INTRODUCTION
The current use of herbs as complementary and/or alternative medicines has become increasingly popular worldwide. Herbs have been shown to be beneficial in several studies, including in treating type 2 diabetes mellitus (DM). DM is a chronic metabolic disease characterized by elevated blood glucose levels, which over time, causes serious damage to the heart, blood vessels, eyes, kidneys, and nerves. The International Diabetes Federation estimates that at least 537 million people aged 20–79 suffer from DM, which is expected to continue to rise until 2,045. In addition, the number of children and adolescents (i.e., up to 19 years old) living with diabetes also increases yearly. Meanwhile, Indonesia is ranked 5th among the countries with the highest incidence rates worldwide, with the percentage of DM sufferers reaching 19.5 million in 2021 [1]. The high incidence of DM requires an effective treatment option.
Mangosteen rind is one of Indonesia’s natural resources. Research on the mangosteen rind has been growing rapidly. The mangosteen plant has been proven to contain various kinds of medicinal substances, especially the skin of the fruit. This shows that this sample has great potential to be developed as a medicinal ingredient. According to the results of research by Ratwita et al. [2], the alpha-mangostin and xanthone compounds found in the mangosteen rind could be efficacious as antidiabetics by increasing the expression of GLUT-4 and PPAR-g. Meanwhile, based on the research of Husen et al. [3], the gamma-mangostin compound significantly decreased plasma BUN and creatinine and significantly improve renal proximal tubular cell damage in diabetic rats. The ethanol extract of mangosteen rind (EEMR) could also inhibit pancreatic lipase and α-amylase enzymes [4], which were considered to be closely related to the antidiabetic effect. In addition, EEMR also showed other effects related to DM in the form of decreasing blood glucose, increasing insulin tolerance and production, improving the structure and diameter of pancreatic beta cells, increasing glycogen and hepatic lipids, increasing HDL and total protein levels, and repairing oxidative damage [5–8]. EEMR can also be used to stop hyperlipidemia and obesity, which are both risk factors for diabetes [9].
SGLT-2 is one of the latest targets for the treatment of DM. SGLT-2 inhibitors, through a non-insulin-dependent mechanism of action, have other advantages, including lowering blood pressure and body weight [10]. This makes research on the effect of EEMR as an antidiabetic mellitus agent through this pathway interesting to study.
MATERIALS AND METHODS
In silico
Homology modeling and ligand preparation
Comparative modeling of hSGLT-2 was carried out. First, the sequence of protein hSGLT-2 (UniProtKB ID: P31639) was retrieved from the UniProt database (https://www.uniprot.org/). Then the structure of hSGLT-2 was predicted using I-TASSER [11]. In the final step, the resulting model with the highest C-score and lowest root-mean-square deviation (RMSD) value was chosen as the receptor structure for the docking study.
The ligand structures were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/compound/) database. alpha-mangostin (CID: 5281650), gamma-mangostin (CID: 464078), and xanthone CIDf: f?7020 structures were saved as 3D SDF format data. Then the minimization structures of ligands were performed using the Avogadro program [12] by setting the GAFF force field and number of steps per update of 100 and the algorithm of conjugate gradients. The final ligand structures were saved in PDB data format.
Molecular docking
Molecular docking was performed by using AutoDock 4.2 [13]. Firstly, the target pocket of the hSGLT-2 receptor was predicted using the RaptorX server (http://raptorx.uchicago.edu/BindingSite) [14]. Then the grid box was set to the predicted binding site with a center_x = 68.565, center_y = 71.303, center_z = 75.611, a size of 20 × 20 × 20, and a grid spacing of 1 Å. The default protocol was applied to other parameters. Visualization of docking results was performed using Discovery Studio Visualizer.
Molecular dynamic (MD) simulation
MD simulations on hSGLT-2 complexes, obtained from the best docking conformation, were performed using the graphics processing unit version of the PMEMD engine provided with the Amber 18 package [15]. The ff14SB force field [16] was used to parameterize the protein, while ligand parameterization was done using the GAFF force field. All systems were immersed in a truncated octahedral box using TIP3P water molecules with a buffer setting of 8 Å between the atom in the complex and the edge of the box. The Na+ and Cl- as counterions were added to neutralize the system. The minimization of the complex system was performed in two stages, with bad contacts removed using the pmemd module in Amber18. In the first stage, 10,000 steps of minimization was implemented, comprising 2,000 steps of the steepest descent method followed by 8,000 steps of the conjugated gradient method with the position restraints of 10 kcal mol−1 Å−2. The second stage of minimization was carried out unrestrained. The SHAKE algorithm was used to constrain all bonds involving hydrogen atoms. The system was heated gradually from 0 to 310 K and equilibrated for 250 ps at 310 K and 1 atm pressure [17]. Finally, full MD simulations for 50 ns were performed in the NPT ensemble at a time step of 2.0 fs. The RMSD, root-mean-square fluctuation (RMSF), and Rg (radius of gyration) of protein (residue 23–547) were calculated using the CPPTRAJ program in AmberTools [18]. The graphs were plotted using XMGRACE [19].
