INTRODUCTION
The protein-coding genes have been extensively investigated and are considered the most thoroughly examined components of the genome and transcriptome. However, it is essential to note that they constitute less than 2% of the genome. Merely comprehending the roles and activities of messenger RNAs (mRNAs) is insufficient to provide a comprehensive picture of the cellular machinery. To have a more thorough understanding of the intricate control of biological processes, it is imperative to investigate the substantial portion of the genome and transcriptome that remains unexplored and understudied. These constituents are responsible for the modulation of gene expression. Noncoding RNAs (ncRNAs) represent a significant constituent inside biological systems. They are categorized into many classes based on their dimensions, configurations, and purposes.
RNAs can carry out a variety of tasks, including catalyzing enzymatic activities (e.g., ribozymes), regulating protein synthesis (e.g., miRNAs), increasing protein production [e.g., long noncoding RNA (lncRNAs)], altering the protein that is produced by an mRNA (e.g., riboswitches), and many more [1]. Vast amounts of data are now easily accessible thanks to the development of contemporary sequencing technologies and the accompanying drop in sequencing cost [2]. This enables us to have a comprehensive understanding of the functioning of the cellular machinery and the emergence of disease, which in turn pave the way to finding a cure for these diseases.
Loss of balance in these relationships can lead to many disorders, including cancers, cardiovascular disorders, and neurological diseases [3,4]. This review discusses several of these disorders and the RNAs involved in them. These relationships can potentially be utilized as therapeutic targets to help combat these illnesses [3]. Ever-increasing rates of cancer, cardiovascular diseases, and relatively weakly addressed neurological disorders are the prime focus of this review. This review not only highlights the different RNA and their roles, but also brings to the attention of the scientific community the interplay between them and their further cascading implications. This kind of cohesive, comprehensive, and informative documentation of RNA involving the aforementioned diseases is not available thus far, and therefore, it will significantly help scientists in the future to identify markers [5].
ROLES OF miRNA, IncRNA, and mRNA IN GENE EXPRESSION AND THEIR BIOGENESIS
Role in gene expression
Regulation of gene expression has a much more significant impact on organism complexity than gene count or genome size. The control of gene expression plays a critical role in determining a cell’s identity [6], functions [7], interactions [8], and ultimately, its fate [9].
Different facets of gene expression are regulated by interactions among mRNA, lncRNAs, and miRNAs. mRNAs directly affect the proteome [10], the physicochemical characteristics of the cell, and the protein-coding process. The miRNAs act to quiet the target mRNA and stop it from being translated into proteins [11]. They can accomplish this by either signaling for the target mRNA’s destruction or by impeding its translation [12]. This is influenced by the miRNA’s target (often the 3′ untranslated region (UTR) of mRNA) and how similar their sequences are.
On the other hand, lncRNAs bind to miRNAs, preventing them from either starting gene transcription or silencing mRNAs, both of which result in the expression of proteins [13]. Figure 2 provides a general understanding of these connections.
MicroRNAs prevent mRNA translation. As a result, miRNA target proteins are downregulated. mRNA’s 3’ UTR (UTR) is where miRNAs find their targets. The miRNA-RISC complex breaks down the mRNA if the target is complementary. The complex prevents the translation of the mRNA if the sequence complementarity is lower [14].
Biogenesis of miRNA, lncRNA, and mRNA
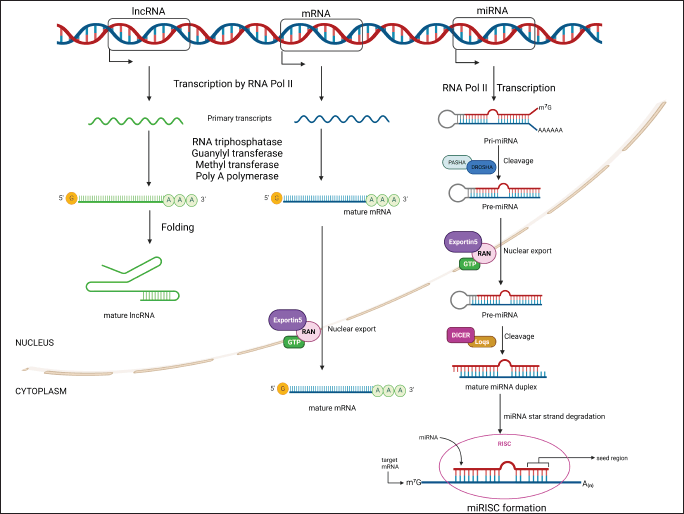
A basic overview is given in Figure 1 below.
Biogenesis of miRNA
miRNA can be produced from specific miRNA genes (in the intergenic area) or as an intragenic component of other gene transcripts (known as mirtrons) [12], employing either their own promoters or those of the host gene [15]. RNA Polymerase II transcribes them to create pri-miRNA, which folds into a stem-loop structure. The expression of miRNAs can be upregulated or downregulated because of a positive or negative feedback loop during transcription [16].
Endonuclease Drosha RNase III and the dsRNA binding protein DGCR8 (DiGeorge syndrome critical region gene 8), also referred to as Pasha [12], cleave the transcribed pri-miRNA in the nucleus. The 5′ phosphate is left unharmed when it cleaves both strands of the hairpin structure, leaving a roughly 2-nucleotide overhang at the 3′ end [17]. The resulting structure is known as pre-miRNA. Instead of using this mechanism, mirtrons go through splicing to create the pre-miRNA [18]. With the aid of Exportin-5 and Ran-GTP, the pre-miRNA is then exported from the nucleus to the cytoplasm [19].
In the cytoplasm, the endonuclease Dicer [20] recognizes the 3′ overhang and 5′ phosphate and cuts the pre-miRNA. Dicer contains a helicase, dsRNA binding domain, a PAZ domain, and two RNase III domains. When the pre-miRNA is loaded into the Dicer, the loop in pre-miRNA is recognized by the helicase domain, while the strands are cleaved by the two RNaseIII domains. This separates the loop and leaves a ds-miRNA:miRNA* complex [21]. One of these strands is the actual miRNA, whereas the other strand, the “miRNA*” is a short-lived, opposing arm of the actual miRNA.
The RISC has been loaded with the miRNA-miRNA* combination (RNA-induced silencing complex). Agronaute-2 protein, Dicer, siRNAs, ADAR1, TRBP, PARN, RNA helicase, Hsp90, TSN, and eIF1A are all components of RISC. Pre-miRNA uptake and the release of Dicer are both affected by Ago-2. To cleave the pre-3′ miRNA’s arm before Dicer processes it, it also has endonuclease activity [22]. Dicer cuts the miRNA* strand, and a loaded RISC complex is now available. Following this, Ago-2 leads the complex to the target mRNA, which is identified by a conserved region of the miRNA [23]. If the miRNA is highly complementary to the target mRNA, the target mRNA is either cleaved or the translation of the mRNA is suppressed [14].
Biogenesis of lncRNA
Depending on the lncRNA itself, the type of cell, and its stage, different lncRNA biogenesis pathways exist. The DNA can contain intergenic regions (lincRNAs), promoter upstream transcripts (PROMPTs), antisense regions (NATs), and enhancers, among other places where they can be translated (eRNAs). Some lncRNA biogenesis resembles that of mRNAs in many ways. There are specific lncRNA genes, and RNA polymerase II is responsible for their transcription. They are produced in regions with chromatin states that are comparable to those of mRNAs [24]. They also go through 3′ poly-A tail, 5′ end m7G capping, and splicing.
However, a number of additional lncRNAs go through unusual processing. Some are subjected to RNase-P cleavage for 3′ end maturation [25], others are capping at the 5′ end [26], some are capping both the 5′ and 3′ ends [27], or they may fold into circular structures that stop their degradation [28]. After their biogenesis, they can either be transferred to the cytoplasm or enriched within the nucleus itself [29]. The NXF1 (nuclear RNA export factor-1) pathway is used for the transport. They interact with different RNA-binding proteins in the cytoplasm, which helps them get to their target organelles [30].
 | Figure 1. Basic overview of miRNA-lncRNA-mRNA interaction. [Click here to view] |
Biogenesis of mRNA
The TATA box region of the gene promoter, which is situated around 25 nucleotides upstream from the transcription start point, is where transcription factor TFIID attaches to begin transcription. TBP oversees identifying the area (TATA-binding protein). The DNA strands become separated as a result. By clearing a path for their binding, RNA Polymerase II and TFIIH form a transcription initiation complex. The TFIIH’s DNA helicase begins to unwind the DNA strands. A protein kinase found in TFIIH phosphorylates the RNA polymerase II C-terminal domain (CTD), improving the enzyme’s ability to bind to the gene and begin transcription. During RNA transcript elongation, the transcription factors are eliminated [31].
RNA alterations take place concurrently with transcription in eukaryotes [32]. This includes 3′ polyadenylation [33], mRNA splicing [34], and 5′ end m7G capping [35]. The mRNA can endure in the cytoplasm thanks to these changes. As the RNA transcript is created from the 5′ end, guanylyl transferase performs the capping process first. Following the transcription and subsequent removal of the introns is splicing by spliceosomes. By using Poly-A polymerase, the 3′ end is polyadenylated last.
ROLE IN DISEASES
Role in cancers
Gastric cancer
With over a million new cases each year, gastric cancer is the most common cancer that results in mortality worldwide. It is the fourth and seventh most diagnosed cancer in men and women, respectively [36]. Numerous genetic variables are also known to play a substantial role in the onset and progression of gastric cancer in addition to the viral causes. These may be inherited or may be dysregulated because of environmental or lifestyle factors [37].
By homophilic interaction with the cytoskeletal elements and plasma membranes of neighboring cells, E-cadherins, calcium-dependent adhesion molecules, help form adherent junctions between those cells [38]. As a result, they are crucial in reducing cell invasion and migration in tumors [39]. Premature stop codons, or PTCs, are inserted by about 80% of all CDH1 germline mutations [40] (premature termination codons). The CDH1 protein is subsequently prematurely terminated as a result. As a result, the CDH1 mRNA in the PTC is drawn to and degraded by the NMD machinery, severely inhibiting the expression of E-cadherin (Table 1).
