarding the various features of AMR by combining obtained data on patients with artificial intelligence aid tools like machine learning (ML) [9].
The main goal of ML is to develop algorithms that can build prediction models with little to no human involvement from a training dataset. According to recent research, a variety of bacterial strains can be predicted to exhibit AMR using ML approaches (reviewed in [10]). Furthermore, rather than using conventional research methods, ML might be used to identify the various mechanisms causing antibiotic resistance [11]. However, there are currently just a few studies looking into the use of ML to forecast AMR; in Jordan, for example, there have been no recent studies looking into the use of ML to combat AMR.
It is possible to better understand the major factors that affect the spread of AMR by including socio-demographic factors, such as age and sex, in the ML models. Age and sex have various epidemiological and etiological effects that are equivalent to the selection pressure exerted by antibiotics on the most common clinical microorganisms. This is primarily because bacterial pathogens interact with hosts with varying degrees of resistance to infection. For example, natural immunity to infection decreases with age, and immunological phenotypes differ according to sex [12]. Theoretically, this variation affects the evolution of pathogenic bacteria [13]. The effects of age and sex on the prevalence and severity of AMR have not been thoroughly studied. Understanding the distribution of antibiotic susceptibility according to age and sex in specific populations could assist in the development of more effective treatment regimens.
AMR is a prevalent issue in Jordan. There are still several unexplored regions in the entire country. As no research has been done in Al-Salt (the center Western region) to date, the primary goal of this study is to determine the prevalence of antimicrobial resistance (AMR), the kind of bacteria that demonstrates profound resistance, and the common infection of a significant Jordanian population in Al-Salt city. Second, our goal is to predict the AMR for various antibiotic categories by using two ML models, namely random forests (RFs) and categorization regression trees (CARTs), on microbiology reports. Our method is to assess the two models’ respective performances. Furthermore, we have examined the significance of patient age and sex as AMR risk factors. These variables function as surrogate markers of the selection pressure that antibiotics exert on the most common bacterial infections, which might result in a variety of diseases and epidemiological ramifications.
METHODS
Data collection and justification
In this study, we conducted a retrospective analysis of AMR data for the period (2020–2022) obtained from Al-Hussein/Salt Hospital, the sole healthcare facility serving the Al-Salt region in Jordan. Hence, the statistics pertain to the total population of 180,090 individuals residing within the city, with 51% being male and 49% being female.
Antibiotic susceptibility data were obtained by cultivating the bacteria for identification in a clinical laboratory and testing them for commonly used antibiotics using the VITEK 2 system. This procedure can take up to 72 hours or more. This information is required to treat bacterial infections. The minimum inhibitory concentration of an antibiotic needed to halt growth or kill a pathogen in the laboratory is usually provided to doctors with cut-offs for medication susceptibility as resistant or susceptible. Data are currently allocated to the Jordanian electronic health record program (Hakeem) which manages and reports health data in Jordan.
The data points obtained in this study (n = 2893) encompass positive bacterial culture results and antibiotic resistance profiles. These data points were collected in a manner that ensured the representation of all patients in a consecutive manner, without any specific group being targeted for sampling. The dataset comprised various attributes, including sex (categorized as female or male), age (measured in years), organism quantity (categorized in an ordered manner), diagnosis, Gram staining (classified as positive or negative), antimicrobial substances, date of sample collection, bacteria (identified by species), source of the clinical sample, and antibiotic susceptibility results (classified as sensitive or resistant). In the intermediate group, a total of 39 observations were recorded for the antibiotics utilized in the investigation. The hospital’s established guidelines consider these intermediate as susceptible. Nevertheless, the analysis employed resistant (positive) cases as a more prudent methodology.
A total of 27 distinct sources were utilized to acquire the clinical samples in the original dataset. The aforementioned action resulted in a substantial proliferation of classes inside the analysis. The classes in question generated a larger number of cases and resulted in a greater degree of data imbalance, as indicated by their sample size. To address the issue at hand, we have consolidated “similar sources” into a single class with the aim of minimizing the number of classes. As an illustration, the anatomical features of fingers, hands, and arms are categorized collectively as a single class, referred to as “arms.”
Prediction algorithms
Multiple experiments were conducted utilizing multi-label classification techniques to tackle the challenge of predicting AMR. The aim was to assess the susceptibility of an individual patient, who had acquired a specific bacterial strain, to the simultaneous administration of multiple antibiotics. Our decision has been made to implement a conservative approach. Initially, a resampling procedure was implemented on the original dataset to focus on investigating the frequently tested drugs. This was done while ensuring that no data points were missing, particularly those about patients’ reports with incomplete or unreported AMR results for the antibiotics under consideration. Following the procedure of resampling, the prediction algorithms applied on an array including 489 observations provide a comprehensive analysis of the outcomes, disregarding any instances of missing data. By implementing this approach, we have effectively secured the reliability and accuracy of our predictions. Antibiotics that are under evaluation include trimethoprim/sulfamethoxazole (TMP/SMX), nitrofurantoin, fosfomycin, ciprofloxacin, gentamicin, amikacin, meropenem, and imipenem. Antibiotics were classified as target labels, also referred to as dependent variables, yet the remaining features in the dataset (as discussed in the previous section) were considered independent variables.
