INTRODUCTION
Inflammation is the body’s defense mechanism against bacterial infections and physical or chemical tissue damage. It is regulated by proinflammatory mediators like inducible nitric oxide synthase (iNOS) and cyclooxygenase-2. Inflammation is also indicated by the increase in proinflammatory cytokines like tumor necrosis factor-alpha (TNF-α), interleukin (IL)-1β, and IL-6 [1,2]. According to Wang and He [3], proinflammatory cytokines can regulate adipocyte proliferation and apoptosis, inhibiting lipid synthesis, promoting lipolysis, and decreasing blood lipids through autocrine and paracrine mechanisms. It activates the immune cell and triggers cytokine release and further production [4]. Uncontrolled cytokine is known to be responsible for cytokine storms, which accelerate inflammation [5].
Probiotics are naturally occurring living organisms found in food products and can help restore the composition of the human gut microbiota and maintain the gut microbial population [6,7]. Probiotic bacteria can significantly reduce the pro-inflammatory circulating markers [8]. According to Rocha-Ramírez et al. [9], Lactobacillus strain probiotics can be used to stimulate the inflammatory response and the immune system. Lactobacillus plantarum is a probiotic bacteria with anti-inflammation potential due to its ability to survive gastric acid and grow in the gastrointestinal tract [10].
Lactobacillus plantarum Su-ls29 is an indigenous Indonesian probiotic bacteria isolated from traditional Indonesian food, “Cassava Tape.” Lactobacillus plantarum Su-ls29 is a potential probiotic bacteria. The potential probiotic bacteria have characteristics of tolerance to bile and acid, have adhesion properties in the digestive tract, and have antimicrobial activity against pathogenic bacteria [11,12]. Lactobacillus plantarum Su-ls29 can inhibit the growth of Escherichia coli, Bacillus cereus, Listeria monocytogenes, Salmonella enterica, Pseudomonas aeruginosa, Staphylococcus aureus, Edwardsiella ictaluri, Aeromonas sobria, Plesiomonas shigelloides, Klebsiella pneumonia, Aeromonas hydrophilla, and Streptococcus agalactiae [11]. It is a Gram-positive bacteria, does not form a spore, and can produce lactic acid from carbohydrate fermentation [13]. Furthermore, it can prevent inflammatory diseases by modulating Th1 and Th2 cell ratio through the stimulation of various inflammatory cytokines like TNF-α, IL-1β, IL-6, IL-10, IL-12, and interferon-gamma [10]. Existing studies have not reported the anti-inflammatory activities of the effect of growth media. Environmental conditions in growth media may affect the activities of bacteria. According to Huang et al. [14], extracellular metabolites produced by bacteria can be used as antibacterial against pathogenic bacteria. Moreover, the extracellular metabolite product could be used as an anti-inflammation.
Based on previous findings, there are no studies on the identification and potential of metabolites from L. plantarum Su-ls29 extract as anti-inflammatory. Furthermore, metabolite profiling is often carried out to identify the profile of active compounds contained in biological materials [15]. Liquid chromatography-mass spectrophotometry (LC-MS) and gas chromatography-mass spectrophotometry (GC-MS) are tandem techniques that can be used for metabolite profiling. Previous studies revealed that these methods have high resolution, sensitivity, and reproducibility; hence, they can be used to analyze the metabolites in test samples, such as amino acids, organic acids, fatty acids, and carbohydrates [16]. The analytical technique of metabolites produced from probiotic bacteria using ultra-high-performance liquid chromatography-MS/MS (UHPLC-MS/MS) and GC-MS instruments have high reproducibility and sensitivity [17,18].
The active compounds obtained from the profiling can be seen by their mechanism of action based on interaction with several proteins that play a role in inflammation using computational chemistry with an in silico approach. A previous study reported that in silico analysis of the interaction model between protein and inhibitor compounds can help develop structure-based drugs [19].
This study compares the anti-inflammatory activities of L. plantarum Su-ls29 extracellular metabolite grown in man ragosa sharpe broth (MRSB) and skimmed milk media based on nitric oxide (NO), TNF-α, IL-1β, and IL-6 proinflammatory cytokine production. It also identifies the extracellular metabolite compound contents in the two samples using UHPLC-MS/MS and compares the inhibitory interactions of ligands with anti-inflammatory potentials to proinflammatory cytokines using an in silico approach.
MATERIALS AND METHODS
The RAW 264.7 macrophage cells were obtained from the Center Pharmaceutical and Medical Technology Laboratory, National Research and Innovation Agency, Republic of Indonesia. Furthermore, the bacterial isolate of L. plantarum used in this study was collected from the Research Center for Applied Microbiology laboratory, National Research and Innovation Agency, Republic of Indonesia. Lipopolysaccharide (LPS), 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT), HPLC grade solvents, and other reagents were purchased from Sigma-Aldrich (Saint Louis, Missouri, USA). Dulbecco’s modified eagle’s medium (DMEM) and fetal bovine serum (FBS) were purchased from HyClone (Logan, Utah, USA). De Man Ragosa Sharpe Agar (MRSA) and de MRSB were purchased from Himedia (Marg, Mumbai, India).