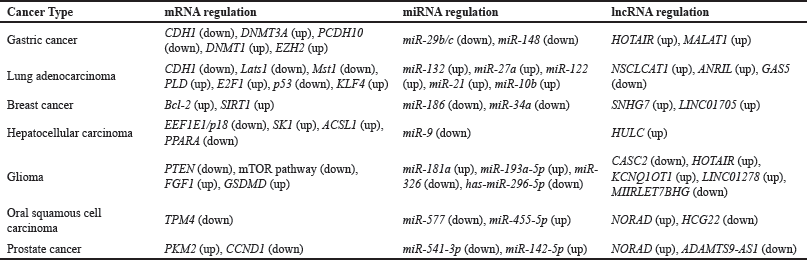 | Table 1. mRNAs, miRNAs, and lncRNAs involved in cancers. [Click here to view] |
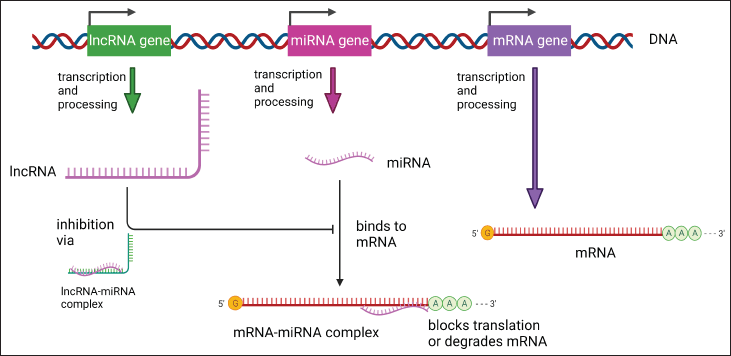 | Figure 2. Overview of biogenesis of miRNA, lncRNA, and mRNA. [Click here to view] |
The microRNA miR-29b/c is reported to function as a suppressor of tumor metastasis in gastric cancers. It acts by targeting the DNMT3A gene, which codes for DNA Methyltransferase 3 Alpha, which is involved in the methylation of cytosine in the DNA. miR-29b/c is complementary to the 3′ UTR of the DNMT3A mRNA, hence involved in its downregulation. On the other hand, DNMT3A also suppressed miR-29b/c by methylating its promoter. This forms a negatively regulating feedback loop between the two [41]. DNMT3A is involved in promoting gastric tumor cell migration. Knockdown of DNMT3A also leads to an overexpression of the CDH1 gene, thus promoting cell tumor migration.
miR-29b/c is also involved in the regulation of the CDH1 gene. It was reported that miR-29b/c prevents promoter methylation of the CDH1 gene, leading to suppression of its downregulation [42]. Higher expression of CDH1 leads to a lower ability of the tumor cells to migrate. Another gene playing a notable role in gastric cancer is the PCDH10 gene, which codes for Protocadherin-10. Protocadherins are the largest subgroup within the cadherin superfamily. In addition, they aid in the prevention of stomach cancer. In addition, it has been stated that PCDH10 mediates the tumor-suppressive effects of p53. Substantial upregulation of the lncRNA HOTAIR has been observed in gastric tumors. It functions by lowering PCDH10 gene expression by increasing methylation of the PCDH10 promoter [43]. In addition, both the transcriptional and translational levels of p53’s expression are considerably decreased by HOTAIR [44]. The connection between HOTAIR, miR-148b, and DNMT1 controls the methylation process. HOTAIR behaves as a competitive endogenous RNA (ceRNA) because it serves as a miRNA sponge for miR-148 [45].
In addition, MALAT1 suppresses the expression of the PCDH10 gene to encourage cell invasion and migration in gastric cancer [46]. This is accomplished by the Polycomb Repressive Complex 2 (PRC2), whose active member, EZH2 (Enhancer of Zeste Homolog 2) [47], catalyzes the trimethylation of histone 3 Lys27 (H3K27me3) [48]. MALAT1 also interacts with EZH2 to partially repress CDH1 in gastric cancers [49], but not in renal cell carcinoma cell lines [50].
Lung cancer
Lung malignancies are to blame for over half of all cancer-related fatalities [51]. The CDH1 gene also has a substantial impact on lung cancer, like in the case of gastric malignancies. As is well known, CDH1 inhibits cancer cell invasion and migration by improving the cell’s adhesion to nearby cells through adherens junctions. A lncRNA NSCLCAT1 was reported to increase cell invasion and migration in nonsmall cell lung cancers (NSCLCs). NSCLC accounts for 85% of all lung cancers. E-cadherins coding the CDH1 gene are the target of the NSCLCAT1 lncRNA, leading to the downregulation of CDH1 [52]. It is reported that in NSCLC tissues, there is an upregulation of lncRNA NSCLCAT1, and mRNAs TAZ and YAP1 [53]. These were related to a downregulation of mRNAs CDH1, Lats1, and Mst1. These genes are a part of the hippo signaling pathway, which is involved in the inhibition of cell proliferation, limiting cell size, and promotion of apoptosis [54].
Numerous malignancies, including lung cancer, have been associated with the upregulation of the Phospholipase D (PLD) gene [55]. PLD is believed to lower apoptosis and boost cell growth. In lung and other malignancies, several miRNAs, including miR-132, miR-27a, miR-122, miR-21, and miR-10b, have been associated with cancer metastasis, cell survival, and cell proliferation [56]. PLD is involved in the hydrolysis of phosphatidylcholine to phosphatidic acid, which is known to be a key up-regulator of the mTOR (mammalian target of rapamycin) signaling pathway[57]. mTOR acts as a regulator for cellular metabolism, proliferation, growth, and survival [58]. PLD inhibition is linked to a 13.6-fold increase in the expression of lncRNA ANRIL. This increased more when the lung tumor cells were exposed to more PLD inhibitors [59]. Higher expression of ANRIL was reported to increase apoptotic cell death in tumor tissues. It also increases the expression of ATG6/BECLIN-1 [60], ATG3 [61] (autophagy-related gene 3), ATG5 [62] (autophagy-related gene 5), and LC3B [63].
A lncRNA known as GAS5 (Growth Arrest Specific 5) is markedly downregulated in NSCLC. In lung malignancies, GAS5 functions as a tumor suppressor. The transcription factor E2F1 (E2F transcription factor 1), which is involved in the progression of the cell cycle, apoptosis, and DNA-damage response [64], is downregulated by GAS5 expression while the expression of the tumor suppressor p53 is upregulated [65]. It has been demonstrated that miR-10b expression and Klf4 (Krüppel-like factor 4) expression are related. Klf4 is a conserved transcription factor with zinc fingers that is involved in cell growth and proliferation [66]. It is one of four transcriptional factors that are employed to create iPSCs [67] (induced pluripotent stem cells). In tumor cells and stem cells, Klf4 is overexpressed. Through an unidentified relationship, higher Klf4 expression is induced by higher miR-10b expression, which increases NSCLC cell invasion and proliferation [68].
Breast cancer
When compared to paracancerous tissues (tissues less than 2 cm away from the boundary of the tumor), lncRNA SNHG7 (small nucleolar RNA host gene 7) has been found to be substantially overexpressed in breast cancers [69]. They are known to encourage the growth and spread of the tumor, serving as an oncogene [69]. The stage of the tumor and the spread of the cancer to the lymph nodes and organs were found to be directly linked with the expression of SNHG7. According to research, cancer was found to be more aggressive when SNHG7 expression was higher, which decreased the patient’s chance of survival [70]. It functions by absorbing miR-186 like a sponge. Cell migration and proliferation are known to be inhibited by microRNA-186 [71].
Higher SNHG7 was associated with increased chemoresistance of breast cancer cells to chemotherapeutic agents such as adriamycin [72] and paclitaxel [73]. In addition, miR-34a is absorbed by SNHG7 [74]. The presence of SNHG7 was observed to negatively correlate with miR-34a expression in chemoresistant cells, indicating that miR-34a is one of lncRNA SNHG7’s targets [75]. MiR-34a is responsible for downregulating Bcl-2 (B-cell Lymphoma 2) and SIRT1 (Sirtuin 1), which leads to inhibited proliferation and migration of cancerous cells [76]. Its sequestering by SNHG7, thus, contributes to the disease.
Other than SNHG7, other lncRNA also interact with miR-186 to regulate the development of breast cancer. One such lncRNA is LINC01705 [77]. It also acts by sponging miR-186, leading to the advancement of cancer. Thus, the proliferation and metastasis of cancer are higher in cells with a higher expression of LINC01705. A decrease in the expression of LINC01705 led to an increase in tumor cell apoptosis, though the mechanism is not clear. Localization studies show that the lncRNA is mostly found in the cytoplasm, indicating that it may perform its regulatory roles post-transcriptionally [78].
Hepatocellular carcinoma
Liver malignancies express a lot of the lncRNA highly elevated in liver cancer (HULC) [79]. In hepatocellular carcinoma (HCC), it is the lncRNA that is most overexpressed [80]. In addition, it has been suggested that it is elevated in a number of different malignancies, including osteosarcoma [81], gastric cancer [82], and pancreatic cancer [83]. In various cancer types, HULC has a role in promoting cell proliferation, cell survival, colony formation, cell migration, cell invasion, and angiogenesis [84].
The Eukaryotic translation elongation factor epsilon-1 (EEF1E1) is also sometimes referred to as p18 [85]. In malignant tissues, HULC levels and EEF1E1 levels are inversely correlated [86]. It follows that the suppression of the EEF1E1 gene is one way that HULC upregulates cancer. The enzyme Sphingosine Kinase 1 (SK1) is crucial for the development of cancer [87]. It is involved in the regulation of sphingolipid levels in the cell. The expression of SK1 was found to be correlated positively with the expression of HULC. It leads to higher levels of Sphingosine-1-Phosphate in HCC and upregulates angiogenesis in these tissues.
An important part of the commencement of the metabolism of long-chain fatty acids is played by the enzyme ACSL1 [85,88]. Positive correlations between the cellular levels of HULC and ACSL1 were discovered [89]. The ACSL1 gene is induced as a result of a decrease in the transcription factor PPARA [85,88]. A hallmark of cancer is the buildup of cholesterol and triglycerides within the cell, which is caused by an increase in ACSL1 expression [90].
MiR-9 is silenced by HULC through epigenetic regulation. Mir-9-3p was identified in HCC cell lines as well as in clinical samples, to act as a tumor suppressor. It does that by targeting the expression of the TAZ protein. Inhibition of TAZ protein leads to reduced cell proliferation and reduced phosphorylations of Erk1/2 (Extracellular Signal-Regulated Kinase 1/2), AKT (Serine/Threonine Kinase Family), and β-catenin [91]. This is yet another way in which HULC contributes to the HCC.
Glioma
LncRNA cancer susceptibility candidate 2 (CASC2) is downregulated in numerous malignancies, including glioma [92]. phosphatase and tensin homolog (c), a recognized tumor suppressor, is upregulated by it. In addition, CASC2 directly inhibits miR-181a, which likewise inhibits the Akt signaling pathway [93]. It also contributes to the downregulation of miR-193a-5p expression, which lowers the therapeutic effectiveness of temozolomide [94]. This is accomplished by CASC2 activating the mTOR pathway.
The lncRNA HOTAIR is suggested to be involved in glioma, enhancing cell cycle progression via PRC2 [95]. Knockdown of this lncRNA leads to an upregulation in the expression of miR-326, which in turn suppresses fibroblast growth factor-1 (FGF1) expression, and blocks PI3K/AKT and MEK1/2 pathways [96]. This leads to an increase in apoptosis and cell cycle arrest, while reducing cell proliferation, cell migration, and cell invasion.
The gene known as Gasdermin D (GSDMD) [97] is crucial for the process of pyroptosis, which in turn affects tumor formation and therapeutic response [98]. When compared to nontumor brain cells, it is found to be elevated in gliomas [99]. The miRNA has-miR-296-5p is one of the main downregulators. The lncRNAs KCNQ1OT1 and LINC01278 are indicative of the development of the disease and have a favorable correlation with GSDMD. Another lncRNA that downregulates the GSDMD gene is MIIRLET7BHG, and the expression of this lncRNA is favorably connected with better clinical results for the patient [100].
Oral squamous cell carcinoma
Oral squamous cell carcinoma (OSCC) is thought to be caused by the lncRNA NORAD (ncRNA Activated by DNA damage). According to reports, it is overexpressed in OSCC. One of the targets of NORAD is miR-577, which has been shown to be downregulated in OSCC [101]. Numerous malignancies, including OSCC, have been reported to have dysregulated expression of the TPM4 (Tropomyosin 4) gene [102]. It has been demonstrated that miR-577 adversely downregulates TPM4 in OSCC. Therefore, NORAD affects the development and maintenance of cancer by miR-577-mediated downregulation of the TPM4 gene [103].