The dataset has been divided into two subsets, namely the training set and the testing set, with proportions of 80% and 20%, respectively. This approach allows for the training of models on a designated subset of the data, while evaluating their performance on separate and unseen samples.
To address our multi-label classification robustness, we have opted to employ two separate classifiers, namely RF [14] and decision trees (CARTs) [15]. This study focuses on the development of multi-output classifiers to effectively tackle the task of antibiotic labeling. The classification of each antibiotic label is approached as an independent binary problem. Both multi-output classifiers are trained using the available training data. The procedure involves gaining an understanding of specific models for each antibiotic label to predict either resistance (1) or susceptibility (0). Following this, the test data was employed to evaluate the efficacy of the trained models.
RESULTS
Features of the patients and the isolated bacteria
There were 2,893 antimicrobial susceptibility tests reported. Table 1 describes the patient characteristics and microorganisms recovered and shows the data points from the various characteristics that were investigated. Of the cases reported, 2,091 (72.2%) were female. The patient’s ages ranged from less than one week to 102 years, with a median of 35 years (standard deviation: 31.2 years). The mean age in male and female groups is almost identical (mean age in males = 36.6 ± 25.4 and females = 36.3 ± 25.1). The calculated p-value for the observed difference between the two means was determined to be 0.94. This finding suggests a resemblance between the two cohorts. Figure 1a shows the AMR distribution according to patient age. Notably, patients with gram-negative infections caused by opportunistic pathogens, such as Enterococcus spp., A. baumannii, and Citrobacter freundii (often acquired in hospitals), had a higher mean age. Escherichia coli was the most prevalent bacterium found in clinical samples (n = 1,454, 50.3%), followed closely by Klebsiella pneumoniae (n = 362, 12.5%), S. aureus (n = 283, 9.8%), and Streptococcus infections, including Streptococcus agalactiae, S. pneumoniae, and Streptococcus viridans, which together accounted for 6.0% of cases. S. agalactiae is the most common streptococcal species. Enterobacter species, including E. faecium, A. baumannii, and P. aeruginosa were frequently isolated pathogens and primary contributors to AMR. The infection rate of each bacterial species differed according to sex (Fig. 1b).
Clinical diagnoses were heterogeneous. Urinary tract infections (UTIs) were present in 1,614 patients (50.8%), followed by gastro intestinal infections (17.9%) and respiratory infections (6.6%), including pneumonia, pharyngitis, and upper and lower respiratory infections. Sepsis and other bloodstream infections accounted for 7.8% of cases, while the “Other” categories included screening swabs of wounds, surgical site infections, otitis media, and otitis externa. Resistance to antibiotics was prevalent in Gram-negative bacteria (n = 2400, 83%). Antibiotics with high resistance rates are listed in Table 2.
ML predictions of antibiotic resistance
The prediction testing outcomes for each drug derived using the CART and RF algorithms are displayed in Table 2. Based on the outcomes of the prediction testing, it is evident that the RF algorithm exhibited superior performance compared to the CART algorithm.
The study patients’ (n = 489) characteristics are shown in Table 3. The patients ranged in age from under a week to 106 years. A total of 260 patients had ages greater than the sample mean, which came out to be 35.99 years old. Seventy-seven percent (n = 381) of the individuals that were affected were female. UTI was the most frequently observed diagnosis among these individuals, accounting for 68.7% (n = 336) of the cases. The bacteria that have been tested are E. coli, K. pneumoniae, Proteus mirabilis, and other species of Enterobacter. Escherichia coli bacteria were detected in a total of 401 individuals, accounting for 82% of the patient group.
Figure 2 depicts the correlation between the AMR for antibiotics that underwent testing and the clinical data, encompassing diagnosis, bacterial species, quantity, and demographic data, notably sex and age. The type of bacteria and the patient’s age are significant variables in predicting an AMR. Sex has been found to be a notable determinant of AMR for ciprofloxacin, gentamicin, and carbapenem antibiotics. The used model demonstrated a high level of efficacy in accurately forecasting the multidrug resistance (MDR) profile of the bacterial isolates that underwent testing, with a particular focus on E. coli and K. pneumoniae (Table 2).