Preparing RAW 264.7 macrophage cells and L. plantarum Su-ls29
The RAW 264.7 cell culture was handled according to the American Type Culture Collection procedure (Manassas, VA, USA). RAW 264.7 cells were stored in an LN2 Static Cryogenic Liquid Nitrogen Storage Tank (Thermo Scientific, USA) at −180°C ± 0.5°C. Frozen cells were removed from a dry ice pack, and immediately, cells were placed below −70°C ± 0.5°C, preferably in liquid nitrogen vapor, until ready to use. Cell viability was calculated at 2.5 × 105 cells/ml using a hemocytometer (Marienfeld, Germany). Thawing was carried out before being used in the RAW 264.7 cell experiment. The tube containing RAW 264.7 cells was removed from the liquid nitrogen tube, then thawed at 37°C ± 0.5°C. 1 ml of cell fluid was pipetted into a centrifuge tube; 10 ml of DMEM media was added slowly and then centrifuged at 125 g for 5 minutes. The pellet formed was suspended with 6 ml of culture medium and 10% FBS. The suspended cells were pipetted and placed into the culture flask, then incubated at 37°C ± 0.5°C in a 5% CO2 incubator (Memmert, Germany) for 24 ± 0.05 hours; the medium was replaced every 3 days to obtain a sufficient number of cells with a density of 80%. The number of cells was counted using a hemocytometer (Marienfeld, Germany).
Lactobacillus plantarum Su-ls29 was grown on MRSA and stored at 4°C ± 0.5°C. Lactobacillus plantarum Su-ls29 was cultivated aerobically in MRSB medium at 30°C ± 0.5°C, then the bacterial culture was centrifuged at 1,000 g for 4 minutes, and the pellet was washed. The pellet was diluted with NaCl 0.9% (w/v) and used as an inoculum starter. The bacterial suspension was made in MRSB with an optical density value of 0.700 ± 0.02 (108 CFU/ml). The cells were counted using a hemocytometer (Marienfeld, Germany).
Preparing L. plantarum Su-ls29 extracellular metabolite
De Man, Rogosa, and Sharpe broth media
The bacterial culture of L. plantarum Su-ls29 1% (v/v) was prepared in 100 ml of MRSB media and incubated for 24 ± 0.05 hours at 37°C ± 0.5°C. The test sample was centrifugated at 10.000 g for 10 minutes at 4°C ± 0.5°C, and the supernatant was freeze-dried (Kubota, Japan) [18]. The sample in the form of dry powder was stored at frozen condition (−4°C ± 0.5°C).
Skimmed milk media
Skim milk with a concentration of 25% (w/v) was heated using a water bath for 1 hour at 90°C. After the dissolved skimmed milk was cooled to 37°C ± 0.02°C, 1% (v/v) suspension of L. plantarum Su-ls29 was inoculated in a laminar flow cabinet and incubated at 37°C ± 0.5°C for 48 ± 0.05 hours. The liquid inoculum was freeze-dried to obtain dry powder [17]. The dry powder was stored at frozen condition (−4°C ± 0.5°C).
Cell viability assay
Cell viability of the bacterial metabolite L. plantarum Su-ls29 was tested using the MTT method [20]. RAW 264.7 cell was cultured in DMEM media containing 10% FBS at 37°C ± 0.5°C, under 5% CO2 and 1% Penicillin-Streptomycin. Meanwhile, 2 × 104 cells/ml of RAW 264.7 were cultured in a 96-well plate (Thermo Scientific, USA) and incubated for 24 ± 0.05 hours. The cells were counted using a hemocytometer. The L. plantarum Su-ls29 extract concentrations in both growth media were 20, 100, and 500 μg/ ml. The extract concentration was given to the cell and incubated for 24 ± 0.05 hours. The cell was washed, added to 100 μl MTT 0.5 mg/ml, and incubated for 4 ± 0.05 hours. The formazan crystal was incubated in sodium 10% dodecyl sulfate for one night at 570 nm using a microplate reader (Benchmark; Bio-Rad Laboratories, Inc., USA). The experiment was repeated three times. The experimental procedure refers to the study of Soonthornsit et al. [21]. The parameter tested was the percent value of cell viability. The percentage of cell viability between treatments was analyzed by one-way analysis of variance (ANOVA) with Tukey’s post hoc test. Measurements were carried out at room temperature (25°C ± 0.5°C). Cell control and medium control were used as controls in this study.
NO production test
The determination of NO production was carried out according to research by Liu et al. [22]. A 2 × 104 cells/ml RAW 264.7 was cultured in a 96-well plate with DMEM 100 μl containing samples of 3.125, 6.25, 12.5, 25, and 50 μg/ml and 1 μg/ml LPS at 37°C ± 0.5°C for 24 ± 0.05 hours. The cells were counted using a hemocytometer. The NO content in the cultured cell was measured using the Griess assay. A 50 μl aliquot of the supernatant was mixed with 50 μl Griess reagent, containing 0.75% sulfanilamide in 0.5 N HCl and 0.075% N-1-naphthyl ethylene diamine in H2O, on a 96-well plate at a room temperature for 10 minutes. The absorbance was measured at a wavelength of 540 nm using a microplate reader (Benchmark; Bio-Rad Laboratories, Inc., USA) [23]. The sodium nitrite concentration was measured as a standard, and the NO concentration of each group was calculated by subtracting its value from the standard curve. The experiment was repeated three times. The parameter tested was the NO concentration. The NO concentration was analyzed by one-way ANOVA with Tukey’s post-hoc test. Measurements were carried out at room temperature (25°C ± 0.5°C).
Cytokine production measurement
A 2 × 104 cells/ml RAW 264.7 cell was incubated in a 96-well plate for 24 ± 0.05 hours with samples and 1 μg/ml LPS was added. As control of 2 × 104 cells/ml, RAW 264.7 cells was only added with 1 μg/ml LPS. The levels of TNF-α, IL-1β, and IL-6 in the media were determined using a commercial Enzyme-Linked Immunosorbent Assay (ELISA) kit (Genie, Ireland), and the procedures followed the protocol instruction [23,24].