The HLA Complex Group 2 lncRNA HCG22 plays a role in a variety of malignancies, including OSCC [104]. Better patient survival is correlated with higher HCG22 expression [105]. The proliferation, invasion, and migration of OSCC cells are inhibited by HCG22 overexpression. Targeting miR-425-5p [106] achieves this. MiR-455-5p is sponged by HCG22, acting as a ceRNA to reduce its availability in the cell. Malignancies, including OSCC, HCC [107], colorectal cancer [108], and cervical cancer [109], to mention a few, have higher expression of miR-425-5p.
Prostate cancer
Prostate cancer too, is affected by the lncRNA NORAD [110]. While blocking apoptosis, it is known to stimulate cell migration and proliferation [111]. It is thought that NORAD also encourages pancreatic cancer bone metastases. As a ceRNA, NORAD targets miR-541-3p. MiR-541-3p’s decreased availability makes (Pyruvate Kinase M2 (PKM2’s) gene expression more prominent. PKM2 is typically present in embryonic and malignant cells and is strongly associated with carcinogenesis, tissue repair, and regeneration [112].
Extracellular vesicles with PKM2 promote metastasis of pancreatic cancer [113]. This gene also affects the internalization and the release of the extracellular vesicles [114]. This is done by PKM2-mediated phosphorylation of synaptosomal-associated protein 23 (SNAP-23), and the formation of soluble N-Ethylmaleimide-sensitive fusion actor attachment protein receptor (SNARE) complex [115]. This leads to the promotion of extracellular vesicle release [116]. These vesicles also contained more ATP, making them easier to be internalized by bone marrow stromal cells [117], which leads to a significant increase in bone metastasis of prostate cancer.
LncRNA that is downregulated in prostate cancer is a Disintegrin and Metalloproteinase with Thrombospondin Motifs 9-Antisense 1 (ADAMTS9-AS1). Apoptosis is known to be influenced by the lncRNA. MiR-142-5p expression and ADAMTS9-AS1 regulation are adversely correlated. It is known that this miRNA affects the Cyclin D1 (CCND1) gene. Several tumor suppressors are activated because of CCND1. As a result, increased CCND1 suppression by miR-142-5p [118] and low expression of ADAMTS9-AS promote prostate cancer.
Role in cardiovascular diseases
Acute ischemic stroke
Blood flow to the brain is reported to abruptly halt during an acute ischemic stroke (AIS) [119,120]. As a result, the brain receives significantly less oxygen and glucose. It is increasingly prevalent among middle-aged and older individuals around the world as well as among aging populations. A lncRNA called potassium voltage-gated channel subfamily Q Member 1 Opposite Strand 1 (KCNQ1OT1) has been linked to atherosclerosis and heart attacks, both are major risk factors for stroke [121]. In the plasma of AIS patients, KCNQ1OT1 expression was said to be greater. This lncRNA’s expression is inversely linked with stroke severity [122]. It was discovered that KCNQ1OT1 silencing in mice resulted in a noticeably smaller volume of cerebral infarction [123]. This could be a possible avenue for treatment.
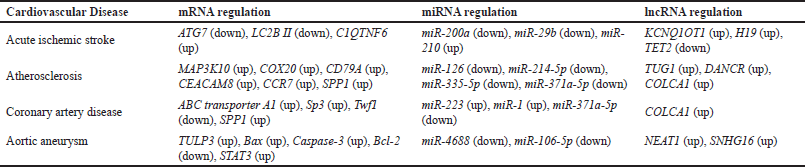 | Table 2. mRNAs, miRNAs, and lncRNAs involved in cardiovascular diseases. [Click here to view] |
The expression of miR-200a and the expression of the lncRNA KCNQ1OT1 are (Table 2) negatively correlated. Each of them has a negative feedback mechanism that reduces the other’s expression. MiR-200a expression rose when KCNQ1OT1 was knocked down, which increased cell survival and decreased autophagy [124]. This outcome was consistent with the reported decline in the levels of the stress-induced autophagy-related proteins ATG7 [125] and LC3B II [126].
According to reports, ischemic stroke patients’ brains, neutrophils, and plasma had higher levels of the lncRNA H19 [127]. It functions for miR-29b as a ceRNA sponge. Through the control of C1q tumor necrosis factor-related Protein 6 (C1QTNF6), miR-29b is known to contribute to an increase in inflammatory response [128]. The gene for C1QTNF6 is the target of this miRNA. The C1QTNF6 gene encourages TNF release, which aggravates inflammation [129]. Therefore, the increased H19 causes miR-29b to be downregulated, which increases the expression of the C1QTNF gene [130].
MiR-210 is a master hypoxamiR (miRNAs that are stimulated by hypoxia) [131], which has been shown in human studies to be a blood biomarker for AIS and other cardiovascular diseases and cancers [132]. It has been proven in animal studies to be very overexpressed in the brain after the stroke. Suppression of its action by miR-210-LNA (locked nucleic acid oligonucleotides) leads to reduced inflammation and a neuroprotective effect [133]. Tet Methylcytosine Dioxygenase 2 (TET2) is an important regulator of proinflammatory cytokines [134]. It has been shown in rats that miR-210 binds to the 3′ UTR of TET2 and downregulates its expression, contributing to the disease [135].
Atherosclerosis
In the medium to large-sized arteries, atherosclerosis is a fat-storage disease with an immunoinflammatory response [136]. Plaque, which is comprised of cholesterol, lipids, and other substances, builds up on the artery walls as a result of this condition. Inflammatory cytokines TNF-α and IL-6 were reduced in mice after the administration of miR-126 [137]. This shows that miR-126 plays a role in the atherosclerotic process’ detrimental regulation of the inflammatory response. MAP3K10, a component of the MAPK signaling pathway, has been found to be another miR-126 target [138]. The process of inflammation causes an upregulation of this pathway. Because miR-126 inhibits the pathway, it plays a role in reducing inflammation [139].
The lncRNA Taurine Upregulated Gene 1 (TUG1) is another ncRNAs involved with atherosclerosis. TUG1 has been found to be strongly expressed in those with atherosclerosis. Its involvement in the illness was confirmed by the fact that it was expressed excessively in the aortic plaques [140]. During hypoxic conditions, the lncRNA is increased in vascular smooth muscle cells (VSMCs) [141]. TUG1 was also discovered to be an up-regulator of proliferation and a down-regulator of apoptosis [142], which may quicken plaque development and heighten disease severity.
In atherosclerotic patients, the lncRNA differentiation antagonizing nonprotein coding RNA (DANCR) is markedly overexpressed. For miR-214-5p [143], DANCR functions as a ceRNA sponge. This miRNA inhibits the production of the cytochrome C oxidase assembly protein (COX20) gene by targeting it. According to reports, COX20 may help a cell become more resistant to oxidative damage and apoptosis [144]. DANCR also acts as a sponge for miR-335-5p [145]. MiR-335-5p is known to have an inhibitory effect on plaque formation in atherosclerosis [146]. This miRNA also targets CD79A, Carcinoembryonic Antigen-Related cell adhesion molecule 8 (CEACAM8), and chemokine receptor 7 (CCR7) genes, which are linked to play a role in the disease [147].
Human Coronary Artery Endothelial Cells, or HCAECs, can be stimulated to induce an inflammatory response using oxidized low-density lipoprotein (OxLDL) [148]. The resulting transcriptome change causes these cells to proliferate less. The lncRNA colorectal cancer associated 1 (COLCA1), which is overexpressed, is one of the main variations in expression. According to reports, COLCA1 alters cell biology via miR-371a-5p. This miRNA is linked to cancer [149] and inflammatory response [150]. COLCA1 and miR-371a-5p share a ceRNA relationship. miR-371a-5p targets the 3’ UTR of secreted phosphoprotein 1 (SPP1) mRNA. SPP1 gene has been shown to play an important role in atherosclerosis, and coronary heart disease [151]. It is overexpressed in VSMCs which are a part of plaques formed due to atherosclerosis [152]. As a result, COLCA1 increases the expression of the SPP1 gene, which in turn promotes the development of atherosclerosis by decreasing the availability of miR-371a-5p [153].
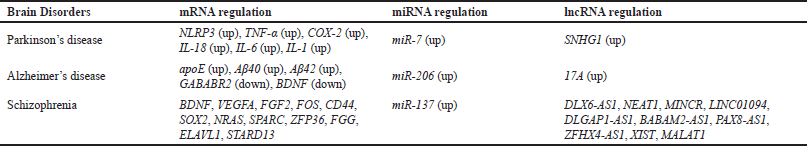 | Table 3. mRNAs, miRNAs, and lncRNAs involved in brain disorders. [Click here to view] |
Coronary artery disease
A buildup of cholesterol deposits (plaques) in the coronary arteries, which carry blood to the heart, is known as coronary artery disease (CAD). Over time, this build-up causes arteries to constrict, reducing the amount of oxygen that can reach the heart muscles [154]. The coronary arteries’ atherosclerosis is to blame for it.
MiR-223 was identified to be upregulated in patients with CAD, which is also a marker for atherosclerosis [155]. MiR-223 is implicated in the regulation of several genes responsible for inflammation [156]. It also plays a role in the homeostasis of high-density lipoprotein-cholesterol (HDL-C) by inhibiting cholesterol synthesis. MiR-223 indirectly promotes the expression of ATP-binding cassettes (ABC) transporter A1 via Sp3, leading to an increase in cholesterol efflux from the cell [157]. This leads to higher deposits in the coronary arteries, contributing to the disease.
MiR-1 is very highly expressed in the heart but is found to be downregulated in patients suffering from cardiovascular diseases [158] including CAD. It is reported to target Twf1 (Twinfilin) gene [158]. Twf1 is a cytoskeletal regulatory protein that binds to actin monomers to stop actin from assembling. Aortic constriction and other stimuli like α-adrenergic stimulation can cause miR-1 to be downregulated, which causes a high enough level of Twf1 protein overexpression to cause cardiac hypertrophy, a factor in the development of CAD [159].
Several hypoxamiRs are known to play a role in CAD too. One of the important hypoxamiR is miR-146b. Expression of tumor necrosis factor receptor associated factor 6 (TRAF6) is induced in hypoxic conditions in rat cardiomyocytes. The 3′ UTR region of TRAF6 gets bound by miR-146b, leading to a higher expression of IL-6 and CCL2 monocyte chemoattractant protein 1 (MCP-1) [160]. This miR-146b-TRAF6-IL-6/MCP-1 axis plays a role in cardiac dysfunction and failure.
Aortic aneurysm
Aortic aneurysms are bulges in the aorta’s wall [157]. Thoracic aortic aneurysms [161] (TAA) are termed if they develop in the portion of the aorta that passes through the chest cavity, while abdominal aortic aneurysms (AAA) [162] are called if they develop in the portion of the aorta that passes through the belly. The bulge may be saccular [163] (spherical) or fusiform [164] (tube-shaped). They have the potential to enlarge, rupture, or explode, resulting in severe injury or even death [165].