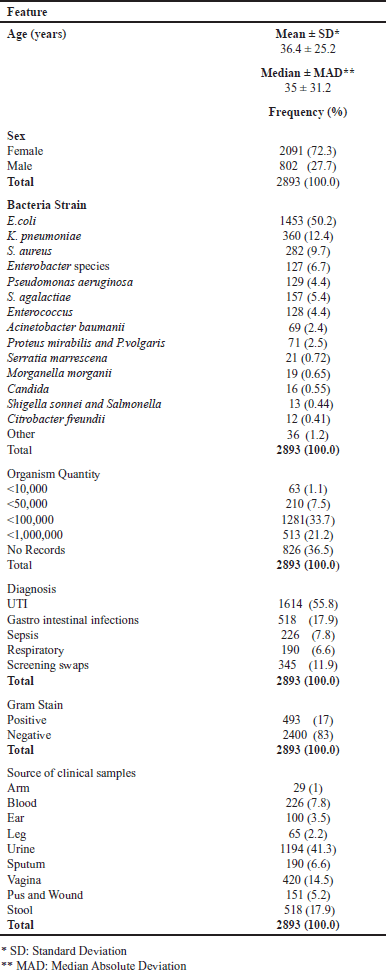 | Table 1. Summary statistics of the dataset. [Click here to view] |
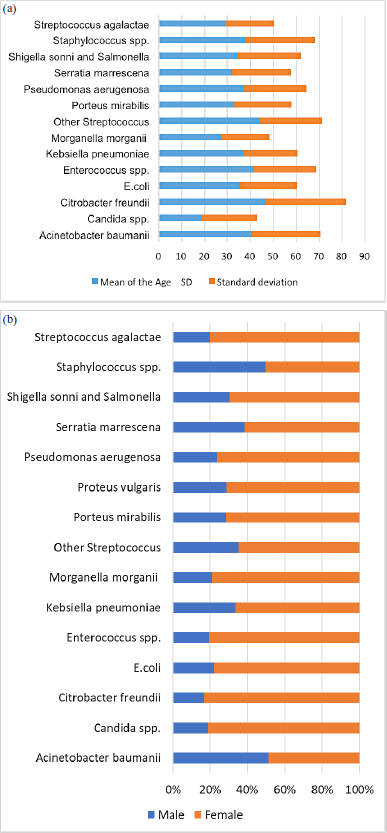 | Figure 1. (a) Age distribution of study patients according to infecting bacterial species. (b) Distribution of bacterial species according to the patients’ sex (n = 2893). [Click here to view] |
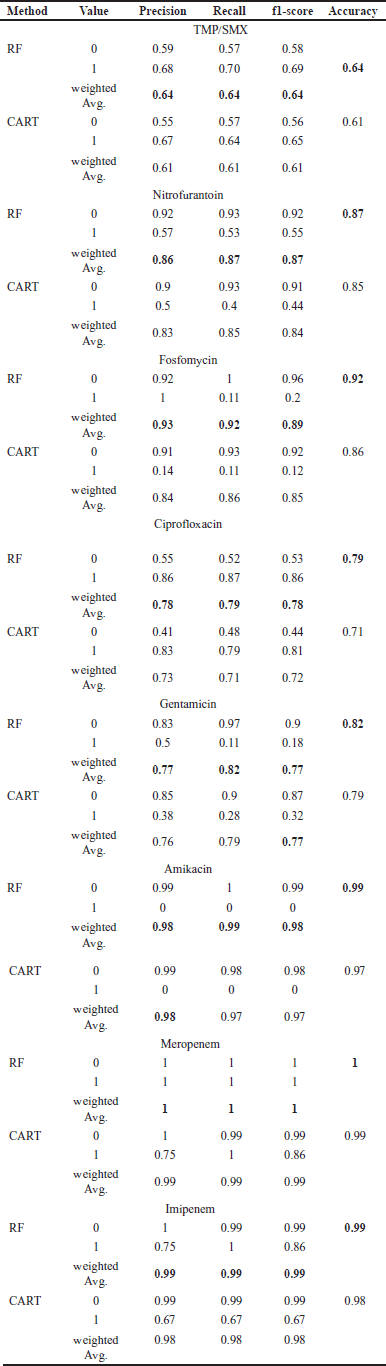 | Table 2. The prediction results for each antibiotic are based on CART and RF algorithms. [Click here to view] |
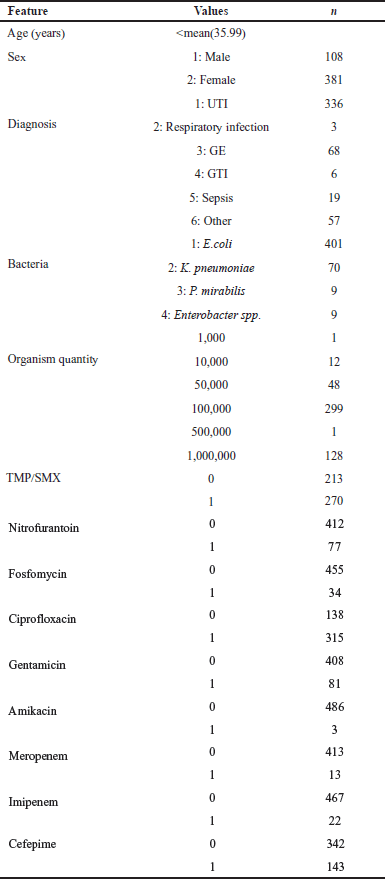 | Table 3. Summary statistics of the resampled dataset utilized in ML algorithms to predict AMR. [Click here to view] |
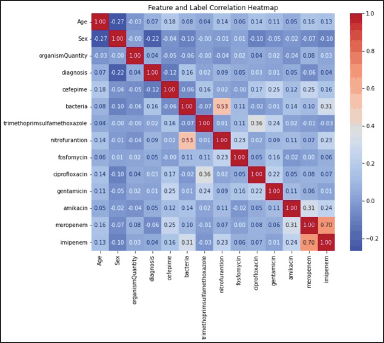 | Figure 2. A heatmap representation as determined by RF illustrates clinical and demographic features associated with AMR to various classes of antibiotics. The dataset used in this analysis was obtained from Al-Hussein/Salt Hospital and comprised 489 observations. [Click here to view] |
DISCUSSION
The present study provides evidence that AMR is a prevalent issue in an important population in Jordan, hence highlighting it as a public health concern. The results enabled us to evaluate the usefulness of commonly given antibiotics for the treatment of infection as well as the occurrence of AMR bacteria among different age and sex groups. The aforementioned outcomes are of great significance in informing the public health initiative on the appropriate administration of antibiotic medication.