In vivo
Antihyperglycemic
The animal study protocol was approved by the Institutional Review Board (or Ethics Committee) of the HEALTH POLYTECHNIC MINISTRY OF HEALTH BANDUNG (protocol code 02/KEPK/EC/II/2022 and February 22, 2022). Preexperimental fasting was performed on rats with Wistar ages 6–8 weeks (150–200 g) before the experiment began and it was feasible to provide water to the rats on an ad libitum basis. Induction of experimental DM was carried out using a high-fat diet (HFD) with a composition of 40% fat, 32% carbohydrates, 28% protein, and streptozotocin 98% (Sigma-AldrichÒ) as much as 35 mg/kg bw in citrate buffer (pH 4.5, 0.1 M). HFD was fed for 4 weeks; then, streptozotocin was administered in a single dose streptozotocin (STZ). For this study, diabetic mice were defined as having fasting blood glucose levels of more than 200 mg/dl. A glucometer (AutoCheckÒ) was used to measure DM levels one day after STZ administration [20,21]. Rats were used in the study. Those rats were divided into seven groups of six animals each. Every week, their blood sugar levels were measured.
1. The control group was given Na.CMC at 0.3% orally.
2. Group 2 was given EEMR at a dose of 100 mg/kg bw orally.
3. Group 3 was given EEMR at a dose of 200 mg/kg bw orally.
4. Group 4 was given EEMR at a dose of 400 mg/kg bw orally.
5. Group 5 was given metformin at a dose of 76.5 mg/kg bw orally.
6. Group 6 was given pioglitazone at a dose of 2.7 mg/kg bw orally.
7. Group 7 was given empagliflozin at a dose of 2.25 mg/kg bw orally.
RT-qPCR analysis for SGLT 2 gene expression
The rat kidney was subjected to RT-qPCR analysis to determine the level of expression of a specific SGLT-2 gene. Quick-RNA MiniPrep Plus was used for RNA extraction using ReverTra Ace qPCR RT Master Mix with gDNA Remover to perform reverse transcription.
A total of 2 l of total RNA was collected and placed in a 0.2 ml tube. The 4X DN Master Mix 2 l was mixed with Nuclease Free Water 7 l, then incubated at 37°C for 5 minutes. They then added up to 2 l of 5 × RT Master Mix II, spun it down, and incubated it at 37°C for 15 minutes, 50°C for 5 minutes, and 98°C for 5 minutes. cDNA was stored at −20°C to be continued in the qPCR stage using Sybr Green Toyobo using the qTOWER (AnalitykJena) tool. To analyze SGLT-2 gene expression, primers rSGLT-2 (rSlc5a2) with primer/probe sequences of rSGLT-2 F (CAT TGT CTC AGG CTG GCA CTG G) and rSGLT-2 R (GGT GTT CAT TGT GGC AGT GTC C) were used. The PCR reaction was carried out under thermocycling conditions of 95°C for 1 minute, then 95°C for 15 seconds, and 55°C for 1 minute. Each was carried out for 40 cycles. Target transcript amplification was performed as part of the multiplex reaction. b-Actin was used as an internal control with primer/probe sequences of beta-actin F (GAA GTG TGA CGT TGA CAT CC) and b-actin R (GAA AGG GTG TAA AAC GCA GC). SGLT-2 gene expression was calculated as a relative value to the reference gene (housekeeping gene) b-actin, following the method of Schmittgen et al. [22] as in the following equations:
ΔCt test = Ct test target – Ct ref test. (1)
ΔCt calibrator = Ct target calibrator – Ct ref calibrator. (2)
ΔΔCt = ΔCt test – Ct calibrator. (3)
Relative expression of genes = 2.ΔΔCt.
Enzyme-linked immunosorbent assay (ELISA) analysis of SGLT-2 protein expression
The ELISA method was used to determine the level of SGLT-2 expression in blood serum collected on the final day of the study. The blood serum was centrifuged at 2,000–3,000 rpm for 20 minutes after being left at room temperature for 10–20 minutes.
Before the experiment was conducted, the ELISA rat SGLT-2 (Bioassay Technology Laboratory) kit’s reagents were kept at room temperature. A standard stock solution of 12 ng/ml was created by combining 20 l of standard (24 ng/ml) with 120 l of standard diluent. Allow it to standardize for 15 minutes with gentle stirring before making dilution. A duplicate standard point was prepared by diluting the standard stock solution sequentially (12 ng/ml) by 1:2 with the standard diluent to produce a solution of 6, 3, 1.5, and 0.75 ng/ml. The standard diluent functions as a zero standard (0 ng/ml).