NEAT1 is an oncogenic lncRNA which is also reported to be upregulated in AAA [166]. NEAT1 overexpression leads to a reduction in the proliferation of the VSMCs and an increase in the apoptosis rates. Thus, NEAT1 contributes to the disease by promoting apoptosis of VSMCs and suppressing their proliferation for repair. NEAT1 was also shown be regulate the tubby-like protein 3 (TULP3) expression. NEAT1 acts like a microRNA sponge for miR-4688 and sequesteres it. This allows for higher expression of TULP3, as it is a target of miR-4688 [167].
In people with aortic aneurysms, the gene signal transducer and activator of transcription 3 (STAT3) is known to be upregulated [168]. This gene’s downregulation promotes cellular growth and prevents apoptosis. According to a report, the gene is targeted by miR-106b-5p. This miRNA is responsible for upregulating Bcl-2 while downregulating Bax (Bcl-2-Like Protein 4) and Caspase-3. Apoptosis is promoted by Bax and Caspase-3 whereas it is depressed by Bcl-2 [169]. This miR-106b-5p contributes to lowering the apoptotic rate [170]. The lncRNA SNHG16 is a ceRNA that sponges miR-106b-5p, leading to higher STAT3 expression. So SNHG16 promotes the disease by reducing the availability of miR-106b-5p, which leads to an increase in STAT3 expression [171].
Role in brain disorders
The role of miRNA-lncRNA-mRNA interrelationship in various neurological disorders is as follows:
Parkinson’s disease
Parkinson’s disease is a neurological condition that progresses. It makes the muscles tremble and spasm out of control. Dopaminergic loss of neurons in the substantia nigra and deregulation of basal ganglia neurons are two features of the disease [172].
It was discovered that microRNA miR-7 is downregulated and the lncRNA SNHG1 is increased in cultured microglial cells. In addition to SNHG1, microglial activation markers CD11b and Iba-1, as well as higher levels of inflammatory cytokines TNF-α, COX-2, IL-18, IL-6, and IL-1, were also detected. Their expression was correlated, indicating that SNHG1 [173] is at least partially responsible for controlling them.
Analysis of the SNHG1 sequence using miRcode showed (Table 3) a conserved miR-7 target site, and its role in downregulating miR-7 was proved. MiR-7 is a regulator of NLRP3 inflammasome [174]. NLRP3 inflammation is one of the most important contributors to the pathogenesis of the disease. SNHG1 overexpression increases the expression of NLRP3 at the transcript and protein level. This shows that downregulation of SNHG1 reduces the activation of microglia and reduces dopaminergic neuron loss.
Alzheimer’s disease
Alzheimer’s disease is a neurological ailment that worsens with time and causes the neurons to atrophy. Dementia, loss of social and behavioral facets of personality, and memory loss that gets progressively worse are the results of this. The abnormal buildup of a variety of different components in the brain and neurological system may be the cause of this. Apolipoprotein E (apoE), α-synuclein, Amyloid-β (Aβ) peptides, and tau are some of them [175].
According to reports, the disorder has an overexpression of lncRNA 17A. Apoptosis occurred at a noticeably lower rate in cells with reduced 17A expression, according to knockdown studies, demonstrating the importance of 17A in the process. It has also been demonstrated that 17A regulates the expression of the suggestive peptides Aβ40 and Aβ42 for the course of Alzheimer’s disease [176]. When 17A was knocked down, there was a lower amount of these peptides produced, while its overexpression led to their increase, indicating the progression of the disease. Overexpression of 17A also corresponded to lower GABABR 2 (Gamma-Aminobutyric Acid Type B Receptor Subunit 2) protein expression, which is a cell-signaling receptor of the nervous system [177]. 17A overexpression also leads to a decrease in nestin-positive cells, indicating its role in the suppression of neurogenesis [175].
There is evidence that miR-206, a microRNA, is increased in Alzheimer’s disease. MiR-206 inhibits the expression of brain-derived neurotrophic factor (BDNF) by attaching to its 3′ UTR. For context, consider that when anti-miR-206 siRNA was delivered into the brain of Alzheimer’s model mice (Tg2576), the mice displayed improved spatial memory [178].
Schizophrenia
One of the most common, chronic brain conditions is schizophrenia [179]. Hallucinations, psychosis, delusions, mental disarray, and so on are some of the disease’s symptoms. It was discovered that miR-137 and schizophrenia are related. Two distinct (genome-wide association studies (GWAS) that involved participants in Sternberg item recognition paradigm (SIRP) working memory tasks implicated miR-137 [180]. MiR-137 had 26 experimentally confirmed targets. Grey matter density and cognitive function are assumed to be maintained by these genes [181]. In addition to miR-137, there were 3,859 differentially expressed (DE) mRNAs, 2400 DE lncRNAs, and 69 DE miRNAs in patients with disease [182]. 375 of these mRNAs were discovered to be involved in autophagy. There were not many DE RNAs associated with the MAPK, mTOR, and ErbB pathways.
A transcriptome analysis of schizophrenia patients and controls was carried out in a 2021 study. The discovered 12 DE mRNAs (BDNF, VEGFA, FGF2, FOS, CD44, SOX2, NRAS, SPARC, ZFP36, FGG, ELAVL1, and STARD13) are thought to be regulated by the reported 10 DE lncRNAs (DLX6-AS1, NEAT1, MINCR, LINC01094, DLGAP1-AS1, BABAM2-AS1, PAX8-AS1, ZFHX4-AS1, XIST, and MALAT1) [183].
CONCLUSION
In the control of cellular machinery, interactions among mRNAs, lncRNAs, and miRNAs are crucial. Several disorders are also thought to be impacted by these connections. Direct protein coding is provided by the mRNAs. The miRNAs either destroy mRNAs or stop their translation. To stop miRNAs from inhibiting mRNAs, lncRNAs act as a sponge for them. Only recently have researchers begun to examine the intricate interaction patterns of the miRNA-lncRNA-mRNA network. As of now, our knowledge of this network is not complete. The interactome is being investigated, and more information is gradually becoming known. Many illnesses, such as strokes, atherosclerosis, Parkinson’s, and so on, were formerly believed to be inherited genetically or acquired through environmental factors. Nevertheless, we can now see that they can play quite an important role thanks to a greater knowledge of ncRNA-mRNA interaction. Using the creation and validation of powerful techniques like luciferase reporter assays, GWAS, and so on, we can confirm the interactions of these RNAs and validate them in vitro studies before in vivo investigations. The fundamental or traditional machine learning methods of disease-related miRNA tools provide important insights into the history and potential of miRNA tools.
ACKNOWLEDGMENT
The authors thank the management and administration of SRM Institute of Science and Technology for the motivation and support for executing this work.
FINANCIAL SUPPORT
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY STATEMENT
All the data is available with the authors and shall be provided upon request.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Eddy SR. Non–coding RNA genes and the modern RNA world. Nat Rev Genet. 2001 Dec 1;2(12):919–29.
2. Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019 Jul 24;20(11):631–56.
3. Loganathan T, Doss C GP. Non-coding RNAs in human health and disease: potential function as biomarkers and therapeutic targets. Funct Integr Genomics. 2023 Jan 10;23(1):33.
4. Sebastian-Delacruz M, Gonzalez-Moro I, Olazagoitia-Garmendia A, Castellanos-Rubio A, Santin I. The role of lncRNAs in gene expression regulation through mRNA stabilization. Non-Coding RNA. 2021;7:3 [Internet]. 2021 Jan 5 [cited 2023 Jan 30];7(1):3. Available from: https://www.mdpi.com/2311-553X/7/1/3/htm
5. Machida A, Ohkubo T, Yokota T. Circulating microRNAs in the cerebrospinal fluid of patients with brain diseases. Methods in Molecular Biology [Internet]. 2013 [cited 2023 Dec 15];1024:203–9. Available from: https://link.springer.com/protocol/10.1007/978-1-62703-453-1_16
6. Efroni I, Ip PL, Nawy T, Mello A, Birnbaum KD. Quantification of cell identity from single-cell gene expression profiles. Genome Biol [Internet]. 2015 Jan 22 [cited 2021 Nov 8];16(1):1–12. Available from: https://link.springer.com/articles/10.1186/s13059-015-0580-x
7. Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell [Internet]. 2013 Mar 14 [cited 2021 Nov 8];152(6):1251. Available from: /pmc/articles/PMC3640494/
8. Balda MS, Matter K. Epithelial cell adhesion and the regulation of gene expression. Trends Cell Biol. 2003 Jun 1;13(6):310–8.
9. Hafner A, Bulyk ML, Jambhekar A, Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol [Internet]. 2019 Mar 1 [cited 2021 Nov 8];20(4):199–210. Available from: https://www.nature.com/articles/s41580-019-0110-x
10. Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012 Jun 8;46(5):674–90.
11. Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol [Internet]. 2012 Jun 5 [cited 2021 Nov 8];19(6):586–93. Available from: https://www.nature.com/articles/nsmb.2296
12. Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002 Sep 2;21(17):4663–70.
13. Xu F, Zhang J. Long non-coding RNA HOTAIR functions as miRNA sponge to promote the epithelial to mesenchymal transition in esophageal cancer. Biomed Pharmacother. 2017 Jun 1;90:888–96.
14. Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science (1979) [Internet]. 2002 Sep 20 [cited 2021 Jul 21];297(5589):2056–60. Available from: https://science.sciencemag.org/content/297/5589/2056
15. Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, et al. Structure and activity of putative intronic miRNA promoters. RNA. 2010 Mar;16(3):495–505.
16. Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, et al. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008 Sep 15;22(18):2535–49.
17. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003 Sep 25;425(6956):415–9.
18. Westholm JO, Lai EC. Mirtrons: MicroRNA biogenesis via splicing. Biochimie. 2011 Nov;93(11):1897–904.
19. Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003 Dec 15;17(24):3011–6.
20. MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, et al. Structural basis for double-stranded RNA processing by Dicer. Science (1979). 2006 Jan 13;311(5758):195–8.
21. Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003 Aug 1;5(2):337–50.
22. Yang N, Cao Y, Han P, Zhu X, Sun L, Li G. Tools for investigation of the RNA endonuclease activity of mammalian Argonaute2 protein. Anal Chem [Internet]. 2012 Mar 6 [cited 2021 Jul 21];84(5):2492–7. Available from: https://pubs.acs.org/doi/abs/10.1021/ac2032854
23. Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nature Genetics 2002 30:4 [Internet]. 2002 Mar 18 [cited 2021 Jul 21];30(4):363–4. Available from: https://www.nature.com/articles/ng865z
24. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009 458:7235 [Internet]. 2009 Feb 1 [cited 2021 Aug 9];458(7235):223–7. Available from: https://www.nature.com/articles/nature07672
25. Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res [Internet]. 2009 Mar 1 [cited 2021 Aug 9];19(3):347–59. Available from: https://genome.cshlp.org/content/19/3/347.full
26. Wu H, Yin QF, Luo Z, Yao RW, Zheng CC, Zhang J, et al. Unusual processing generates SPA LncRNAs that sequester multiple RNA binding proteins. Mol Cell. 2016 Nov 3;64(3):534–48.
27. Xing YH, Yao RW, Zhang Y, Guo CJ, Jiang S, Xu G, et al. SLERT regulates DDX21 rings associated with Pol I transcription. Cell. 2017 May 4;169(4):664-678.e16.