UHPLC-MS/MS metabolite identification
The metabolite sample was analyzed using UHPLC Vanquish Tandem Q Exactive Plus Orbitrap HRMS (Thermo Scientific, USA). The samples were filtered using a 0.2 μm polytetrafluoroethylene membrane filter. A total of 2.5 μl of the sample was injected through Accucore column C18 (100 × 2.1 mm, 1.5 μm) with a flow rate of 0.2 ml/minute. The eluent was H2O + 0.1% formic acid (A) and acetonitrile + 0.1 % formic acid (B). This study uses the universal gradient elution (0–1 minute (5% B), 1–17 minutes (5–100% B), 17–18.5 minutes (100%B), 18.5–20 minutes (5%B)) as an elution system [25]. The positive ionization was in a mass range of 200–1500 m/z.
Preparation of proteins and ligands
The proteins used in this study were proinflammatory cytokines, namely, IL-1β protein data bank (PDB ID: 4G6M), IL-6 (PDB ID: 4CNI), TNF-α (PDB ID: 6OP0), and INOS (PDB ID: 4NOS). Structures were obtained on the PDB database (http://www.rcsb.org/pdb/). The protein structure was optimized, and the solvent, native ligand, and other nonprotein residues were separated using AutoDock Tools 1.5.7. Protein target active sites were visualized using MGL Tools 1.5.7. The ligand structure was obtained from the PubChem database (http://PubChem.ncbi.nlm.nih.gov) in *.sdf format [26]. Ligands were optimized by adding polar hydrogen atoms and adjusting the protein’s active site grid boxes. Ligands were optimized using AutoDock Tools 1.5.7 and saved in Protein Data Bank, Partial Charge (Q), & Atom Type (T) format.
Molecular docking of proteins and ligands
Docking between proinflammatory cytokines and ligands was performed using AutoDock Vina [27,28]. The target area of the protein active site was determined based on the grid box value using AutoDock Tools 1.5.7. The active site region of the receptor is covered by the grid box value of the protein, which consists of x, y, and z sizes with a space of 1.0. The grid boxes include 4G6M (−8.035; 23.743; −9.208), 4CNI (−1.724; −22.955; −5.019), 6OP0 (−13.444; −1.631; 19.021), and 4NOS (−1.806; 97.733; 20.415) and were saved in *.conf format. Molecular docking was validated using a reference ligand with a root mean square deviation (RMSD) parameter <2 Å [29]. The results were analyzed to obtain the lowest Gibbs free energy and bond interactions as hydrogen bonds, hydrophobic, alkyl, and van der Waals interactions. Pymol 2.5.2 and BIOVIA Discovery Studio 2021 were used to analyze the interactions [30]. Ligands predicted physicochemical properties and absorption, distribution, metabolism, and excretion (ADME) value using Swiss ADME [31].
The molecular dynamic of proteins and ligands
The software used in molecular dynamic simulations was Desmond Package from Schrödinger LLC (2019-4). The simulation was carried out for 100 ns using a solvent model with transferable intermolecular interaction potential 3 Points orthorhombic box. optimized potentials for liquid simulations-2005 was used as a force field in molecular dynamic simulations. A temperature of 300K was used with the constant-temperature, constant-pressure ensemble and 1 atm pressure. Furthermore, trajectory analysis was carried out every 10 ps by calculating the RMSD and the root mean square fluctuation (RMSF) of the protein and ligand over simulation time [32]. Prime molecular mechanics generalized Born surface area (MMGBSA) of total energy was calculated using Prime version 5.4.
Statistical analysis
Statistical analysis of the collected data was carried out using Statistical Package for the Social Sciences 26 software. The statistical analysis used was one-way ANOVA analysis and Tukey’s post-hoc test (α = 0.05). A simple linear regression curve was used to determine cell viability and NO, TNF-α, IL-1β, and IL-6 production. Sample measurements were carried out with three repetitions.
RESULT AND DISCUSSION
The viability of L. plantarum Su-ls29 metabolite extract cell
This study established the relationship between cell viability value and the cytotoxicity of the L. plantarum Su-ls29 extracellular metabolite extract. The L. plantarum Su-ls29 extract concentrations in both growth media were 20, 100, and 500 μg/ ml. When tested with extract samples, the cytotoxicity was determined using three low-high concentrations to ensure nontoxicity to the identified RAW 264.7 cells. The metabolite extract concentration of L. plantarum Su-ls29 below 100 μg/ml in skimmed milk media was nontoxic, while 500 μg/ml exhibited weak toxicity. Meanwhile, the concentration below 500 μg/ml in MRSB was nontoxic, as shown in Figure 1. The toxicity categorization was based on ISO10993-5, where the cell with viability above 80% was categorized as nontoxic and 60%–80% as weak toxicity. The viability value of 40%–60% and below 40% were classified as moderate and toxicity, respectively [33]. The results showed that L. plantarum Su-ls29-MRSB and L. plantarum Su-ls29- skimmed milk at a concentration of 20 μg/ml were in the same group as control cells with cell viability values of 100.3% and 101.9%, respectively. In contrast, control cell viability was 100% (p > 0.05). Lactobacillus plantarum Su-ls29- skimmed milk at a concentration of 500 μg/ml was toxic with a cell viability value of 69.3% (p < 0.05).