The next step was filling each well with a standard volume of 50 l. Both samples and standards were loaded with 40 l of samples and 10 lof antibodies against MBP/MBL. It was covered with a sealer after thoroughly mixing. After that, it was placed in a 37°C incubator for 60 minutes. Wells were cleaned 5times with a washing buffer after the sealer was removed. For each cleaning, at least 0.35 ml of washing buffer was applied to the wells for 30 seconds to 1 minute. Finally, 50 l of substrate solution A and 50 l of substrate solution B were added to each well, and the mixture was stirred until diluted. It was incubated at 37°C for 10 minutes in the dark. Each well should be filled with 50 l of stop solution. The blue color would change to yellow almost immediately. The OD value of each well was immediately measured with a microplate reader at 450 nm within 10 minutes of adding the stop solution.
Statistical analysis
Two-way ANOVA and Tukey’s post hoc follow-up tests were used to determine a significant difference (p < 0.05) between the test groups.
RESULTS
In silico
Homology modeling and ligand preparation
This protein sequence query from UniProt yielded a 31.84% similarity to the vSGLT template (GDP ID: 2XQ2). The iTASSER server was then used to model the hSGLT-2 protein from the selected sequences. The model with the lowest C-score was chosen based on the predicted three-dimensional structure of the resulting protein. The SWISS-MODEL server was then used to examine the protein structure in greater depth. There were 71.36% Ramachandran Favored and 12.34% outliers, according to the results of the analysis of the simulated structure. For the docking process, they compared the MolProbity value of the modeled hSGLT-2 protein to the vSGLT template, which had a value of 2.46, and the modeled protein had a value of 4.07.
Molecular docking
The docking method was used to predict the conformation of a ligand on the active site of a receptor, which is usually a protein. Before docking, first predict the active center of the receptor, which in this case used the RaptorX server. Based on the analysis of the active site of the receptor, it was known that the residues that played a role in the ligand binding process were as follows: A73, S74, N75, I76, G77, H80, L149, Y150, K154, V286, W289, Y290, I397, Q457, S460, S392, and S393. Based on this information, these residues were put into a grid box for the docking process.
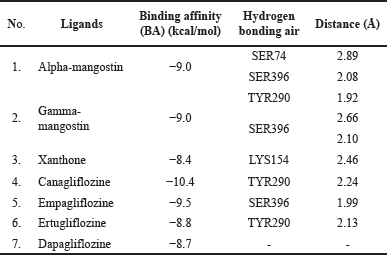
The hSGLT-2 protein obtained was then analyzed for binding with alpha-mangostin, gamma-mangostin, xanthone, and their comparison ligands. The comparison ligand used in the docking process was obtained on the PubChem website (https://pubchem.ncbi.nlm.nih.gov/) with the codes canagliflozin (24812758), empagliflozin (11949646), ertugliflozin (44814423), and dapagliflozin (9887712). The analysis results using the AutoDock Vina 4.2 server can be seen in Table 1 and illustrated in Figure 1.
Observations on alpha-mangostin, gamma-mangostin, and xanthones showed that the BA and hydrogen bonding values for the hSGLT-2 receptor amino acids were almost similar to those of the comparison ligands, so it can be assumed that these compounds had almost similar activity to their comparisons. However, alpha-mangostin and gamma-mangostin with the same BA value (−9.0) were better than xanthone (−8.4). The docking results were then visualized using the Chimera 1.14 program. The ligand pose with the lowest affinity energy value binding to the residue at the active center was selected for further analysis using the Discovery Studio 2020 software.
 | Table 1. Molecular docking of hSGLT-2 receptors with AutoDock Vina software. [Click here to view] |
MD simulation
The description of RMSD (root-mean-square deviation) and RMSF (root-mean-square fluctuation) of alpha-mangostin, gamma-mangostin, dan xanthone ligands through MD simulation analysis of the values is shown in Figure 2.
The RMSD value of alpha-mangostin was found to be higher than that of gamma-mangostin and xanthone in this research. Meanwhile, the high RMSF value for each of these ligands is indicated by amino acid residues 248–261, 347–357, and 510–523, which are thought to have a coiled structure. This is also supported by the analysis of Rg (radius of gyration) in Figure 3, which shows that the Rg ligand alpha-mangostin has a higher value. So, we can conclude that alpha-mangostin is more flexible than the other two ligands during the simulation for 50 ns.