Bacterial species were a major determinant of AMR patterns. Different antibiotic exposures and biological variables (such as the outer membrane of Gram-negative bacteria) may account for the variability in AMR frequencies of diverse bacterial species. The majority of Gram-negative bacteria that cause AMR infections, especially in female patients, are E. coli, followed by K. pneumoniae. In addition, women seemingly experienced infections at a younger age than men did, probably due to the frequent association between E. coli-K. pneumoniae and female genital infection [16]. The risk of UTIs significantly increases with age [17], which may explain why our analysis revealed age as a predictor of AMR development, particularly for antibiotics used to treat UTIs.
The majority of the identified bacteria were Enterobacteriaceae, which typically demonstrate intrinsic resistance to several classes of antibiotics, caused by the production of β-lactamases, and due to acquiring fluoroquinolone, carbapenems, and aminoglycoside antimicrobial-resistance genes [8]. These bacterial species are usually called “high-risk clones” owing to their multidrug-resistant (MDR) characteristics. The misuse of antibiotics, including their use as preventive measures, may lead to a gradual increase in AMR rates in this category of bacteria [18].
Infections caused by S. aureus and Streptococcus, namely S. agalactiae, have been identified as significant contributors to AMR. These bacterial strains are particularly prevalent within the Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species group reported in this study, which encompasses Enterobacter species, E. faecium, A. baumannii, and P. aeruginosa, making them the most often seen bacteria in this group. In addition, both pathogens are a major cause of nosocomial infections. The findings of our study are consistent with previous research conducted in Jordan, which has identified elevated rates of antibiotic resistance in Gram-positive bacterial infections, particularly MRSA and Coagulase-negative Staphylococci [7]. Factors that contribute to the escalation of these infections include advancing age, prolonged hospital stays, excessive or inappropriate administration of broad-spectrum antibiotics, and an increased prevalence of invasive medical devices and procedures [19].
The results of this study demonstrated that the prediction of AMR can be effectively achieved using medical records, even when the available patient data is restricted. Other studies lacked varied datasets that included patients of both sexes of different ages with different illnesses, bacterial species, and antibiotic treatments. These variables greatly hinder the development of predictive models. We developed a prediction system that produced accurate outcomes despite the heterogeneity of the data. However, a larger sample size would have provided greater statistical power for the prediction analysis. We choose to utilize two distinct classifiers: The RF and CART. This choice was primarily motivated by their demonstrated efficacy in classifying medical data, as evidenced by previous studies [20]. Because the dataset was imbalanced, we have resampled the data while focusing on the antibiotics, which are frequently tested and encompassing different classes. In the resampled dataset, the key exploratory variable used to predict AMR in our models was E coli bacterial species. However, this allowed the prediction model to have better statistical power.
As can be seen from the prediction results, the RF performed better than the CART. This is expected because RF is an ensemble method that combines multiple decision trees to improve predictive accuracy and reduce overfitting, and here we used the default 100 trees for the ensembles, while CART represents a single decision tree classifier. It is worth noting that some of the antibiotic classes, such as amikacin, meropenem, and imipenem, are extremely class-imbalanced. However, solving such a problem as recommended by [21,22] is out of the scope of this paper. In general, the results indicate a strong capability for ML methods to predict AMR for multiple antibiotics simultaneously.
Our study revealed accuracy values of 0.64–0.99 based on the application of the RF classifier, which is even higher than values reported in studies focused on single bacterial species, such as MRSA [23], vancomycin-resistant Enterococcus, fluoroquinolone-resistant Gram-negative bacteria [24], and carbapenem-resistant Enterobacteriaceae [25]. However, the results of previous studies involving heterogeneous data and several types of bacteria were comparable with our findings [26]. The applied model simultaneously evaluated MDR in relation to several drugs in the bacterial isolates that underwent testing. In comparison to conventional statistical calculation methods such as the MDR index proposed by Krumperman (1983) [27], our approach provides a comprehensive depiction of the MDR profile without the need for significant human intervention.