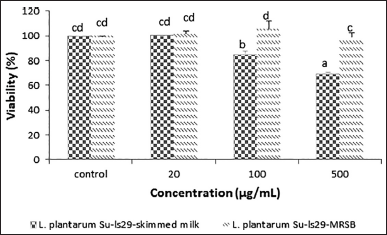 | Figure 1. RAW 264.7 macrophage cell viability of L. plantarum Su-ls29-skimmed milk and L. plantarum Su-ls29-MRSB metabolite extracts in various concentrations (p < 0.05). [Click here to view] |
The effect of L. plantarum Su-ls29 on NO production of RAW 264.7 cell
Nitric oxide produced by the multistep reaction of L-arginine catalyzed by iNOS is typically employed as a sign of inflammation [34,35]. Macrophage cell RAW 264.7 was previously induced with LPS. LPS can activate macrophages, inducing local inflammation and modulating iNOS expression [36]. The initial filtering result showed that the 100 μg/ml L. plantarum Su-ls29-skimmed milk inhibited a more significant NO production than LPS, as indicated by p < 0.05. Meanwhile, the NO production of L. plantarum Su-ls29-MRSB was not significantly different, as shown in Figure 2.
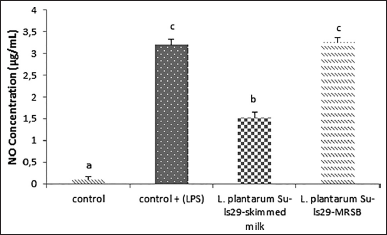 | Figure 2. NO production of L. plantarum Su-ls29-skimmed milk and L-plantarum Su-ls29-MRSB in concentration: 100μg/ml (p < 0.05). [Click here to view] |
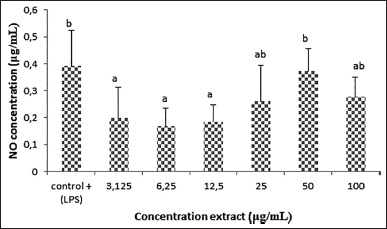 | Figure 3. NO production of RAW 264.7 macrophage cells in various concentrations of L. plantarum Su-ls29-skimmed milk (p < 0.05). [Click here to view] |
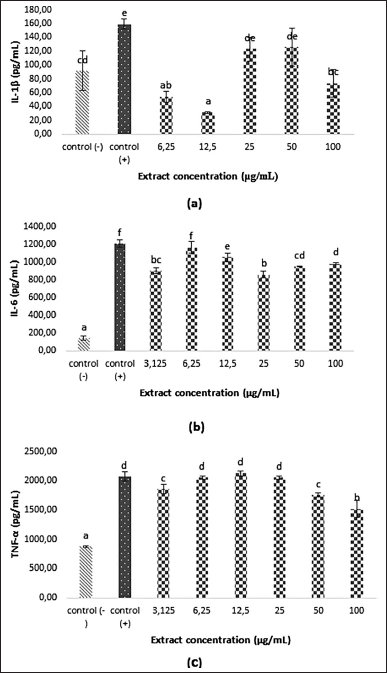 | Figure 4. IL-1β (A), IL-6 (B), and TNF-α (C) expressions of LPS-induced RAW 264.7 cell in various extract concentrations (p < 0.05). [Click here to view] |
Lactobacillus plantarum Su-ls29 cultured in skimmed milk exhibits the best anti-inflammatory activities, indicating the need for further studies on the best anti-inflammatory concentration. Figure 3 presents the NO production based on L. plantarum Su-ls29-skimmed milk concentration. The study showed that the extracellular metabolite extract of L. plantarum Su-ls29-skimmed milk with 6.25 μg/ml concentration exhibited the most significant NO reduction (0.17 ± 0,07 μg/ml; p < 0.05). The result indicates that extracellular metabolite extract of L. plantarum Su-ls29-skimmed milk can inhibit the iNOS expression and NO synthesis, decreasing NO production. The best concentration of extracellular metabolite extract of L. plantarum Su-ls29-skimmed milk in reducing NO was previously unknown. Therefore, this study used several extract concentrations to determine the optimal concentration of extracellular metabolite extract of L. plantarum Su-ls29-skimmed milk, which can reduce NO production. The production of cytokine expression and NO production was compared to LPS addition as the positive control. The result of this study is consistent with previous reports that L. plantarum KSFY02 could decrease NO production by 23 ± 0.62 μmol/l [37]. In addition, L. plantarum Su-ls29 fermentation in skimmed milk media can reduce NO production by 1.66 ± 0.1 μg/ml.
The probiotic bacteria can reduce cytokines, which play a role in the formation of inflammatory cells and a decrease in the immune system [38]. Liu et al. [39] reported that short-chain fatty acids produced by Lactobacillus can induce an immune response through the tolerogenic process of dendritic cells. This process will encourage CD4+T cells to differentiate into regulatory T cells (Treg) so that they can inhibit cytokine production by neutrophils and macrophages. Tolerogenic dendritic cells will produce anti-inflammatory cytokines, namely, IL-10 and transforming growth factor-β.
The effect of L. plantarum Su-ls29 extracts on IL-1β, IL-6, and TNF-α expression.
Macrophage activation could increase IL-1β, IL-6, and TNF-α production [40]. Excessive cytokine production could hamper immune response and overreaction, triggering inflammation [34,35]. This study analyzed IL-1β, IL-6, and TNF-α production to determine whether the inflammation inhibition mechanism passes through cytokine. Figure 4 shows that the L. plantarum Su-ls29-skimmed milk could significantly inhibit the production of IL-1β, IL-6, and TNF-α (p < 0.05). Meanwhile, the collected data were compared to the LPS addition as the positive control. The cytokine production value was estimated to be lower than the positive control. Furthermore, the most optimum concentration in reducing IL-1β, IL-6, and TNF-α was 12.2 μg/ml (IL-1β level: 31.33 ± 1.05 pg/ml), 25 μg/ml (IL-6 level: 859.93 ± 36.34 pg/ml), and 100 μg/ml ( TNF-α level: 1521.48 ± 138.03 pg/ml). The extracellular metabolite extract of L. plantarum Su-ls29-skimmed milk can inhibit the inflammatory activity of RAW264.7 cells by reducing the expression and secretion of inflammation mediators of IL-1β, IL-6, and TNF-α. This study is consistent with the report of Le et al. [41] that L. plantarum FB003 fermented in phenol-rich kimchi mustard can reduce the production of IL-1β, IL-6, and TNF-α.