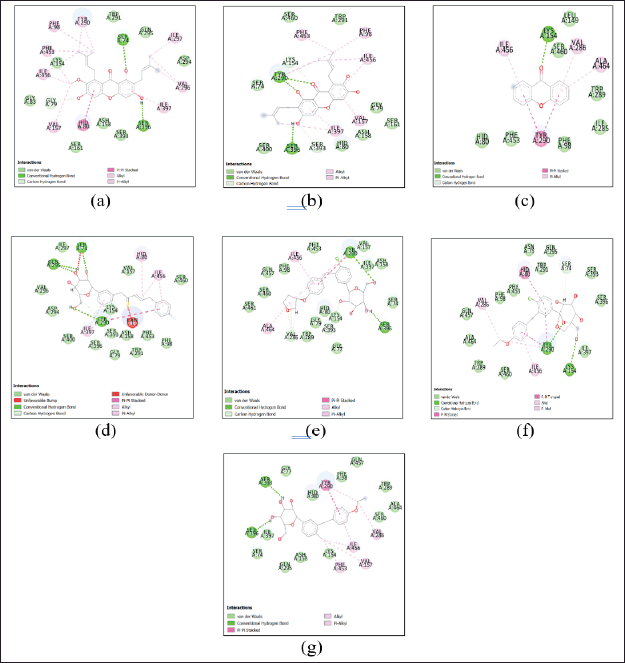 | Figure 1. Interaction between alpha-mangostin (a), gamma-mangostin (b), xanthone (c), canagliflozine (d), empagliflozine (e), ertugliflozine (f), and dapagliflozine (g) with amino acids at the SGLT-2 receptor. [Click here to view] |
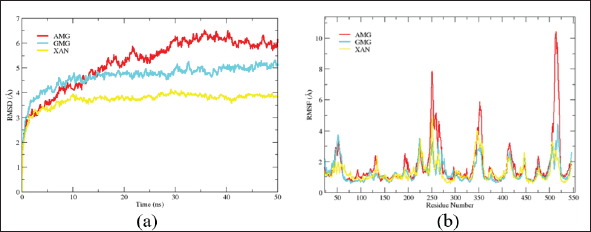 | Figure 2. RMSD (a) and RMSF (b) curves of alpha-mangostin (red), gamma-mangostin (blue), and xanthone (yellow) ligands. [Click here to view] |
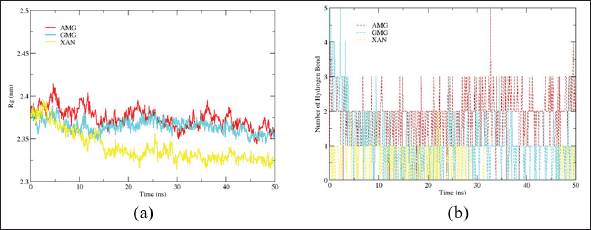 | Figure 3. Rg curves (a) and the number of hydrogen bonds (b) of alpha-mangostin (red), gamma-mangostin (blue), and xanthone (yellow) ligands. [Click here to view] |
Alpha-mangostin was expected to be more stable at the active site of the protein due to its more stable hydrogen bonding than gamma-mangostin and xanthone compounds. This is supported by the binding free energy values of these three ligands, as shown in Table 2.
Alpha-mangostin showed a lower or negative binding free energy value than the other two ligands. This means that the ligand has the ability to bind more securely. This stability was also tested by analyzing the presentation of the hydrogen bonds of the three ligands during a 50 ns simulation using Amber 18 software. The results of the presentation are shown in Figure 4.
The hydrogen bond stability of alpha-mangostin, simulated and displayed in the 2D form, shows the numbers 0.1, 32.8, and 50 ns. This is shown by Figure 5, which shows that the hydrogen bonding of alpha-mangostin, mainly through TRY290, persists for the given simulation time.
In vivo
Antihyperglycemic
The results of observations on the blood glucose levels of rats for 1 month of testing showed that EEMR affected reducing blood glucose (antihyperglycemia) in the fourth week, in line with the observations shown by the comparison drug and significantly different (p < 0.05) from the control group. The antihyperglycemic effect of EEMR increases with increasing dose (Fig. 6).
 | Table 2. Binding free energy of alpha-mangostin, gamma-mangostin, and xanthone compounds. [Click here to view] |
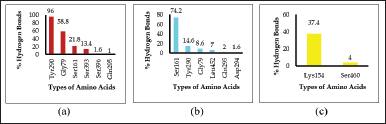 | Figure 4. Percentage of hydrogen bonds between alpha-mangostin (a), gamma-mangostin (b), and xanthone (c) ligands and hSGLT-2 receptor amino acids. [Click here to view] |
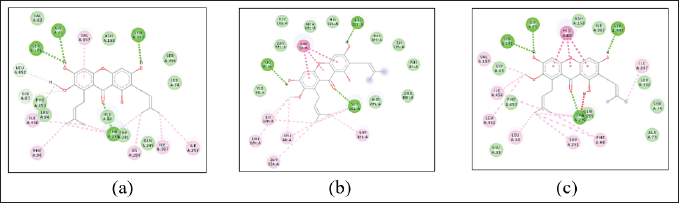 | Figure 5. Simulation of alpha-mangostin hydrogen bond stability to the hSGLT-2 receptor amino acid at 0.1 (a), 32.8 (b), and 50 ns (c). [Click here to view] |
Metformin and pioglitazone showed similar results in lowering the blood glucose levels of test animals. Meanwhile, empagliflozin gave the best effect through its mechanism specifically on the kidneys as an SGLT-2 inhibitor.
RT-qPCR analysis for SGLT 2 gene expression
For this research, RT-qPCR analysis was used to measure the amount of SGLT-2 gene expression in the renal tubules (Fig. 7).