In the present study, the degree of resistance to TMP/SMX is substantial. The prevalence of TMP/SMX resistance has been observed in medical isolates originating from Jordan [28]. Mainly, it has been observed that the prevalence of E. coli resistance to TMP/SMX in Jordan exceeds the stated rates observed internationally. Furthermore, our analysis revealed a significant association between sex and a heightened likelihood of resistance to TMP/SMX, which aligns with previous research findings [29].
The creation of carbapenem resistance is influenced by a range of characteristics, as indicated by the prediction analysis. These aspects encompass age, gender, and the utilization of diverse classes of antibiotics. The regions of the Arabian Peninsula and Jordan have been recognized as areas that possess a potential susceptibility to the genesis and dissemination of carbapenem resistance [30]. The high incidence of MDR strains, specifically in E. coli, imposes substantial limitations on the therapeutic options that are now accessible. The findings of this study are consistent with other studies, suggesting that Gram-negative bacteria that produce carbapenemase demonstrate resistance to a wide range of beta-lactam antibiotics, including cefepime [31]. Furthermore, it is common for these bacteria to possess genetic elements that provide resistance to fluoroquinolones and/or aminoglycosides, including amikacin [25]. Therefore, it is possible that well-established medications like fosfomycin could be considered as last-resort options, despite concerns over their effectiveness. The observed negative correlation between resistance to fosfomycin and the other antibiotics examined is of particular significance. However, the Jordanian Food and Drug Administration (JFDA) recommends the use of fosfomycin as part of an oral regimen for acute uncomplicated cystitis only. E. coli and K. pneumoniae had the highest prevalence of resistance in our dataset, supporting these recommendations. Numerous empirical trials have shown that fosfomycin exhibits favorable action against enteric gram-negative bacteria such as Enterobacter and K. pneumoniae [32]. However, the JFDA has not approved fosfomycin for the treatment of Klebsiella infections, adhering to European and US recommendations. The findings of this study align with previous research indicating a role for gender in carbapenems resistance [33].
The growing resistance exhibited by bacterial isolates towards gentamicin, cefepime, and ciprofloxacin in this cohort is a subject of apprehension due to its potential ramifications in restricting the array of available treatment choices like the treatment of infections caused by ESBL-producing bacteria. Moreover, these antibiotics have been identified as significant indicators of MDR profile in the utilized models. The prevalence of gentamicin resistance has been observed in medical isolates originating from Jordan [7]. The model predicts an increased likelihood of acquiring MDR to gentamicin, in conjunction with at least six other antibiotics. Gentamicin is commonly employed as a primary antibacterial agent in the treatment of UTIs. Ciprofloxacin is a commonly employed antibacterial drug that exhibits a wide range of action against many bacterial pathogens, making it a valuable therapeutic option for the treatment of numerous bacterial infections. However, ciprofloxacin among other fluoroquinolones has been documented as the most antibiotic commonly utilized for the treatment of UTIs without a prescription in Jordan [34]. The over utilization of antibiotics may result in an escalation of antibiotic resistance.
Patient age was a significant predictor of AMR in the model. Increased resistance to all antibiotics tested in our cohort may be anticipated with age, the importance of age may indicate that resistant microorganisms continue to affect those who are already vulnerable, such as older adults with various health issues, particularly those with poor physical and mental health who are vulnerable to AMR infections. However, the association between age and AMR remains poorly understood. Studies of the Jordanian population suggest that age plays a variable role in antibiotic use [35]. In addition, the AMR bacteria isolated in this study displayed notable variations with age. Some researchers have attributed this pattern to the mode of action of antibiotics [36]. Understanding the antibiotic susceptibility patterns according to age in a region or country can contribute to higher cure rates.
In this cohort, the sex variable significantly predicted resistance to TMP/SMX, ciprofloxacin, and carbapenems. The issue of sexual dimorphism in AMR has received little attention, and previous studies have not adequately demonstrated the direct effects of sex on antibiotic resistance in infectious settings. To explain this pattern, we first considered antibiotics for treating infections in men. We hypothesized that the extensive use of ciprofloxacin for the treatment of UTIs and chronic bacterial prostatitis in men would result in a greater rate of male resistance, which requires attention. UTIs were the main cause of infection in our study. According to previous studies, men with UTIs are more likely to develop ciprofloxacin resistance [37]. P. aeruginosa and S. aureus were the two ciprofloxacin-resistant bacteria found in this cohort. These species were responsible for most of the disparities in the frequency of AMR UTIs in males and females [16] because in older men, AMR- UTIs are associated with urinary tract catheters, resulting in more complicated UTIs [17,38]. Therefore, the prevalence of specific causative agents, choice of antibiotics, and AMR rates in pathogens that frequently cause UTIs among Jordanians require further research. Several triggers in women’s and men’s health should be considered because they might otherwise help increase the rates of bacterial resistance. Psychological and physiological factors are involved in this process. The strong evidence of sex bias in infectious disease severity is primarily caused by differences in immune responses according to sex, associated with several male-predominant infections [39]. Men possess immunological traits such as sex chromosomal complements, immune cell receptors, and sex steroids that make them more prone to severe infections [12]. From a social perspective, lifestyle habits such as smoking and drinking, which are more prevalent in men than in women, make men more susceptible to severe bacterial illnesses that are generally caused by higher AMR strains. According to the WHO, men in developing countries are less likely than women to use the healthcare system [40]. Traditionally, men must exhibit traits such as power and strength and conceal their illnesses to avoid being stigmatized by weakness. Therefore, men are more likely to self-medicate and disregard treatment guidelines than women are.