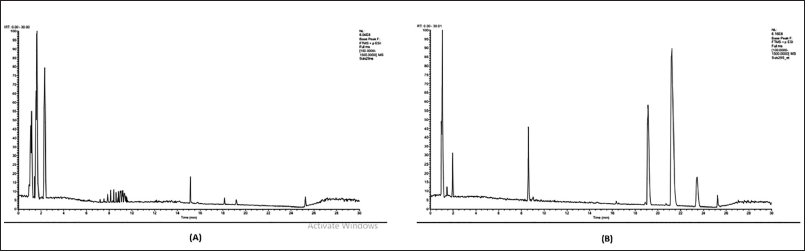 | Figure 5. Separation chromatogram of L. plantarum Su-ls29-MRSB (A) and L. plantarum Su-ls29-skimmed milk (B). [Click here to view] |
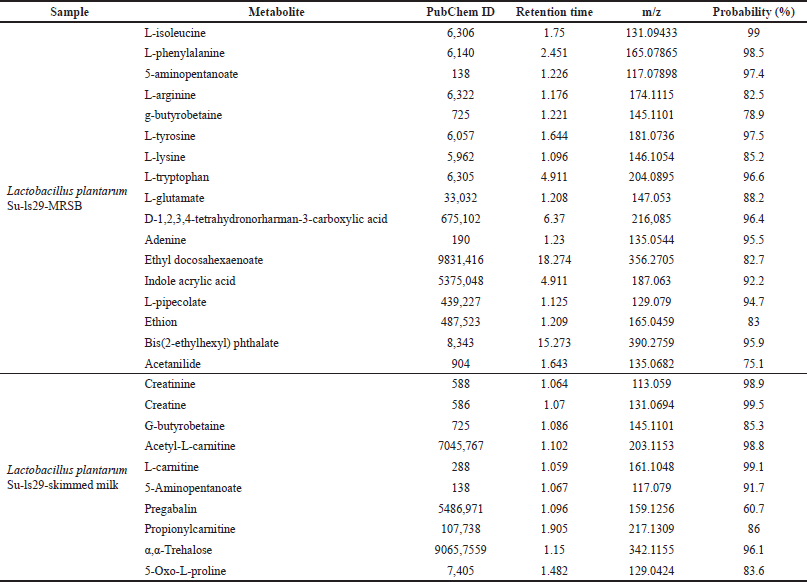 | Table 1. Metabolite compounds of L. plantarum Su-ls29 extract. [Click here to view] |
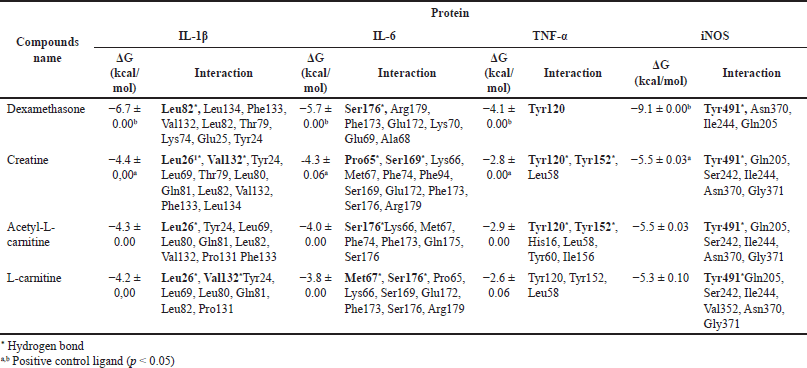 | Table 2. Gibbs free energy and proinflammatory protein metabolites of L.plantarum Su-ls29. [Click here to view] |
The metabolite of L. plantarum Su-ls29 extract
The extracellular metabolites of L. plantarum Su-ls29-MRSB and L. plantarum Su-ls29-skimmed milk were identified using UHPLC-MS/MS with mzCloud Mass Spectral Library. Data analysis identified 17 metabolite compounds in L. plantarum Su-ls29-MRSB and 10 in L. plantarum Su-ls29-skimmed milk extracts, as shown in Figure 5 and Table 1.
Several metabolites produced by the extracellular metabolite extract of L. plantarum Su-ls29-skimmed milk were reported to have anti-inflammatory activities. Creatine could also reduce IL-6 and IL-10 levels in rats’ blood plasma [42]. Meanwhile, daily injections of 10.5 mg and 3 g·kg−1·day−1 of diet-based creatine were reported to suppress tumor growth [43]. According to acetyl-L carnitine contains anti-inflammatory activities by reducing TNF-α, IL-1β, iNOS, and C-reactive protein of atherosclerotic rats. 300 mg/kg/day of L-carnitine was reported to effectively lower IL-1β and IL-6 in the plasma and serum of diabetic rats [44]. Furthermore, pregabalin metabolite was reported to be neuropathic pain in adults [45]. This study indicates that creatine, acetyl-L-carnitine, and L-carnitine were anti-inflammatory in L. plantarum Su-ls29-skimmed milk. Creatine can be produced from L-arginine, methionine, and glycine [46]. L-arginine and glycine were contained in skim milk [47]. Creatine synthesis involves two enzymes, namely, glycine amidinotransferase and guanidinoacetate N-methyltransferase. Creatine was spontaneously converted into creatinine [48].