A decrease in the expression of the SGLT-2 gene was observed among the test animals compared to the control group. However, the expression of SGLT-2 only decreased significantly (p0.05) at EEMR doses of 200 and 400 mg/kg bw, as well as a comparison drug (empagliflozin 2.25 mg/kg bw).
ELISA analysis of SGLT-2 protein expression
The results of observations on EEMR administration of SGLT-2 protein expression only showed a significant difference (p < 0.05) at the highest dose (400 mg/kg bw) compared to the control group. However, it was not better than the decreasing effect shown by empagliflozin (Fig. 8).
DISCUSSION
In silico
Protein modeling based on homology is crucial in predicting protein structure, especially for a protein whose structure has not been elucidated or registered in the Protein Data Bank database. To obtain good structural modeling results, the similarity (% identity) between the protein sequences owned and the template must be high. According to Xu et al. [23], the value of % identity between query sequences and templates above 25% is believed to be able to provide fairly good structure prediction results. In this case, the hSGLT-2 protein query sequence obtained from UniProt has a percentage identity of 32.84% with the vSGLT template (GDP ID: 2XQ2). This significantly different value was due to the fact that the protein modeling was done using the entire sequence of the hSGLT protein, so most of the residues, particularly those at the N- and C-termini, were in an unfavorable position. However, because the active center of the predicted protein model was in the safe zone of the Ramachandran Plot, the modeled protein was used for the next docking process.
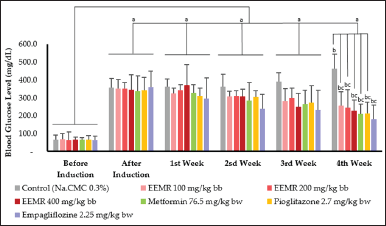 | Figure 6. Antihyperglycemic effect of each test preparation. The data were presented as mean ±SD with n = 10 (a = p < 0.05 for the preinduction group; b = p < 0.05 for the postinduction group; c = p < 0.05 for the control group). EEMR: ethanol extract of mangosteen rind. [Click here to view] |
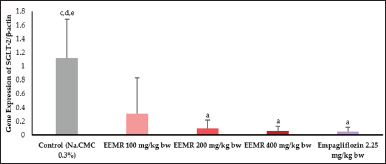 | Figure 7. Results of RT-qPCR analysis on SGLT-2 expression in the kidneys. The data were presented as mean ±SD with n = 3 (a = p < 0.05 for the control group; b = p < 0.05 for the EEMR group of 100 mg/kg bw; c = p < 0.05 for the EEMR group of 200 mg /kg bw; d = p < 0.05 for the EEMR group of 400 mg/kg bw; e = p < 0.05 for the empagliflozin group 2.25 mg/kg bw). EEMR: ethanol extract of mangosteen rind. [Click here to view] |
Furthermore, the ability of alpha-mangostin, gamma-mangostin, and xanthone ligands to bind to the hSGLT-2 receptor was analyzed through molecular docking imaging. In this case, the better the BA of a ligand, the better its binding strength to the protein (receptor). This method is often applied to determine the activity of a drug design [24,25].
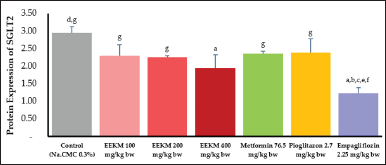 | Figure 8. ELISA analysis results on the protein expression of SGLT-2. The data were expressed as mean ± SD with n = 5 (a = p < 0.05 for the control group; b = p < 0.05 for the 100 mg/kg bw EEMR group; c = p < 0.05 for the 200 mg EEMR group /kg bw; d = p < 0.05 for the EEMR group of 400 mg/kg bw; e = p < 0.05 for the metforminute group 76.5 mg/kg bw; f = p < 0.05 for the pioglitazone 2.7 group mg/kg bw; g = p < 0.05 against the empagliflozin group 2.25 mg/kg bw). EEMR. [Click here to view] |
Although alpha-mangostin, gamma-mangostin, and xanthone were able to bind to the hSGLT-2 receptor, each of these ligands showed different BA values, so they were predicted to have different levels of binding strength. Alpha-mangostin and gamma-mangostin showed BA values (−9.0) and were able to show hydrogen bonds with two types of hSGLT-2 amino acids. Meanwhile, xanthones showed BA values (−8.4) and had hydrogen bonds with only 1 type of amino acid receptor. Based on this prediction, alpha-mangostin and gamma-mangostin were considered to have the best binding to the hSGLT-2 receptor. A MD simulation was carried out to strengthen this assumption. MD predicts the movement of each atom in a protein or other molecular system over a period of time based on a general model of physics [26]. Through this analysis, it is expected that the overall ability of the ligand to bind to the hSGLT-2 receptor will be able to be described comprehensively.
MD analysis of the hSGLT-2 receptor was conducted using Amber 18 software. Some of the things assessed were RMSD, RMSF, Rg, binding free energy value, hydrogen bond presentation, and stability. RMSD (Root-Mean-Square Deviation) aimed to analyze the movement of atoms during the simulation (against time) and RMSF (Root-Mean-Square Fluctuation) aimed to analyze the movement/flexibility of atoms in the protein main chain for each amino acid residue, while Rg (radius of gyration) aimed to see changes in the overall protein structure during the simulation (against time). RMSD, RMSF, and Rg all indicate how easily and how stable an atom or ligand could be moved around the system [27].