28. Zhang XO, Wang H Bin, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014 Sep 25;159(1):134–47.
29. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet [Internet]. 2016 Jan 1 [cited 2021 Aug 9];17(1):47–62. Available from: https://pubmed.ncbi.nlm.nih.gov/26666209/
30. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol [Internet]. 2021 Feb 1 [cited 2021 Aug 9];22(2):96–118. Available from: https://pubmed.ncbi.nlm.nih.gov/33353982/
31. Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. From DNA to RNA. 2002 [cited 2021 Aug 9]; Available from: https://www.ncbi.nlm.nih.gov/books/NBK26887/
32. Turowski TW, Tollervey D. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip Rev RNA [Internet]. 2015 Jan 1 [cited 2021 Nov 8];6(1):129–39. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/wrna.1263
33. Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev [Internet]. 1997 Nov 1 [cited 2021 Nov 8];11(21):2755–66. Available from: http://genesdev.cshlp.org/content/11/21/2755.full
34. Lee Y, Rio DC. Mechanisms and regulation of alternative Pre-mRNA splicing. Annu Rev Biochem [Internet]. 2015 Jun [cited 2021 Nov 8];84:291–323. Available from: https://www.annualreviews.org/doi/abs/10.1146/annurev-biochem-060614-034316
35. Ramanathan A, Robb GB, Chan SH. mRNA capping: biological functions and applications. Nucleic Acids Res [Internet]. 2016 Sep 19 [cited 2021 Nov 8];44(16):7511–26. Available from: https://academic.oup.com/nar/article/44/16/7511/2460195
36. Sexton RE, Al Hallak MN, Diab M, Azmi AS. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev 2020 39:4 [Internet]. 2020 Sep 7 [cited 2021 Aug 5];39(4):1179–203. Available from: https://link.springer.com/article/10.1007/s10555-020-09925-3
37. Vogelaar IP, van der Post RS, Bisseling TM, van Krieken JHJ, Ligtenberg MJ, Hoogerbrugge N. Familial gastric cancer: detection of a hereditary cause helps to understand its etiology. Hereditary Cancer Clin Pract 2012 10:1 [Internet]. 2012 Dec 12 [cited 2021 Aug 5];10(1):1–6. Available from: https://hccpjournal.biomedcentral.com/articles/10.1186/1897-4287-10-18
38. Meng W, Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol [Internet]. 2009 Dec 1 [cited 2021 Aug 5];1(6):a002899. Available from: http://cshperspectives.cshlp.org/content/1/6/a002899.full
39. Suriano G, Oliveira MJ, Huntsman D, Mateus AR, Ferreira P, Casares F, et al. E-cadherin germline missense mutations and cell phenotype: evidence for the independence of cell invasion on the motile capabilities of the cells. Hum Mol Genet [Internet]. 2003 Nov 15 [cited 2021 Aug 5];12(22):3007–16. Available from: https://academic.oup.com/hmg/article/12/22/3007/606571
40. Oliveira C, Seruca R, Carneiro F. Genetics, pathology, and clinics of familial gastric cancer [Internet]. 2016 Jul 27 [cited 2021 Aug 5];14(1):21–33. Available from: https://journals.sagepub.com/doi/abs/10.1177/106689690601400105
41. Gong J, Li J, Wang Y, Liu C, Jia H, Jiang C, et al. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis [Internet]. 2014 Feb 1 [cited 2021 Aug 5];35(2):497–506. Available from: https://academic.oup.com/carcin/article/35/2/497/2462815
42. Alizadeh M, Safarzadeh A, Beyranvand F, Ahmadpour F, Hajiasgharzadeh K, Baghbanzadeh A, et al. The potential role of miR-29 in health and cancer diagnosis, prognosis, and therapy. J Cell Physiol [Internet]. 2019 Nov 1 [cited 2021 Aug 5];234(11):19280–97. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/jcp.28607
43. Seo SI, Yoon JH, Byun HJ, Lee SK. HOTAIR induces methylation of PCDH10, a tumor suppressor gene, by regulating DNMT1 and sponging with miR-148b in gastric adenocarcinoma. Yonsei Med J [Internet]. 2021 Feb 1 [cited 2021 Aug 5];62(2):118. Available from: /pmc/articles/PMC7859685/
44. Zheng H, Min J. Role of long noncoding RNA HOTAIR in the growth and apoptosis of osteosarcoma cell MG-63. Biomed Res Int. 2016;2016:5757641. doi: https://doi.org/10.1155/2016/5757641. Epub 2016 Aug 31. PMID: 27660759; PMCID: PMC5021870
45. Wang G, Li Z, Tian N, Han L, Fu Y, Guo Z, et al. miR-148b-3p inhibits malignant biological behaviors of human glioma cells induced by high HOTAIR expression. Oncol Lett [Internet]. 2016 Aug 1 [cited 2021 Aug 5];12(2):879–86. Available from: https://pubmed.ncbi.nlm.nih.gov/27446363/
46. Qi Y, Ooi HS, Wu J, Chen J, Zhang X, Tan S, et al. MALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10. Oncotarget [Internet]. 2016 Mar 15 [cited 2021 Jun 20];7(11):12693–703. Available from: /pmc/articles/PMC4914315/
47. He LJ, Cai MY, Xu GL, Li JJ, Weng ZJ, Xu DZ, et al. Prognostic significance of overexpression of EZH2 and H3k27me3 proteins in gastric cancer. Asian Pac J Cancer Prevention [Internet]. 2012 [cited 2021 Aug 6];13(7):3173–8. doi: http://dx.doi.org/10.7314/
48. Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010 Sep 3;7(3):299–313.
49. Qi Y, Zhang X, Kang Y, Wu J, Chen J, Li H, et al. Genome-wide transcriptional profiling analysis reveals annexin A6 as a novel EZH2 target gene involving gastric cellular proliferation. Mol Biosyst [Internet]. 2015 Jun 16 [cited 2021 Aug 6];11(7):1980–6. Available from: https://pubs.rsc.org/en/content/articlehtml/2015/mb/c5mb00233h
50. Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res [Internet]. 2015 Apr 1 [cited 2021 Aug 6];75(7):1322–31. Available from: https://cancerres.aacrjournals.org/content/75/7/1322
51. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin [Internet]. 2021 Jan 12 [cited 2021 Aug 6];71(1):7–33. Available from: https://europepmc.org/article/med/33433946
52. Zhao W, Zhang LN, Wang XL, Zhang J, Yu HX. Long noncoding RNA NSCLCAT1 increases non-small cell lung cancer cell invasion and migration through the Hippo signaling pathway by interacting with CDH1. Federation of American Societies for Experimental Biology [Internet]. 2018 [cited 2021 Aug 6]; Available from: www.fasebj.org
53. Fulford A, Tapon N, Ribeiro PS. Upstairs, downstairs: spatial regulation of Hippo signalling. Curr Opin Cell Biol. 2018 Apr 1;51:22–32.
54. Nakatani K, Maehama T, Nishio M, Goto H, Kato W, Omori H, et al. Targeting the Hippo signalling pathway for cancer treatment. J Biochem [Internet]. 2017 Mar 1 [cited 2021 Aug 6];161(3):237–44. Available from: https://academic.oup.com/jb/article/161/3/237/2662317
55. Cho JH, Han JS. Phospholipase D and its essential role in cancer. Mol Cells [Internet]. 2017 [cited 2021 Aug 6];40(11):805. Available from: /pmc/articles/PMC5712509/
56. Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int [Internet]. 2015 Apr 2 [cited 2021 Nov 10];15(1):1–6. Available from: https://cancerci.biomedcentral.com/articles/10.1186/s12935-015-0185-1
57. Sun Y, Chen J. mTOR signaling: PLD takes center stage. Cell Cycle [Internet]. 2008 Oct 15 [cited 2021 Aug 6];7(20):3118–23. Available from: https://pubmed.ncbi.nlm.nih.gov/18927511/
58. Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017 Mar 9;168(6):960–76.
59. Kang YH, Kim D, Jin EJ. Down-regulation of phospholipase D stimulates death of lung cancer cells involving up-regulation of the long ncRNA ANRIL. Anticancer Res. 2015 May;35(5):2795–803. PMID: 25964559.
60. Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011 18:4 [Internet]. 2011 Feb 11 [cited 2021 Aug 6];18(4):571–80. Available from: https://www.nature.com/articles/cdd2010191
61. Yang L, Zhang X, Li H, Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst [Internet]. 2016 Jul 19 [cited 2021 Aug 6];12(8):2605–12. Available from: https://pubs.rsc.org/en/content/articlehtml/2016/mb/c6mb00114a
62. Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, et al. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:2300. doi: https://doi.org/10.1038/ncomms3300. PMID: 23939249; PMCID: PMC3753544.
63. Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, Pawelek JM. Human cancer biology punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res [Internet]. 2012 [cited 2021 Aug 6];18(2):370–9. Available from: http://clincancerres.aacrjournals.org/
64. Denechaud PD, Fajas L, Giralt A. E2F1, a novel regulator of metabolism. Front Endocrinol (Lausanne). 2017 Nov 10;0(NOV):311.
65. Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog [Internet]. 2015 Jul 1 [cited 2021 Aug 6];54(S1):E1–12. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/mc.22120
66. Ghaleb AM, Yang VW. Krüppel-like factor 4 (KLF4): What we currently know. Gene [Internet]. 2017 May 5 [cited 2021 Aug 6];611:27. Available from: /pmc/articles/PMC5391259/
67. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007 Nov 30;131(5):861–72.
68. Liu Y, Li M, Zhang G, Pang Z. MicroRNA-10b overexpression promotes non-small cell lung cancer cell proliferation and invasion. Eur J Med Res [Internet]. 2013 Nov 12 [cited 2021 Aug 6];18(1). Available from: https://pubmed.ncbi.nlm.nih.gov/24216130/
69. Zhou Y, Tian B, Tang J, Wu J, Wang H, Wu Z, et al. SNHG7: A novel vital oncogenic lncRNA in human cancers. Biomedicine & Pharmacotherapy. 2020 Apr 1;124:109921.
70. Gao YT, Zhou YC. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 7 (SNHG7) promotes breast cancer progression by sponging miRNA-381. Eur Rev Med Pharmacol Sci [Internet]. 2019 [cited 2021 Aug 7];23(15):6588–95. Available from: https://pubmed.ncbi.nlm.nih.gov/31378900/
71. Sun WJ, Zhang YN, Xue P. miR-186 inhibits proliferation, migration, and epithelial-mesenchymal transition in breast cancer cells by targeting Twist1. J Cell Biochem [Internet]. 2019 Jun 1 [cited 2021 Aug 7];120(6):10001–9. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/jcb.28283
72. Zhang H, Zhang XY, Kang XN, Jin LJ, Wang ZY. LncRNA-SNHG7 enhances chemotherapy resistance and cell viability of breast cancer cells by regulating miR-186. Cancer Manag Res [Internet]. 2020 [cited 2021 Aug 7];12:10163–72. Available from: https://pubmed.ncbi.nlm.nih.gov/33116871/
73. Yu Z, Wang Y, Wang B, Zhai J. Metformin affects paclitaxel sensitivity of ovarian cancer cells Through autophagy mediated by long noncoding RNASNHG7/miR-3127-5p Axis. https://home.liebertpub.com/cbr [Internet]. 2020 Jun 9 [cited 2021 Aug 7]; Available from: https://www.liebertpub.com/doi/abs/10.1089/cbr.2019.3390
74. Chen Z, He M, Chen J, Li C, Zhang Q. Long noncoding RNA SNHG7 inhibits NLRP3dependent pyroptosis by targeting the miR34a/SIRT1 axis in liver cancer. Oncol Lett [Internet]. 2020 Jul 1 [cited 2021 Aug 7];20(1):893–901. Available from: http://www.spandidos-publications.com/10.3892/ol.2020.11635/abstract
75. Li ZH, Yu NS, Deng Q, Zhang Y, Hu YY, Liu G, et al. LncRNA SNHG7 mediates the chemoresistance and stemness of breast cancer by sponging miR-34a. Front Oncol. 2020 Nov 24;0:2506.
76. Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med [Internet]. 2013 May 24 [cited 2023 Dec 15];13(2):109–17. Available from: https://link.springer.com/article/10.1007/s10238-012-0186-5
77. Zhu L, Wang X. Integrative network analysis identified master regulatory long non-coding RNAs underlying the squamous subtype of pancreatic ductal adenocarcinoma. Proceedings—2020 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2020. 2020 Dec 16;2936–42.
78. Du C, Zhang JL, Wang Y, Zhang YY, Zhang JH, Zhang LF, et al. The long non-coding RNA LINC01705 regulates the development of breast cancer by sponging miR-186-5p to mediate TPR expression as a competitive endogenous RNA. Front Genet. 2020 Jul 31;0:779.
79. Klec C, Gutschner T, Panzitt K, Pichler M. Involvement of long non-coding RNA HULC (highly up-regulated in liver cancer) in pathogenesis and implications for therapeutic intervention. [Internet]. 2019 Mar 4 [cited 2021 Aug 7];23(3):177–86. Available from: https://www.tandfonline.com/doi/abs/10.1080/14728222.2019.1570499
80. Panzitt K, Tschernatsch MMO, Guelly C, Moustafa T, Stradner M, Strohmaier HM, et al. Characterization of HULC, a novel gene with striking up-regulation in Hepatocellular Carcinoma, as noncoding RNA. Gastroenterology. 2007 Jan 1;132(1):330–42.
81. Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol [Internet]. 2015 [cited 2021 Aug 7];8(3):2994. Available from: /pmc/articles/PMC4440118/
82. Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep [Internet]. 2014 Jan 1 [cited 2021 Aug 7];31(1):358–64. Available from: http://www.spandidos-publications.com/10.3892/or.2013.2850/abstract
83. Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Medical Oncol 2014 31:12 [Internet]. 2014 Nov 21 [cited 2021 Aug 7];31(12):1–7. Available from: https://link.springer.com/article/10.1007/s12032-014-0346-4
84. Ye H, Lin J, Yao X, Li Y, Lin X, Lu H. Overexpression of long non-coding RNA NNT-AS1 correlates with tumor progression and poor pognosis in osteosarcoma. Cellular Physiology and Biochemistry [Internet]. 2018 Mar 1 [cited 2021 Aug 7];45(5):1904–14. Available from: https://www.karger.com/Article/FullText/487966
85. Han R, Feng P, Pang J, Zou D, Li X, Geng C, et al. A novel HCC prognosis predictor EEF1E1 is related to immune infiltration and may be involved in EEF1E1/ATM/p53 signaling. Front Oncol. 2021 Jul 2;0:2590.
86. Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, et al. Elevation of highly up-regulated in liver cancer (HULC) by Hepatitis B Virus X protein promotes hepatoma cell proliferation via Down-regulating p18*. J Biol Chem [Internet]. 2012 Jul 27 [cited 2021 Aug 7];287(31):26302–11. Available from: http://www.jbc.org/article/S0021925820736410/fulltext
87. Heffernan-Stroud LA, Obeid LM. Sphingosine Kinase 1 in cancer. Adv Cancer Res. 2013 Jan 1;117:201–35.
88. Cristiano L. EEF1E1 (eukaryotic translation elongationfactor 1 epsilon 1). Atlas Genet Cytogenet Oncol Haematol. 2020;24(11):387–95.
89. Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in Hepatoma cells through an miR-9–mediated RXRA signaling pathway. Cancer Res [Internet]. 2015 Mar 1 [cited 2021 Aug 7];75(5):846–57. Available from: https://cancerres.aacrjournals.org/content/75/5/846
90. Yu X, Zheng H, Chan MTV, Wu WKK. HULC: an oncogenic long non-coding RNA in human cancer. J Cell Mol Med. 2017 Feb 1;21(2):410–7.
91. Higashi T, Hayashi H, Ishimoto T, Takeyama H, Kaida T, Arima K, et al. miR-9-3p plays a tumour-suppressor role by targeting TAZ (WWTR1) in hepatocellular carcinoma cells. Br J Cancer [Internet]. 2015 Jun 30 [cited 2023 Dec 15];113(2):252–8. Available from: https://www.nature.com/articles/bjc2015170
92. Palmieri G, Paliogiannis P, Sini MC, Manca A, Palomba G, Doneddu V, et al. Long non-coding RNA CASC2 in human cancer. Crit Rev Oncol Hematol [Internet]. 2017 Mar 1 [cited 2022 May 24];111:31–8. Available from: https://pubmed.ncbi.nlm.nih.gov/28259293/
93. Liao Y, Shen L, Zhao H, Liu Q, Fu J, Guo Y, et al. LncRNA CASC2 interacts with miR-181a to modulate glioma growth and resistance to TMZ through PTEN pathway. J Cell Biochem [Internet]. 2017 Jul 1 [cited 2022 May 24];118(7):1889–99. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/jcb.25910
94. Jiang C, Shen F, Du J, Fang X, Li X, Su J, et al. Upregulation of CASC2 sensitized glioma to temozolomide cytotoxicity through autophagy inhibition by sponging miR-193a-5p and regulating mTOR expression. Biomed Pharmacother [Internet]. 2018 Jan 1 [cited 2022 May 24];97:844–50. Available from: https://pubmed.ncbi.nlm.nih.gov/29136760/
95. Ebrahimpour A, Sarfi M, Rezatabar S, Tehrani SS. Novel insights into the interaction between long non-coding RNAs and microRNAs in glioma. Mol Cell Biochem [Internet]. 2021 Jun 1 [cited 2022 May 31];476(6):2317–35. Available from: https://link.springer.com/article/10.1007/s11010-021-04080-x
96. Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J, et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget [Internet]. 2015 Sep 9 [cited 2022 May 31];6(26):21934. Available from: /pmc/articles/PMC4673137/
97. Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature [Internet]. 2015 Sep 16 [cited 2022 Jun 8];526(7575):666–71. Available from: https://www.nature.com/articles/nature15541
98. Ju X, Yang Z, Zhang H, Wang Q. Role of pyroptosis in cancer cells and clinical applications. Biochimie. 2021 Jun 1;185:78–86.
99. Liu J, Gao L, Zhu X, Geng R, Tao X, Xu H, et al. Gasdermin D is a novel prognostic biomarker and relates to TMZ response in Glioblastoma. Cancers (Basel) [Internet]. 2021 Nov 10 [cited 2022 Jun 8];13(22):5620. Available from: https://www.mdpi.com/2072-6694/13/22/5620/htm
100. Zi H, Tuo Z, He Q, Meng J, Hu Y, Li Y, et al. Comprehensive bioinformatics analysis of Gasdermin family of Glioma. Comput Intell Neurosci. 2022;2022:1–19.
101. Xiong X, Liu J, He Q, Dai R, Zhang H, Cao Z, et al. Long non-coding RNA NORAD aggravates acute myocardial infarction by promoting fibrosis and apoptosis via miR-577/COBLL1 axis. Environ Toxicol [Internet]. 2021 Nov 1 [cited 2022 Jun 10];36(11):2256–65. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/tox.23339
102. Zhao X, Jiang M, Wang Z. TPM4 promotes cell migration by modulating F-actin formation in lung cancer. Onco Targets Ther [Internet]. 2019 [cited 2022 Jun 10];12:4055. Available from: /pmc/articles/PMC6554522/
103. Qi C, Liu J, Guo P, Xu Y, Hu J, Han X. LncRNA NORAD facilitates oral squamous cell carcinoma progression by sponging miR-577 to enhance TPM4. Biol Direct [Internet]. 2022 Dec 1 [cited 2022 Jun 10];17(1):1–10. Available from: https://biologydirect.biomedcentral.com/articles/10.1186/s13062-021-00299-2
104. Razavi H, Katanforosh A. Identification of novel key regulatory lncRNAs in gastric adenocarcinoma. BMC Genomics [Internet]. 2022 Dec 1 [cited 2022 Jun 10];23(1):1–14. Available from: https://link.springer.com/articles/10.1186/s12864-022-08578-6
105. Li X, Xiao X, Chang R, Zhang C. Comprehensive bioinformatics analysis identifies lncRNA HCG22 as a migration inhibitor in esophageal squamous cell carcinoma. J Cell Biochem [Internet]. 2020 Jan 1 [cited 2022 Jun 10];121(1):468–81. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/jcb.29218
106. Fu Y, Liu Y, Nasiroula A, Wang Q, Cao X. Long noncoding RNA HCG22 inhibits the proliferation, invasion and migration of oral squamous cell carcinoma cells by downregulating miR4255p expression. Exp Ther Med [Internet]. 2022 Mar 1 [cited 2022 Jun 10];23(3):1–10. Available from: http://www.spandidos-publications.com/10.3892/etm.2022.11171/abstract
107. Fang F, Song T, Zhang T, Cui Y, Zhang G, Xiong Q, et al. MiR-425-5p promotes invasion and metastasis of hepatocellular carcinoma cells through SCAI-mediated dysregulation of multiple signaling pathways. Oncotarget [Internet]. 2017 Mar 7 [cited 2022 Jun 10];8(19):31745–57. Available from: https://www.oncotarget.com/article/15958/text/
108. Cristóbal I, Madoz-Gúrpide J, Rojo F, García-Foncillas J. Potential therapeutic value of miR-425-5p in metastatic colorectal cancer. J Cell Mol Med [Internet]. 2016 Nov 1 [cited 2022 Jun 10];20(11):2213–4. Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/jcmm.12902
109. Sun L, Jiang R, Li J, Wang B, Ma C, Lv Y, et al. MicoRNA-425-5p is a potential prognostic biomarker for cervical cancer. Ann Clin Biochem [Internet]. 2017 Jan 1 [cited 2022 Jun 10];54(1):127–33. Available from: https://pubmed.ncbi.nlm.nih.gov/27166306/
110. Soghli N, Yousefi T, Abolghasemi M, Qujeq D. NORAD, a critical long non-coding RNA in human cancers. Life Sci. 2021 Jan 1;264:118665.
111. Zhang H, Guo H. Long non-coding RNA NORAD induces cell proliferation and migration in prostate cancer. Journal of International Medical Research [Internet]. 2019 Aug 1 [cited 2022 Jun 10];47(8):3898–904. Available from: https://journals.sagepub.com/doi/full/10.1177/0300060519862076
112. Zahra K, Dey T, Ashish, Mishra SP, Pandey U. Pyruvate kinase M2 and cancer: the role of PKM2 in promoting tumorigenesis. Front Oncol [Internet]. 2020 Mar 2 [cited 2022 Jun 10];10:159. Available from: https://www.frontiersin.org/articles/10.3389/fonc.2020.00159/full
113. Dai J, Escara-Wilke J, Keller JM, Jung Y, Taichman RS, Pienta KJ, et al. Primary prostate cancer educates bone stroma through exosomal pyruvate kinase M2 to promote bone metastasis. J Exp Med [Internet]. 2019 Dec 12 [cited 2022 Jun 10];216(12):2899. Available from: /pmc/articles/PMC6888980/
114. Ronquist KG, Sanchez C, Dubois L, Chioureas D, Fonseca P, Larsson A, et al. Energy-requiring uptake of prostasomes and PC3 cell-derived exosomes into non-malignant and malignant cells. J Extracell Vesicles [Internet]. 2016 Jan 1 [cited 2022 Jun 10];5(1). Available from: /pmc/articles/PMC4783432/
115. Agliardi C, Meloni M, Guerini FR, Zanzottera M, Bolognesi E, Baglio F, et al. Oligomeric α-Syn and SNARE complex proteins in peripheral extracellular vesicles of neural origin are biomarkers for Parkinson’s disease. Neurobiol Dis. 2021 Jan 1;148:105185.