To the best of our knowledge, this is the first study to assess sex and age as a predictor of AMR in Jordan. However, most patients were female. Therefore, the differences in infectious complications between male and female patients must be considered under the same conditions, such as UTI. One significant drawback of this comparison is the lack of comprehensive surveillance to track the infection dynamics in Jordan. Most studies have focused only on a few types of infections. The absence of several diseases, bacterial species, and antibiotic types in most investigations in the field supports the validity of our model. These results highlight the need for surveillance to reverse the increased risk of AMR in male patients.
One limitation of our study was its observational nature, which prevented further causal inferences. The limited number of covariates in the model might have confounded the results. However, the adjusted models provided consistent results with good predictive ability. Including additional covariates such as illness type, site of infection, bacterial species, age, and sex can enhance our understanding of the pathways of bacterial resistance and potential interactions.
CONCLUSION
We attempted to answer the question of AMR by using two distinct ML classifiers: RF and CART with observational data. The advantage of our approach is that it employs cross-validation using an ML method to confirm our predictive models. Moreover, we implemented a reverse association using antibiotics as the dependent variable to overcome the challenge of data imbalance when resampling the data, to maintain the statistical power of the models as much as possible. These results provide a foundation for further research on the roles of age and sex in bacterial resistance. In general, the findings of our study indicate that antibiotic resistance poses a considerable challenge in the context of Jordan, with a particular emphasis on Gram-negative bacterial infections. Furthermore, our results imply that the success rate of specific medicines in treating infections may vary depending on the patient’s sex or age. This study emphasizes the significance of surveillance of antibiotic resistance patterns and the formulation of novel approaches to address the rise and dissemination of antibiotic-resistant microorganisms.
ACKNOWLEDGMENTS
The authors thank Jordan’s AL-Hussein/Salt Hospital for allowing access to medical records from the microbiology laboratory section.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
This research received no specific grants from any funding agency in the public, commercial, or not-for-profit sectors.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study was approved by the Scientific and Administrative Committee of Scientific Research at Al-Balqa University and Jordan’s Al-Hussein/Salt Hospital—Approval number/ID: 35/4/1205. This material is the original work of the authors and has not been published elsewhere.
DATA AVAILABILITY
The dataset is constantly updated and a subset can be made available upon request.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
SUPPLEMENTARY MATERIALS
The supplementary material can be accessed at the journal's website:[https://japsonline.com/admin/php/uploadss/4252_pdf.pdf]
REFERENCES
1. WHO global priority pathogens list of antibiotic-resistant bacteria—Combat AMR [Internet]. [cited 2023 Aug 24]. Available from: https://www.combatamr.org.au/news-events/who-global-priority-pathogens-list-of-antibiotic-resistant-bacteria
2. Ayobami O, Willrich N, Reuss A, Eckmanns T, Markwart R. The ongoing challenge of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis in Europe: an epidemiological analysis of bloodstream infections. Emerg Microbes Infect [Internet]. [cited 2023 Aug 23];9(1):1180–93. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7448851/
3. Ibrahim ME. Prevalence of Acinetobacter baumannii in Saudi Arabia: risk factors, antimicrobial resistance patterns and mechanisms of carbapenem resistance. Ann Clin Microbiol Antimicrob [Internet]. 2019 Jan 3 [cited 2023 Aug 24];18:1. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6317247/
4. Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Aguilar GR, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet [Internet]. 2022 Feb 12 [cited 2023 Feb 27];399(10325):629–55. Available from: https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(21)02724-0/fulltext
5. Jordan: National Action Plan for Combating Antimicrobial Resistance in the Hashemite Kingdom of Jordan [Internet]. [cited 2023 Oct 30]. Available from: https://www.who.int/publications/m/item/jordan-national-action-plan-for-combating-antimicrobial-resistance-in-the-hashemite-kingdom-of-jordan
6. Jordan_0.pdf [Internet]. [cited 2023 Nov 4]. Available from: https://www.healthdata.org/sites/default/files/files/Projects/GRAM/Jordan_0.pdf
7. Al-Tamimi M, Albalawi H, Isied W, Musallam A, Qazzaz F, Azab M, et al. Gram-positive bacterial infections and antibiotics resistance in Jordan: current status and future perspective. Jordan Med J [Internet]. 2022 Jul 31 [cited 2023 Jan 6];56(1). Available from: https://jjournals.ju.edu.jo/index.php/JMJ/article/view/219
8. Nairoukh YR, Mahafzah AM, Irshaid A, Shehabi AA. Molecular characterization of multidrug resistant uropathogenic E. Coli isolates from Jordanian Patients. Open Microbiol J [Internet]. 2018 Jan 31 [cited 2023 Jan 6];12:1–7. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5806174/
9. Alkhawaldeh I, Al-Jafari M, Abdelgalil M, Tarawneh A, Hassanat A. A machine learning approach for predicting bone metastases and its three-month prognostic risk factors in hepatocellular carcinoma patients using SEER data. Ann Oncol. 2023 Jun 27;34:140.