In this research, it was shown that the values of RMSD, RMSF, and Rg of alpha-mangostin were higher than those of gamma-mangostin and xanthone. Furthermore, it was suspected that alpha-mangostin has the ability to move more flexibly for a certain period of time. Meanwhile, the binding free energy value of the ligand was more negative than the other two ligands. The value of binding free energy is a description of the sum of all intermolecular interactions that exist between the ligand and the target/receptor spontaneously at a certain temperature [28,29]. So, it can be concluded that the energy required to break the bond between alpha-mangostin and the hSGLT-2 receptor is greater than those of gamma-mangostin and xanthone.
The presentation of the hydrogen bonds of the three ligands during the 50 ns simulation using Amber 18 software showed that alpha-mangostin and gamma-mangostin ligands were able to bind to six types of hSGLT-2 amino acids, while xanthones could only bind to two types of amino acids. Alpha-mangostin, through its binding to the amino acid TRY290, showed the most stable binding with a percentage of 96%. So, based on this observation of molecular docking and MD, it can be concluded that alpha-mangostin is the ligand/compound that binds best to the hSGLT-2 receptor compared to gamma-mangostin and xanthone contained in EEMR.
In vivo
Tests on the effect of antidiabetes mellitus with the in vivo method were carried out on research animals in the form of Wistar rats aged 6–8 weeks (150–200 g). Each test animal was induced by DM with a combination of a high-fat diet (HFD) and low-dose STZ (35 mg/kg bw). This induction model simulates the natural disease progression and metabolic characteristics typical of individuals at higher risk of developing type 2 diabetes due to insulin resistance and obesity [30].
STZ is one of the antibiotics that is considered capable of causing DM conditions by the specific necrosis effect on pancreatic cells. STZ induction causes a decrease in insulin levels in the blood and an increase in blood glucose concentrations. Glucose oxidation is affected by STZ, reducing insulin biosynthesis and secretion. STZ enters pancreatic cells via GLUT2 glucose transport, causing a decrease in GLUT2 expression, which results in a decrease in peripheral insulin receptor sensitivity and causes an increase in insulin resistance and blood glucose levels [31]. The state of DM in animal modeling induced by STZ was more stable in tests, especially in rats [7].
Based on research by Taher et al. [5], EEMR at 100 and 200 mg/kg bw could decrease blood glucose levels and repair the islets of Langerhans . This is in line with the results of this study, which showed that EEMR from the lowest to the highest dose (100, 200, and 400 mg/kg bw) could have an antihyperglycemic effect. The higher the dose given, the better the effect. The ability of EEMR to lower blood glucose levels was considered due to its xanthone-derived compounds, namely, alpha-mangostin and gamma-mangostin, through various mechanisms of action [32–34].
RT-qPCR analysis on EEMR administration for one month showed that EEMR with doses of 200 and 400 mg/kg bw could decrease SGLT-2 gene expression in line with the effect given by empagliflozin and significantly different (p < 0.05) from the test control group. The SGLT-2 receptor, which was specifically located only in the proximal renal tubule, has a role in glucose reabsorption in about 90% [35].
Based on the results of the research, blood serum obtained on the last day of observation was analyzed using the ELISA method to quantitatively detect the presence of SGLT-2. This method is an immunological test that is considered sensitive to measure antibodies, antigens, proteins, glycoproteins, and hormones in biological samples, specifically by using a special kit [36].
SGLT-2 is a parameter that plays a role in the treatment of DM. At the SGLT-2 receptor, decreased expression or inhibition of its action can decrease renal tubular glucose reabsorption and result in a reduction in blood glucose without stimulating insulin release [10,37]. One drug that works by this mechanism is empagliflozin. According to research by Revelian et al. [38] and Chung et al. [39], empagliflozin could cause a decrease in SGLT-2 gene expression in human kidney-2 cell lines (HK-2 cells) and the kidneys of DM animal models. In line with the findings in this study, empagliflozin had the best effect on SGLT-2 protein expression. Meanwhile, only EEMR with a dose of 400 mg/kg bw showed a significantly different effect (p < 0.05) compared to the control group (Na.CMC 0.3%).
In conclusion, based on the results of observations on the expression of SGLT-2, the role of EEMR in the treatment of DM is expressed through the mechanism of decreasing the expression of the SGLT-2 gene and protein. Further research in the form of activity tests on its combination with other extracts/compounds, safety tests, and clinical trials on the use of EEMR is expected to supplement these findings, allowing EEMR to be widely used as an alternative in treating diabetes.