116. Wei Y, Wang D, Jin F, Bian Z, Li L, Liang H, et al. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat Commun [Internet]. 2017 Jan 9 [cited 2022 Jun 10];8:14041. Available from: /pmc/articles/PMC5228053/
117. Hu CY, Chen J, Qin XH, You P, Ma J, Zhang J, et al. Long non-coding RNA NORAD promotes the prostate cancer cell extracellular vesicle release via microRNA-541-3p-regulated PKM2 to induce bone metastasis of prostate cancer. J Exp Clin Cancer Res [Internet]. 2021 Dec 1 [cited 2022 Jun 10];40(1):1–16. Available from: https://jeccr.biomedcentral.com/articles/10.1186/s13046-021-01891-0
118. Zhou Z, Wu X, Zhou Y, Yan W. Long non-coding RNA ADAMTS9-AS1 inhibits the progression of prostate cancer by modulating the miR-142-5p/CCND1 axis. J Gene Med [Internet]. 2021 May 1 [cited 2022 Jun 10];23(5):e3331. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/jgm.3331
119. van der Worp HB, van Gijn J. Clinical practice. Acute ischemic stroke. N Engl J Med [Internet]. 2007 Oct 9 [cited 2021 Aug 8];357(6):572–9. Available from: https://www.nejm.org/doi/full/10.1056/nejmcp072057
120. Phipps MS, Cronin CA. Management of acute ischemic stroke. BMJ [Internet]. 2020 Feb 13 [cited 2021 Aug 8];368. Available from: https://www.bmj.com/content/368/bmj.l6983
121. Arslan S, Berkan Ö, Lalem T, Özbilüm N, Göksel S, Korkmaz Ö, et al. Long non-coding RNAs in the atherosclerotic plaque. Atherosclerosis [Internet]. 2017 Nov 1 [cited 2021 Aug 8];266:176–81. Available from: http://www.atherosclerosis-journal.com/article/S0021915017313321/fulltext
122. Wolska M, Jarosz-Popek J, Junger E, Wicik Z, Porshoor T, Sharif L, et al. Long non-coding RNAs as promising therapeutic approach in ischemic stroke: a comprehensive review. Molecular Neurobiology 2020 58:4 [Internet]. 2020 Nov 24 [cited 2021 Aug 8];58(4):1664–82. Available from: https://link.springer.com/article/10.1007/s12035-020-02206-8
123. Li X, Dai Y, Yan S, Shi Y, Han B, Li J, et al. Down-regulation of lncRNA KCNQ1OT1 protects against myocardial ischemia/reperfusion injury following acute myocardial infarction. Biochem Biophys Res Commun. 2017 Sep 30;491(4):1026–33.
124. Yang H, Wang H, Zhang X, Yang Y, Li H. Upregulated LINC00319 aggravates neuronal injury induced by oxygenglucose deprivation via modulating miR200a3p. Exp Ther Med [Internet]. 2021 Aug 1 [cited 2021 Aug 8];22(2):1–10. Available from: http://www.spandidos-publications.com/10.3892/etm.2021.10276/abstract
125. Xie C, Ginet V, Sun Y, Koike M, Zhou K, Li T, et al. Neuroprotection by selective neuronal deletion of Atg7 in neonatal brain injury. Autophagy [Internet]. 2016 Jan 1 [cited 2021 Aug 8];12(2):410–23. Available from: https://www.tandfonline.com/doi/abs/10.1080/15548627.2015.1132134
126. Huang X, Bai HM, Chen L, Li B, Lu YC. Reduced expression of LC3B-II and Beclin 1 in glioblastoma multiforme indicates a down-regulated autophagic capacity that relates to the progression of astrocytic tumors. Journal of Clinical Neuroscience. 2010 Dec 1;17(12):1515–9.
127. Wang J, Zhao H, Fan Z, Li G, Ma Q, Tao Z, et al. Long noncoding RNA H19 promotes neuroinflammation in ischemic stroke by driving histone deacetylase 1-Dependent M1 microglial polarization. Stroke [Internet]. 2017 Aug 1 [cited 2022 Jun 15];48(8):2211–21. Available from: https://www.ahajournals.org/doi/abs/10.1161/STROKEAHA.117.017387
128. Wang J, Zhu M, Ye L, Chen C, She J, Song Y. MiR-29b-3p promotes particulate matter-induced inflammatory responses by regulating the C1QTNF6/AMPK pathway. Aging (Albany NY) [Internet]. 2020 Jan 1 [cited 2022 Jun 15];12(2):1158. Available from: /pmc/articles/PMC7053628/
129. Zhai Y, Pang Y. Systemic and ovarian inflammation in women with polycystic ovary syndrome. J Reprod Immunol. 2022 Jun 1;151:103628.
130. Li G, Ma X, Zhao H, Fan J, Liu T, Luo Y, et al. Long non-coding RNA H19 promotes leukocyte inflammation in ischemic stroke by targeting the miR-29b/C1QTNF6 axis. CNS Neurosci Ther [Internet]. 2022 Jun 1 [cited 2022 Jun 15];28(6):953–63. Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/cns.13829
131. Nallamshetty S, Chan SY, Loscalzo J. Hypoxia: a master regulator of microRNA biogenesis and activity. Free Radic Biol Med. 2013 Sep 9;64:20–30.
132. Zeng L, Liu J, Wang Y, Wang L, Weng S, Tang Y, et al. MicroRNA-210 as a novel blood biomarker in acute cerebral ischemia. Frontiers in Bioscience—Elite [Internet]. 2011 Jan 6 [cited 2023 Dec 13];3 E(4):1265–72. Available from: https://www.imrpress.com/journal/FBE/3/4/10.2741/E330
133. Li Y, Song R, Shen G, Huang L, Xiao D, Ma Q, et al. MicroRNA-210 Downregulates TET2 (Ten-Eleven Translocation Methylcytosine Dioxygenase 2) and Contributes to Neuroinflammation in Ischemic Stroke of Adult Mice. Stroke. 2023 Mar;54(3):857–67.
134. Carrillo-Jimenez A, Deniz Ö, Niklison-Chirou MV, Ruiz R, Bezerra-Salomão K, Stratoulias V, et al. TET2 regulates the neuroinflammatory response in Microglia. Cell Rep. 2019 Oct 15;29(3):697–713.e8.
135. Ma Q, Dasgupta C, Shen G, Li Y, Zhang L. MicroRNA-210 downregulates TET2 and contributes to inflammatory response in neonatal hypoxic-ischemic brain injury. J Neuroinflammation [Internet]. 2021 Dec 1 [cited 2023 Dec 13];18(1):1–14. Available from: https://jneuroinflammation.biomedcentral.com/articles/10.1186/s12974-020-02068-w
136. Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation [Internet]. 2002 Mar 5 [cited 2021 Aug 8];105(9):1135–43. Available from: https://www.ahajournals.org/doi/abs/10.1161/hc0902.104353
137. Chen YJ, Lin TL, Cai Z, Yan CH, Gou SR, Zhuang YD. Assessment of acute pancreatitis severity via determination of serum levels of hsamiR1265p and IL6. Exp Ther Med [Internet]. 2021 Jan 1 [cited 2021 Aug 8];21(1):1–1. Available from: http://www.spandidos-publications.com/10.3892/etm.2020.9458/abstract
138. Hao XZ, Fan HM. Identification of miRNAs as atherosclerosis biomarkers and functional role of miR-126 in atherosclerosis progression through MAPK signalling pathway. Eur Rev Med Pharmacol Sci. 2017 Jun;21(11):2725–33.
139. Kyriakis JM, Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-Year update. [Internet]. 2012 Apr 1 [cited 2021 Aug 8];92(2):689–737. Available from: https://journals.physiology.org/doi/abs/10.1152/physrev.00028.2011
140. Du H, Yang L, Zhang H, Zhang X, Shao H. LncRNA TUG1 silencing enhances proliferation and migration of ox-LDL-treated human umbilical vein endothelial cells and promotes atherosclerotic vascular injury repairing via the Runx2/ANPEP axis. Int J Cardiol. 2021 Sep 1;338:204–14.