10. Farhat F, Athar MT, Ahmad S, Madsen DØ, Sohail SS. Antimicrobial resistance and machine learning: past, present, and future. Front Microbiol [Internet]. 2023 [cited 2024 Jan 20];14. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1179312
11. Baker M, Zhang X, Maciel-Guerra A, Dong Y, Wang W, Hu Y, et al. Machine learning and metagenomics reveal shared antimicrobial resistance profiles across multiple chicken farms and abattoirs in China. Nat Food [Internet]. 2023 Aug [cited 2024 Jan 21];4(8):707–20. Available from: https://www.nature.com/articles/s43016-023-00814-w
12. Schneider-Hohendorf T, Görlich D, Savola P, Kelkka T, Mustjoki S, Gross CC, et al. Sex bias in MHC I-associated shaping of the adaptive immune system. Proc Natl Acad Sci [Internet]. 2018 Feb 27 [cited 2023 Mar 2];115(9):2168–73. Available from: https://www.pnas.org/doi/10.1073/pnas.1716146115
13. Davies NG, Flasche S, Jit M, Atkins KE. Within-host dynamics shape antibiotic resistance in commensal bacteria. Nat Ecol Evol [Internet]. 2019 Mar [cited 2023 Mar 23];3(3):440–9. Available from: https://www.nature.com/articles/s41559-018-0786-x
14. Katuwal R, Suganthan PN, Zhang L. An ensemble of decision trees with random vector functional link networks for multi-class classification. Appl Soft Comput [Internet]. 2018 Sep 1 [cited 2023 Nov 4];70:1146–53. Available from: https://www.sciencedirect.com/science/article/pii/S1568494617305598
15. Steinberg D. CART: classi?cation and regression trees. In: The top ten algorithms in data mining. Chapman and Hall/CRC; 2009.
16. Fagan M, Lindbæk M, Grude N, Reiso H, Romøren M, Skaare D, et al. Antibiotic resistance patterns of bacteria causing urinary tract infections in the elderly living in nursing homes versus the elderly living at home: an observational study. BMC Geriatr [Internet]. 2015 Aug 4 [cited 2023 Mar 2];15(1):98. CrossRef
17. Mortazavi-Tabatabaei SAR, Ghaderkhani J, Nazari A, Sayehmiri K, Sayehmiri F, Pakzad I. Pattern of antibacterial resistance in urinary tract infections: a systematic review and meta-analysis. Int J Prev Med [Internet]. 2019 Oct 9 [cited 2023 Apr 19];10:169. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6826787/
18. Sahin K, Tekin A, Ozdas S, Akin D, Yapislar H, Dilek AR, et al. Evaluation of carbapenem resistance using phenotypic and genotypic techniques in Enterobacteriaceae isolates. Ann Clin Microbiol Antimicrob [Internet]. 2015 Oct 6 [cited 2023 Mar 2];14(1):44. CrossRef
19. Yelin I, Snitser O, Novich G, Katz R, Tal O, Parizade M, et al. Personal clinical history predicts antibiotic resistance of urinary tract infections. Nat Med [Internet]. 2019 Jul [cited 2023 Jan 10];25(7):1143–52. Available from: https://www.nature.com/articles/s41591-019-0503-6
20. Alkhawaldeh I, Al-Jafari M, Abdelgalil M, Tarawneh A, Hassanat A. A machine learning approach for predicting bone metastases and its three-month prognostic risk factors in hepatocellular carcinoma patients using SEER data. Ann Oncol. 2023 Jun 27;34:140.
21. Hassanat A, Altarawneh G, Alkhawaldeh IM, Alabdallat YJ, Atiya AF, Abujaber A, et al. The jeopardy of learning from over-sampled class-imbalanced medical datasets. In: 2023 IEEE Symposium on Computers and Communications (ISCC) [Internet]. 2023 [cited 2023 Oct 18]. pp. 1–7. Available from: https://ieeexplore.ieee.org/document/10218211
22. Hassanat AB, Tarawneh AS, Abed SS, Altarawneh GA, Alrashidi M, Alghamdi M. RDPVR: random data partitioning with voting rule for machine learning from class-imbalanced datasets. Electronics [Internet]. 2022 Jan [cited 2023 Nov 1];11(2):228. Available from: https://www.mdpi.com/2079-9292/11/2/228
23. Janjusevic A, Cirkovic I, Minic R, Stevanovic G, Soldatovic I, Mihaljevic B, et al. Predictors of vancomycin-resistant Enterococcus spp. Intestinal Carriage among High-Risk Patients in University Hospitals in Serbia. Antibiotics [Internet]. 2022 Sep [cited 2023 Jan 11];11(9):1228. Available from: https://www.mdpi.com/2079-6382/11/9/1228
24. Fernández M, Conde S, de la Torre J, Molina-Santiago C, Ramos JL, Duque E. Mechanisms of resistance to chloramphenicol in pseudomonas putida KT2440. Antimicrob Agents Chemother [Internet]. 2012 Feb [cited 2023 Mar 2];56(2):1001–9. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3264264/
25. Lin MY, Ray MJ, Rezny S, Runningdeer E, Weinstein RA, Trick WE, et al. Predicting carbapenem-resistant enterobacteriaceae carriage at the time of admission using a Statewide Hospital Discharge Database. Open Forum Infect Dis [Internet]. 2019 Dec 1 [cited 2023 Jan 11];6(12):ofz483. CrossRef
26. Silva A, Costa E, Freitas A, Almeida A. Revisiting the frequency and antimicrobial resistance patterns of bacteria implicated in community urinary tract infections. Antibiotics [Internet]. 2022 Jun 3 [cited 2023 Jan 10];11(6):768. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9220357/
27. Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983 Jul;46(1):165–70.