In silico metabolite molecular interaction
The in vitro test results for creatine, acetyl-L-carnitine, and L-carnitine for IL-1β, IL-6, and TNF-α proteins were continued in silico tests. Dexamethasone was used as a comparison ligand and can affect cytokine production, such as IL-1 β, IL-6, and TNF-α [49]. In silico testing was also carried out on iNOS, which plays a significant role in the NO production process. Before the molecular docking simulation, a validation step was carried out with the RMSD value of 1.029 Å. This value indicates that the docking results were categorized as valid with the RMSD value of <2.0 Å [50]. In silico testing with molecular docking showed that creatine, acetyl-L-carnitine, and L-carnitine strongly bind with IL-1β, IL-6, and iNOS proteins. In contrast, there was a weak interaction with TNF-α. These results were based on the value of the Gibbs free energy in Table 2. Creatine had the lowest Gibbs free energy compared to the other two ligands. The Gibbs free energy values ??of creatine for proteins IL-1β, IL-6, TNF-α, and iNOS are −4.4, −4.3, −2.8, and −5.5 kcal/mol, respectively. According to Desiraju and Steiner [51] and Sari et al. [52], a free energy value above −4 kcal/mol was included in the strong category. The energy with the lowest value was the best and most stable conformation. Figure 6 shows the two-dimensional interaction between creatine and four proteins. The bond interactions between creatine and protein were hydrogen and van der Waals bonds and creatinine were found to have the best activity against iNOS protein. Hydrogen bonds in the iNOS protein are present in Tyr491, while van der Waals interactions are present in Gln205, Ser242, Ile244, Asn370, and Gly371. The interactions of hydrogen and van der Waals bonds with the amino acids of proinflammatory proteins are shown in Table 2. These bonds play a significant role in changing the value of potential energy [53]. Furthermore, the lowest Gibbs free energy of creatine, acetyl-L-carnitine, and L-carnitine was found in the iNOS protein. This is consistent with the in vitro results where the metabolite extract of L. plantarum Su-ls29-skimmed milk significantly reduces NO production. The test results also showed a decrease in IL-1β and IL-6 levels, where the ligands creatine, acetyl-L-carnitine, and L-carnitine strongly interact with IL-1β and IL-6 proteins based on in-line in silico testing. The TNF levels drop was not statistically significant but consistent with the in silico data.
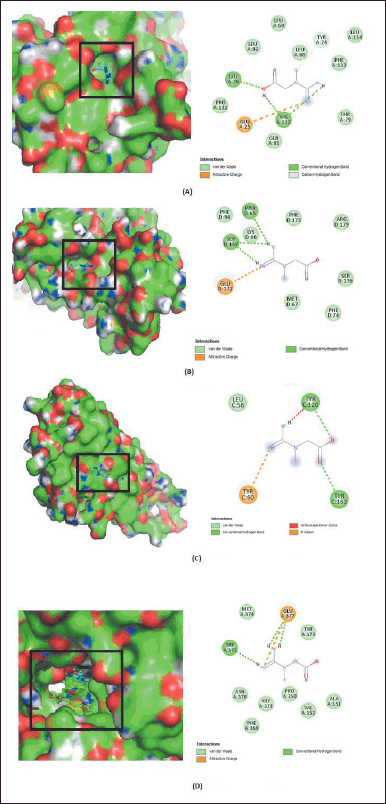 | Figure 6. Two-dimensional interaction between creatine and the proteins IL-1β (A), IL-6 (B), TNF-α (C), and iNOS (D). [Click here to view] |
Based on molecular docking simulation data, creatine showed the best results on the interaction of the iNOS proteins. This study used molecular dynamic simulation to track the creatine ligand. Molecular dynamics aims to see fluctuations in changes in the ligand complex due to thermal and solvent influences on changes in simulation time. The RMSD value of the iNOS and creatine complex can be seen in Figure 7a. The conformational stability of the dynamic molecular simulation starts at 10 to 30 ns and is indicated by a moderate variation in the RMSD value, with an average RMSD value of 1.2 Å. The RMSF analysis and 2D interactions showed fluctuations in the hydrogen bonds of several amino acid residues, namely, Trp194, Trp372, Ile201, and Cys200, as shown in Figure 7b and c. The movement of the unfolded side chains causes the fluctuation of the RMSD value [54]. Prime MM-GBSA calculates the energy of optimized free ligands, receptors, and the complex of the ligand with a receptor [55]. The total binding energy from the MMGBSA analysis was −88.73 kcal/mol, with a total solvation energy of −15.07 kcal/mol. The predicted value of physicochemical properties and ADME are shown in Table 3. Table 3 describes the physicochemical descriptors for predicting ADME parameters, pharmacokinetic properties, drug-like properties, and drug-chemical friendliness of the compounds creatine, acetyl-L-carnitine, and L-carnitine. In the early stages of drug discovery and development, in silico prediction of human oral bioavailability is a relevant tool for screening potential drug candidates [56]. The rule of five (RO5) is a rule that can be used in evaluating drug-likeness and pharmacological activity as an oral drug in humans. A drug that meets the RO5’s criteria was predicted to be absorbed by the human body through the lipid bilayer. Rule parameters include molecular mass below 500 g/mol, having less than five hydrogen bond donors and 10 hydrogen bond acceptors, having molar refractivity between 40–130, and high lipophilicity (logP less than 5,0) [57]. The creatine, acetyl-L-carnitine, and L-carnitine complied with RO5 based on predicted hydrophobicity and bioavailability. The molecular weight value, number of rotatable bonds, H-bond acceptors, H-bond donors and lipophilicity of creatine, acetyl-L-carnitine, and L-carnitine meet RO5 requirements. At the same time, the molar refractivity value ??was only acetyl-L-carnitine, which meets the RO5 value. Based on the original RO5 agreement, using four physicochemical parameters, molecular weight, log P, H-bond donors, and H-bond acceptors can predict the oral bioavailability of a drug compound. The four physicochemical parameters were related to acceptable water solubility and intestinal permeability [57]. The water solubility of creatine had a highly soluble class with a solubility value of 2.51 × 102 mg/ml. In addition, creatine, acetyl-L-carnitine, and L-carnitine had a high category of gastrointestinal permeability.