141. Lin J, Feng X, Zhang J, Tong Z. Long noncoding RNA TUG1 promotes airway smooth muscle cells proliferation and migration via sponging miR-590-5p/FGF1 in asthma. Am J Transl Res [Internet]. 2019 [cited 2021 Aug 8];11(5):3159. Available from: /pmc/articles/PMC6556671/
142. Li FP, Lin DQ, Gao LY. LncRNA TUG1 promotes proliferation of vascular smooth muscle cell and atherosclerosis through regulating miRNA-21/PTEN axis. Eur Rev Med Pharmacol Sci [Internet]. 2018 [cited 2021 Aug 8];22(21):7439–47. Available from: https://pubmed.ncbi.nlm.nih.gov/30468492/
143. Zhang R, Hao Y, Zhang J. The lncRNA DANCR promotes development of atherosclerosis by regulating the miR-214-5p/COX20 signaling pathway. Cell Mol Biol Lett [Internet]. 2022 Dec 1 [cited 2022 Jun 15];27(1):1–15. Available from: https://link.springer.com/articles/10.1186/s11658-022-00310-2
144. Keerthiraju E, Du C, Tucker G, Greetham D. A Role for COX20 in tolerance to oxidative stress and programmed cell death in saccharomyces cerevisiae. Microorganisms [Internet]. 2019 Nov 1 [cited 2022 Jun 15];7(11). Available from: /pmc/articles/PMC6920987/
145. An F, Yin Y, Ju W. Long noncoding RNA DANCR expression and its predictive value in patients with atherosclerosis. Bioengineered [Internet]. 2022 Mar 2 [cited 2022 Jun 15];13(3):6919–28. Available from: https://www.tandfonline.com/doi/abs/10.1080/21655979.2022.2033408
146. Sun D, Ma T, Zhang Y, Zhang F, Cui B. Overexpressed miR-335-5p reduces atherosclerotic vulnerable plaque formation in acute coronary syndrome. J Clin Lab Anal [Internet]. 2021 Feb 1 [cited 2022 Jun 15];35(2):e23608. Available from: /pmc/articles/PMC7891542/
147. Yang X, Wang P, Yan S, Wang G. Study on potential differentially expressed genes in stroke by bioinformatics analysis. Neurol Sci [Internet]. 2022 Feb 1 [cited 2022 Jun 15];43(2):1155–66. Available from: https://link.springer.com/article/10.1007/s10072-021-05470-1
148. Peng K, Jiang P, Du Y, Zeng D, Zhao J, Li M, et al. Oxidized low-density lipoprotein accelerates the injury of endothelial cells via circ-USP36/miR-98-5p/VCAM1 axis. IUBMB Life [Internet]. 2021 Jan 1 [cited 2022 Jun 20];73(1):177–87. Available from: https://pubmed.ncbi.nlm.nih.gov/33249762/
149. Ernst S, Heinzelmann J, Bohle RM, Weber G, Stöckle M, Junker K, et al. The metastatic potential of seminomatous germ cell tumours is associated with a specific microRNA pattern. Andrology [Internet]. 2020 Nov 1 [cited 2022 Jun 20];8(6):1687–98. Available from: https://pubmed.ncbi.nlm.nih.gov/32530514/
150. Jang HY, Lim SM, Lee HJ, Hong JS, Kim GJ. Identification of microRNAs and their target genes in the placenta as biomarkers of inflammation. Clin Exp Reprod Med [Internet]. 2020 Mar 1 [cited 2022 Jun 20];47(1):53. Available from: /pmc/articles/PMC7127901/
151. Barchetta I, Ceccarelli V, Cimini FA, Bertoccini L, Fraioli A, Alessandri C, et al. Impaired bone matrix glycoprotein pattern is associated with increased cardio-metabolic risk profile in patients with type 2 diabetes mellitus. J Endocrinol Invest [Internet]. 2019 May 1 [cited 2022 Jun 20];42(5):513–20. Available from: https://pubmed.ncbi.nlm.nih.gov/30132286/
152. Goikuria H, del Mar Freijo M, Manrique RV, Sastre M, Elizagaray E, Lorenzo A, et al. Characterization of carotid smooth muscle cells during phenotypic transition. Cells [Internet]. 2018 Mar 18 [cited 2022 Jun 20];7(3):23. Available from: https://www.mdpi.com/2073-4409/7/3/23/htm
153. Li MP, Hao ZC, Yan MQ, Xia CL, Wang ZH, Feng YQ. Possible causes of atherosclerosis: lncRNA COLCA1 induces oxidative stress in human coronary artery endothelial cells and impairs wound healing. Ann Transl Med [Internet]. 2022 Mar [cited 2022 Jun 20];10(6). Available from: https://atm.amegroups.com/article/view/91408/html
154. Epstein FH, Fuster V, Badimon L, Badimon JJ, Chesebro JH. The pathogenesis of coronary artery disease and the acute coronary syndromes (2). N Engl J Med [Internet]. 1992 Jan 30 [cited 2021 Aug 9];326(5):310–8. Available from: https://pubmed.ncbi.nlm.nih.gov/1728735/
155. Shan Z, Qin S, Li W, Wu W, Yang J, Chu M, et al. An Endocrine genetic signal between blood cells and vascular smooth muscle cells: role of MicroRNA-223 in smooth muscle function and atherogenesis. J Am Coll Cardiol [Internet]. 2015 Jun 16 [cited 2021 Aug 9];65(23):2526–37. Available from: https://pubmed.ncbi.nlm.nih.gov/26065992/
156. Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature [Internet]. 2008 Feb 28 [cited 2021 Aug 9];451(7182):1125–9. Available from: https://pubmed.ncbi.nlm.nih.gov/18278031/
157. Vickers KC, Landstreet SR, Levin MG, Shoucri BM, Toth CL, Taylor RC, et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc Natl Acad Sci USA [Internet]. 2014 Oct 7 [cited 2021 Aug 9];111(40):14518–23. Available from: https://pubmed.ncbi.nlm.nih.gov/25246565/
158. Kura B, Kalocayova B, Devaux Y, Bartekova M. Potential clinical implications of miR-1 and miR-21 in heart disease and cardioprotection. Int J Mol Sci [Internet]. 2020 Jan 21 [cited 2022 Jun 20];21(3):700. Available from: https://www.mdpi.com/1422-0067/21/3/700/htm
159. Ono K. MicroRNAs and cardiovascular remodeling. In: Santulli G, editor. microRNA: medical evidence advances in experimental medicine and biology [Internet]. Springer New York LLC; 2015 [cited 2022 Jun 20]. p. 197–213. Available from: https://link.springer.com/chapter/10.1007/978-3-319-22671-2_10
160. Chouvarine P, Legchenko E, Geldner J, Riehle C, Hansmann G. Hypoxia drives cardiac miRNAs and inflammation in the right and left ventricle. J Mol Med [Internet]. 2019 Oct 1 [cited 2023 Dec 13];97(10):1427–38. Available from: https://link.springer.com/article/10.1007/s00109-019-01817-6
161. Goldfinger JZ, Halperin JL, Marin ML, Stewart AS, Eagle KA, Fuster V. Thoracic aortic aneurysm and dissection. J Am Coll Cardiol. 2014 Oct 21;64(16):1725–39.
162. Ernst CB. Abdominal Aortic Aneurysm. [Internet]. 2010 Jan 15 [cited 2021 Aug 9];328(16):1167–72. Available from: https://www.nejm.org/doi/full/10.1056/NEJM199304223281607
163. Taylor BV, Kalman PG. Saccular aortic aneurysms. Annals Vascular Surg 1999;13:6 [Internet]. 2014 May 1 [cited 2021 Aug 9];13(6):555–9. Available from: https://link.springer.com/article/10.1007/s100169900297
164. Roberts CS, Roberts WC. Combined thoracic aortic dissection and abdominal aortic fusiform aneurysm. Ann Thorac Surg. 1991 Sep 1;52(3):537–40.
165. Choke E, Cockerill G, Wilson WRW, Sayed S, Dawson J, Loftus I, et al. A review of biological factors implicated in abdominal aortic aneurysm rupture. Eur J Vascular Endovascular Surg. 2005 Sep 1;30(3):227–44.
166. Tian L, Hu X, He Y, Wu Z, Li D, Zhang H. Construction of lncRNA-miRNA-mRNA networks reveals functional lncRNAs in abdominal aortic aneurysm. Exp Ther Med [Internet]. 2018 Nov 1 [cited 2021 Aug 9];16(5):3978–86. Available from: https://pubmed.ncbi.nlm.nih.gov/30344676/
167. Cai B, Yang B, Huang D, Wang D, Tian J, Chen F, et al. STAT3-induced up-regulation of lncRNA NEAT1 as a ceRNA facilitates abdominal aortic aneurysm formation by elevating TULP3. Biosci Rep [Internet]. 2020 Jan 31 [cited 2021 Jul 22];40(1):20193299. Available from: /bioscirep/article/40/1/BSR20193299/221717/STAT3-induced-up-regulation-of-lncRNA-NEAT1-as-a
168. Fernandez-García CE, Tarin C, Roldan-Montero R, Martinez-Lopez D, Torres-Fonseca M, Lindhot JS, et al. Increased galectin-3 levels are associated with abdominal aortic aneurysm progression and inhibition of galectin-3 decreases elastase-induced AAA development. Clinical Science (London) [Internet]. 2017 Nov 15 [cited 2022 Jun 20];131(22):2707–19. Available from: /clinsci/article/131/22/2707/71699/Increased-galectin-3-levels-are-associated-with
169. Chen J, Qian C, Duan H, Cao S, Yu X, Li J, et al. Melatonin attenuates neurogenic pulmonary edema via the regulation of inflammation and apoptosis after subarachnoid hemorrhage in rats. J Pineal Res [Internet]. 2015 Nov 1 [cited 2022 Jun 20];59(4):469–77. Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/jpi.12278
170. Goksu E, Dogan O, Ulker P, Tanriover G, Konuk E, Dilmac S, et al. Pentoxifylline alleviates early brain injury in a rat model of subarachnoid hemorrhage. Acta Neurochir (Wien) [Internet]. 2016 Sep 1 [cited 2022 Jun 20];158(9):1721–30. Available from: https://link.springer.com/article/10.1007/s00701-016-2866-5
171. Yang B, Wang X, Ying C, Peng F, Xu M, Chen F, et al. Long Noncoding RNA SNHG16 facilitates abdominal aortic aneurysm progression through the miR-106b-5p/STAT3 Feedback Loop. J Atheroscler Thromb [Internet]. 2021 Jan 1 [cited 2022 Jun 20];28(1):66–78. doi: http://doi.org/10.5551/jat.52274
172. Filion M, Tremblay L. Abnormal spontaneous activity of globus pallidus neurons in monkeys with MPTP-induced parkinsonism. Brain Res. 1991 Apr 26;547(1):140–4.
173. Cao B, Wang T, Qu Q, Kang T, Yang Q. Long noncoding RNA SNHG1 promotes neuroinflammation in Parkinson’s disease via regulating miR-7/NLRP3 Pathway. Neuroscience. 2018 Sep 15;388:118–27.
174. Mao Z, Liu C, Ji S, Yang Q, Ye H, Han H, et al. The NLRP3 Inflammasome is involved in the pathogenesis of Parkinson’s $$. Neurochem Res [Internet]. 2017 Apr 1 [cited 2021 Aug 10];42(4):1104–15. Available from: https://pubmed.ncbi.nlm.nih.gov/28247334/
175. Mucke L. Alzheimer’s disease. Nature [Internet]. 2009 Oct 14 [cited 2021 Aug 10];461(7266):895–7. Available from: https://www.nature.com/articles/461895a
176. Mayeux R, Honig LS, Tang MX, Manly J, Stern Y, Schupf N, et al. Plasma Aβ40 and Aβ42 and Alzheimer’s disease. Neurology [Internet]. 2003 Nov 11 [cited 2021 Aug 10];61(9):1185–90. Available from: https://n.neurology.org/content/61/9/1185
177. Wang X, Zhang M, Liu H. LncRNA17A regulates autophagy and apoptosis of SH-SY5Y cell line as an in vitro model for Alzheimer’s disease. Biosci Biotechnol Biochem [Internet]. 2019 Apr 3 [cited 2021 Aug 10];83(4):609–21. Available from: https://academic.oup.com/bbb/article/83/4/609/5938020
178. Lee ST, Chu K, Jung KH, Kim JH, Huh JY, Yoon H, et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann Neurol [Internet]. 2012 Aug 1 [cited 2021 Aug 10];72(2):269–77. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/ana.23588
179. Insel TR. Rethinking schizophrenia. Nature 2010 468:7321 [Internet]. 2010 Nov 10 [cited 2021 Aug 10];468(7321):187–93. Available from: https://www.nature.com/articles/nature09552
180. Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nature Genetics 2011 43:10 [Internet]. 2011 Sep 18 [cited 2021 Aug 10];43(10):969–76. Available from: https://www.nature.com/articles/ng.940
181. Wright C, Turner JA, Calhoun VD, Perrone Bizzozero N. Potential impact of miR-137 and its targets in Schizophrenia. Front Genet. 2013;0:58.
182. Li R, Wang Q, Qiu Y, Meng Y, Wei L, Wang H, et al. A potential autophagy-related competing endogenous RNA network and corresponding diagnostic efficacy in Schizophrenia. Front Psychiatry. 2021 Feb 23;0:187.
183. Sabaie H, Moghaddam MM, Moghaddam MM, Ahangar NK, Asadi MR, Hussen BM, et al. Bioinformatics analysis of long non-coding RNA-associated competing endogenous RNA network in schizophrenia. Sci Rep [Internet]. 2021 Dec 24 [cited 2022 Jun 20];11(1):1–13. Available from: https://www.nature.com/articles/s41598-021-03993-3