28. Abughosh Z, Alsadi M, Ayoub F, Alhakim M, Alnawaji T. Over the counter antibiotics? Effects on Escherichia Coli incidence and resistance in uncomplicated urinary tract infections in Jordan. The J Urol. 2016;195:e355.
29. Wesolek JL, Wu JY, Smalley CM, Wang L, Campbell MJ. Risk factors for trimethoprim and sulfamethoxazole-resistant Escherichia Coli in ED patients with urinary tract infections. Am J Emerg Med [Internet]. 2022 Jun 1 [cited 2023 Nov 4];56:178–82. Available from: https://www.sciencedirect.com/science/article/pii/S0735675722002108
30. Hays JP, Safain KS, Almogbel MS, Habib I, Khan MA. Extended spectrum- and carbapenemase-based β-lactam resistance in the Arabian Peninsula-a descriptive review of recent years. Antibiot Basel Switz. 2022 Oct 5;11(10):1354.
31. Jean SS, Harnod D, Hsueh PR. Global threat of carbapenem-resistant gram-negative bacteria. Front Cell Infect Microbiol [Internet]. 2022 Mar 15 [cited 2023 Nov 4];12:823684. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8965008/
32. Anastasia A, Bonura S, Rubino R, Giammanco GM, Miccichè I, Di Pace MR, et al. The use of intravenous fosfomycin in clinical practice: a 5-year retrospective study in a Tertiary Hospital in Italy. Antibiotics [Internet]. 2023 May 27 [cited 2023 Nov 4];12(6):971. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10295113/
33. Said D, Willrich N, Ayobami O, Noll I, Eckmanns T, Markwart R. The epidemiology of carbapenem resistance in Acinetobacter baumannii complex in Germany (2014–2018): an analysis of data from the national Antimicrobial Resistance Surveillance system. Antimicrob Resist Infect Control [Internet]. 2021 Mar 1 [cited 2023 Mar 2];10:45. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7923473/
34. Almaaytah A, Mukattash TL, Hajaj J. Dispensing of non-prescribed antibiotics in Jordan. Patient Prefer Adherence [Internet]. 2015 Sep 30 [cited 2023 Jan 10];9:1389–95. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4599149/
35. Abdelmalek S, AlEjielat R, Rayyan WA, Qinna N, Darwish D. Changes in public knowledge and perceptions about antibiotic use and resistance in Jordan: a cross-sectional eight-year comparative study. BMC Public Health [Internet]. 2021 Apr 19 [cited 2023 Jan 7];21(1):750. CrossRef
36. Garcia A, Delorme T, Nasr P. Patient age as a factor of antibiotic resistance in methicillin-resistant Staphylococcus aureus. J Med Microbiol [Internet]. 2017 [cited 2023 Mar 2];66(12):1782–9. Available from: https://www.microbiologyresearch.org/content/journal/jmm/10.1099/jmm.0.000635
37. Bashir MF, Qazi JI, Ahmad N, Riaz S. Diversity of urinary tract pathogens and drug resistant isolates of Escherichia coli in different age and gender Groups of Pakistanis. Trop J Pharm Res [Internet]. 2008 Sep 11 [cited 2023 Jan 6];7(3):1025–31. Available from: https://www.ajol.info/index.php/tjpr/article/view/14687
38. McGregor JC, Elman MR, Bearden DT, Smith DH. Sex- and age-specific trends in antibiotic resistance patterns of Escherichia coli urinary isolates from outpatients. BMC Fam Pract [Internet]. 2013 Feb 22 [cited 2023 Nov 4];14:25. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3610120/
39. Jacobsen H, Klein SL. Sex differences in immunity to viral infections. Front Immunol [Internet]. 2021 Aug 31 [cited 2023 Jan 5];12:720952. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8438138/
40. Uneven access to health services drives life expectancy gaps: WHO [Internet]. [cited 2023 Mar 2]. Available from: https://www.who.int/news/item/04-04-2019-uneven-access-to-health-services-drives-life-expectancy-gaps-who