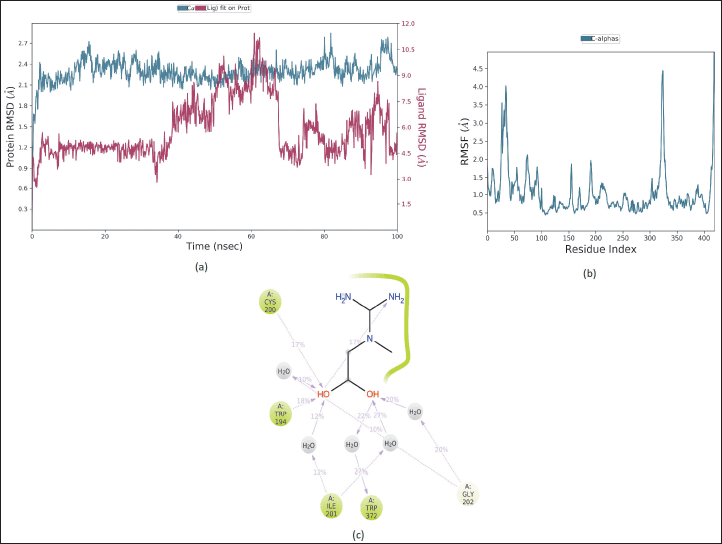 | Figure 7. The RMSD (A), the RMSF (B), and 2D ligan-protein contacts (C) relative to the starting complexes during 100 ns MD trajectory for creatine. [Click here to view] |
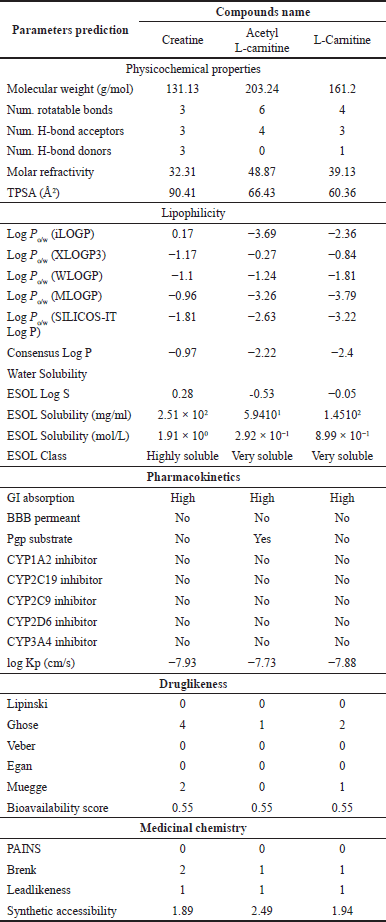 | Table 3. Physicochemical property and ADME prediction value. [Click here to view] |
CONCLUSION
The extracellular metabolite extract of L. plantarum Su-ls29-skimmed milk exhibited more anti-inflammatory activities than L. plantarum Su-ls29-MRSB in terms of NO production. The 6.25 μg/ml concentration showed the most significant NO reduction activities. Furthermore, the L. plantarum Su-ls29-skimmed milk reduced IL-1β, IL-6, and TNF-α productions in RAW 264.7 cells. The most optimum concentration in reducing IL-1β, IL-6, and TNF-α was 12.2 μg/ml, 25 μg/ml, and 100 μg/ml, respectively. This study identified 10 metabolites in the metabolite extract of L. plantarum Su-ls29-skimmed milk and 17 in L. plantarum Su-ls29-MRSB. Furthermore, metabolite creatine, acetyl-L-carnitine, and L-carnitine were anti-inflammatory in L. plantarum Su-ls29-skimmed milk metabolite extract. It is recommended to use the extracellular metabolite extract of L. plantarum Su-ls29-skimmed milk as a source of anti-inflammatory compounds.
ACKNOWLEDGMENT
The authors are grateful to the Advanced Study Laboratory, IPB University, for allowing them to conduct metabolite analysis using UHPLC-MS/MS and the Lab of Theoretical Physics Division, Department of Physics, IPB University, to support computational resources. This research was funded by the “Hibah Pengembangan Inovasi Program Promoting Research and Innovation Through Modern and Efficient Science and Techno Park 2023, number 105.”
AUTHOR CONTRIBUTION
All authors contributed substantially to the study’s concept and design, data collection, analysis, and interpretation. All authors took part in writing and revising this article critically for important intellectual content, adhered to the journal’s procedure, agreed on the final version of the published study, and were responsible for all aspects. All authors meet the definition of authorship given by the International Committee of Medical Journal Editors (ICMJE).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest in this study.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All generated and analyzed data are included in this study.
PUBLISHER NOTES
This journal remains neutral related to the jurisdictional claim of the published institutional affiliation.