CONCLUSION
EEMR at doses of 100, 200, and 400 mg/kg bw can provide an antihyperglycemic effect and decrease SGLT-2 expression. The content of alpha-mangostin as one of the ligands/marker compounds in EEMR is considered the best to play a role in this matter. Based on these findings, it is possible that EEMR can be used to treat DM as an alternative therapy.
ACKNOWLEDGMENTS
The authors would like to thank the Ministry of Research, Technology and Higher Education of the Republic of Indonesia for funding this research and they are grateful to Bandung Institute of Technology (ITB), our research site.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
The animal study protocol was approved by the Institutional Review Board (or Ethics Committee) of the HEALTH POLYTECHNIC MINISTRY OF HEALTH BANDUNG (protocol code 02/KEPK/EC/II/2022 and February 22, 2022).
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. IDF. IDF Diabetes Atlas. 10th ed. International Diabetes Federatio; 2021.
2. Ratwita W. Study on the mechanism of action of alpha mangostin and xanthones as antidiabetic compounds. (Doctoral Program Dissertation), Bandung Institute of Technology, 2018.
3. Husen SA, Salamun Ansori ANM, Hayaza S, Susilo RJK, Winarni D, Darmanto W. Renal protective effects of gamma-mangostin in streptozotocin-induced diabetic mice. Indian J Forensic Med Toxicol. 2020;14(3):1251–6. doi: https://doi.org/10.37506/ijfmt.v14i3.10562
4. Adnyana IK, Abuzaid AS Iskandar EY, Kurniati NF. Pancreatic lipase and α-amilase inhibitory potential of mangosteen (Garcinia mangostana Linn.) pericarp extract. Ijmrhs 2016;5(1):23–8. doi: https://doi.org/10.5958/2319-5886.2016.00006.0
5. Taher M, Zakaria TM, Susanti D, Zakaria ZA. Hypoglycaemic activity of ethanolic extract of Garcinia mangostana Linn. in normoglycaemic and streptozotocininduced diabetic rats. MC Complement Altern Med. 2016;16:135. doi: https://doi.org/10.1186/s12906-016-1118-9
6. Karim N, Jueenduang N, Tangpong J. Anti-Glycemic and anti-hepatotoxic effects of mangosteen vinegar rind from garcinia mangostana against HFD/STZ-induced type II diabetes in mice. Pol J Food Nutr Sci. 2018;68(2):163–9. doi: https://doi.org/10.1515/pjfns-2017-0018
7. Kumar V, Bhatt PC, Kaithwas G, Rashid M, Al-Abbasi FA, Khan JAA,et al. α-mangostin mediated pharmacological modulation of hepatic carbohydrate metabolism in diabetes induced wistar rat. Beni-Suef Univ J Basic Appl Sci. 2016;5:255–76. doi: https://doi.org/10.1016/j.bjbas.2016.07.001
8. Husen SA, Kalqutny SH, Ansori AN, Susilo RJ, Alymahdy AD, Winarni D. Antioxidant and antidiabetic activity of Garcinia mangostana L. pericarp extract in streptozotocin induced diabetic mice. Biosci Res. 2017;14(4):1238–45.
9. Abuzaid AS, Sukandar EY, Kurniati NF, Adnyana IK. Antihyperlipidemic effects of mangosteen (Garcinia mangostana L.) pericarp ethanolic extract in high-carbohydrate wistar rats. J Nat Remed. 2017;17(4):163–73. doi: https://doi.org/10.18311/jnr/2017/11051
10. Hsia DS, Grove O, Cefalu WT. An update on SGLT-2 inhibitors for the treatment of diabetes mellitus. Curr Opin Endocrinol Diabetes Obes. 2017;24(1):73–9. doi: https://doi.org/10.1097/MED.0000000000000311.
11. Yang J, Zhang Y. Protein structure and function prediction using I-TASSER. Curr Protoc Bioinformatics. 2015;52:5.8.1–5. doi: https://doi.org/10.1002/0471250953.bi0508s52
12. Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminform. 2012;4:17. doi: https://doi.org/10.1186/1758-2946-4-17
13. Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J Mol Recog. 1996;9(1):1–5. doi: https://doi.org/10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6
14. Wang S, Li W, Liu S, Xu J. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res. 2016;44(W1):W430–5. doi: https://doi.org/10.1093/nar/gkw306
15. Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, CheathamTE, Cruzeiro VWD,et al. AMBER 2018. San Francisco, CA: University of California; 2018.
16. Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–716. doi: https://doi.org/10.1021/acs.jctc.5b00255
17. Dong C, Jin M, Lingam M, Airapetian VS, Ma Y, van der Holst B. Atmospheric escape from the TRAPPIST-1 planetsand implications for habitability. PNAS. 2018;115(2):260–5. doi: https://doi.org/10.1073/pnas.1708010115
18. Roe DR, Cheatham TE. PTRJ and CPPTRAJ: software for processing and analysis of molecular dynamic trajectory data. J Chem Theory Comput. 2013;9(7):3084–95. doi: https://doi.org/10.1021/ct400341p
19. Turner PJ. XMGRACE, Version 5.1.19. Beaverton, OR: Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology; 2005.
20. Skovsø S. Modeling type 2 diabetes in rats using high fat diet and streptozotocin. J. Diabetes Investig. 2014;5(4):349–58. doi: https://doi.org/10.1111/jdi.12235
21. Miaffo D, Ntchapda F, Mahamad TA, Maidadi B, Kamanyi A. Hypoglycemic, antidyslipidemic and antioxydant effects of vitellaria paradoxa barks extract on high-fat diet and streptozotocin-induced type 2 diabetes rats. Metabolism Open. 2020;9:100071. doi: https://doi.org/10.1016/j.metop.2020.100071
22. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–8. doi: https://doi.org/10.1038/nprot.2008.73
23. Xu X, Dimitrov D, Rahbek C, Wang Z. NCBIminer: sequences harvest from genbank. Ecography. 2015, 38, 426-430. doi: https://doi.org/10.1111/ecog.01055
24. Raval K, Ganatra T. Basics, types and applications of molecular docking: a review. IP Int J Compr Adv Pharmacol. 2022;7(1):12–16. doi: https://doi.org/10.18231/j.ijcaap.2022.003
25. Chaudhary KK, Mishra N. A review on molecular docking: novel tool for drug discovery. JSM Chem. 2016;4(3):1029.
26. Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol. 2002;9:646–52. [PubMed: 12198485]
27. Cheng X, Ivanov I. Molecular dynamics. Methods Mol Biol. . 2013;929:243–84. doi: https://doi.org/10.1007/978-1-62703-050-2_11
28. Wan S, Bhati AP, Zasada SJ, Coveney PV. Rapid, accurate, precise and reproducible ligand–protein binding free energy prediction. Interface Focus. 2020;10:20200007. 102020000720200007 http://doi.org/10.1098/rsfs.2020.0007
29. Hall R, Dixon T, Dickson A. On calculating free energy differences using ensembles of transition paths. Front. Mol. Biosci. 2020;7:106. doi: https://doi.org/10.3389/fmolb.2020.00106
30. Srinivasan K, Viswanad B, Asrat L, Kaul CL, Ramarao P. Combination of high-fat diet-fed and low-dose streptozotocin-treated rat: a model for type 2 diabetes and pharmacological screening. Pharmacol Res. 2005;52(4):313–320. doi: https://doi.org/10.1016/j.phrs.2005.05.004
31. Deeds MC, Anderson JM, Armstrong AS, Gastineau DA, Hiddinga HJ, Jahangir A, et al. Single dose streptozotocin-induced diabetes: considerations for study design in islet transplantation models. Lab Anim. 2011;45:131–40. doi: https://doi.org/10.1258/la.2010.010090
32. Usman F, Shah HS, Zaib S, Manee S, Mudassir J, Khan A, et al. Fabrication and biological assessment of antidiabetic α-mangostin loaded nanosponges: in vitro, in vivo, and in silico studies. Molecules. 2021;26:663. doi: https://doi.org/10.3390/molecules26216633
33. Yani, F, Bellatasie R, Fauziah F. Antidiabetic potential of G. mangostana extract and α-mangostin compounds from mangosteen (Garcinia mangostana Linn.). EAS J Pharm Pharmacol. 2021;3(5):94–105. doi: https://doi.org/10.36349/easjpp.2021.v03i05.001
34. Chen SP, Lin SR, Chen TH, Ng HS, Yim HS, Leong MK, et al. Mangosteen xanthone γ-mangostin exerts lowering blood glucose effect with potentiating insulin sensitivity through the mediation of AMPK/PPARγ. Biomed Pharmacother. 2021;144:112333. doi: https://doi.org/10.1016/j.biopha.2021.112333
35. Fioretto P, Zambon A, Rossato M, Busetto L, Vettor R. SGLT-2 inhibitors and the diabetic kidney. Diabetes Care. 2016;39(2):165–71. https://doi.org/10.2337/dcS15-3006.
36. Alhajj M, Farhana A. Enzyme linked immunosorbent assay. [Updated 2022 Feb 2]. Treasure Island (FL): StatPearls Publishing; 2022.
37. Yu L, Xu Q, Hou P, Zhang H. Decreased expression and function of sodium-glucose co-transporter 2 from a novel c-terminal mutation: a case report. BMC Nephrol. 2016;17:31. doi: https://doi.org/10.1186/s12882-016-0244-4.
38. Ndibalema AR, Kabuye D, Wen S, Li L, Li X, Fan Q. Empagliflozin protects against proximal renal tubular cell injury induced by high glucose via regulation of hypoxia-inducible factor 1-alpha. Diabetes Metab Syndr Obes. 2020;13:1953–67. doi: https://doi.org/10.2147/DMSO.S243170
39. Chung S, Kim S, Son M, Kim M, Koh ES, Shin SJ, et al. Empagliflozin contributes to polyuria via regulation of sodium transporters and water channels in diabetic rat kidneys. Front Physiol. 2019;10:271. doi: https://doi.org/10.3389/fphys.2019.00271