REFERENCES
1. Choi SH, Lee SH, Kim MG, Lee HJ, Kim GB. Lactobacillus plantarum CAU1055 ameliorates inflammation in lipopolysaccharide-induced RAW264.7 cells and a dextran sulfate sodium–induced colitis animal model. J Dairy Sci. 2019;102:6718–25. doi: https://doi.org/10.3168/jds.2018-16197
2. Moradian N, Gouravani M, Salehi MA, Heidari A, Shafeghat M, Hamblin MR, et al. Cytokine release syndrome: inhibition of pro-inflammatory cytokines as a solution for reducing COVID-19 mortality. Eur Cytokine Netw. 2020;31:81–93. doi: https://doi.org/10.1684/ecn.2020.0451
3. Wang T, He C. Pro-inflammatory cytokines: The link between obesity and osteoarthritis. Cytokine Growth Factor Rev. 2018;44:38–50. https://doi.org/10.1016/j.cytogfr.2018.10.002
4. Schaper F, Rose-John S. Interleukin-6: Biology, signaling and strategies of blockade. Cytokine Growth Factor Rev. 2015;26:475–87. doi: https://doi.org/10.1016/j.cytogfr.2015.07.004
5. Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76:16–32. doi: https://doi.org/10.1128/MMBR.05015-11
6. Park C, Brietzke E, Rosenblat JD, Musial N, Zuckerman H, Ragguett R-M, et al. Probiotics for the treatment of depressive symptoms: An anti-inflammatory mechanism? Brain Behav Immun. 2018;73:115–24. doi: https://doi.org/10.1016/j.bbi.2018.07.006
7. Wang X, Zhang P, Zhang X. Probiotics regulate gut microbiota: an Effective method to improve immunity. Molecules. 2021;26(19):6076. doi: https://doi.org/10.3390/molecules26196076
8. Sarkar A, Lehto SM, Harty S, Dinan TG, Cryan JF, Burnet PWJ. Psychobiotics and the manipulation of bacteria–gut–brain signals. Trends Neurosci. 2016;39:763–81. doi: https://doi.org/10.1016/j.tins.2016.09.002
9. Rocha-Ramírez LM, Pérez-Solano RA, Castañón-Alonso SL, Moreno Guerrero SS, Ramírez Pacheco A, García Garibay M, et al. Probiotic Lactobacillus strains Stimulate the inflammatory response and activate human macrophages. J Immunol Res. 2017;2017:4607491. doi: https://doi.org/10.1155/2017/4607491
10. Le B, Yang SH. Efficacy of Lactobacillus plantarum in prevention of inflammatory bowel disease. Toxicol Rep. 2018;5:314–7. doi: https://doi.org/10.1016/j.toxrep.2018.02.007
11. Sulistiani, Novarina I, Inawati, Dinoto A, Julistiono H, Handayani R, et al. Assessment of potential probiotic lactic acid bacteria from tempe and tape. IOP Conf Ser Earth Environ Sci. 2020;572:012026. doi: https://doi.org/10.1088/1755-1315/572/1/012026
12. Tarique M, Abdalla A, Masad R, Al-Sbiei A, Kizhakkayil J, Osaili T, et al. Potential probiotics and postbiotic characteristics including immunomodulatory effects of lactic acid bacteria isolated from traditional yogurt-like products. LWT. 2022;159:113207. doi: https://doi.org/10.1016/j.lwt.2022.113207
13. Bintsis T. Lactic acid bacteria: their applications in foods. J Bacteriol Mycol Open Access. 2018;6:89–94. doi: https://doi.org/10.15406/jbmoa.2018.06.00182
14. Huang CB, Alimova Y, Myers TM, Ebersole JL. Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch Oral Biol. 2011;56:650–4. doi: https://doi.org/10.1016/j.archoralbio.2011.01.011
15. Rai AK, Jini R, Swapna HC, Sachindra NM, Bhaskar N, Baskaran V. Application of native lactic acid bacteria (lab) for fermentative recovery of lipids and proteins from fish processing wastes: bioactivities of fermentation products. J Aquat Food Prod Technol. 2011;20:32–44. doi: https://doi.org/10.1080/10498850.2010.528174
16. Park SE, Yoo SA, Seo SH, Lee KI, Na CS, Son HS. GC–MS based metabolomics approach of Kimchi for the understanding of Lactobacillus plantarum fermentation characteristics. LWT Food Sci Technol. 2016;68:313–21. doi: https://doi.org/10.1016/j.lwt.2015.12.046
17. Chang CY, Pan TM. Identification of bioactive compounds in Lactobacillus paracasei subsp. paracasei NTU 101-fermented reconstituted skimmed milk and their anti-cancer effect in combination with 5-fluorouracil on colorectal cancer cells. Food Funct. 2019;10:7634–44. doi: https://doi.org/10.1039/c9fo01819k
18. Chaudhary A, Verma K, Saharan BS. A GC-MS based metabolic profiling of probiotic lactic acid bacteria isolated from traditional food products. J Pure Appl Microbiol. 2020;14:657–72. doi: https://doi.org/10.22207/JPAM.14.1.68
19. Hernández-Santoyo A, Tenorio-Barajas AY, Altuzar V, Vivanco-Cid H, Mendoza-Barrera C. Protein-protein and protein-ligand docking. In: Ogawa T, editor. Protein Eng. Rijeka, Croatia: IntechOpen; 2013. doi: https://doi.org/10.5772/56376
20. Chang SH, Lin YY, Wu GJ, Huang CH, Tsai GJ. Effect of chitosan molecular weight on anti-inflammatory activity in the RAW 264.7 macrophage model. Int J Biol Macromol. 2019;131:167–75. doi: https://doi.org/10.1016/j.ijbiomac.2019.02.066
21. Zhang L, Chen Juan, Liang R, Liu C, Chen M, Chen Jun. Synergistic anti-inflammatory effects of lipophilic grape seed proanthocyanidin and camellia oil combination in LPS-stimulated RAW264.7 cells. Antioxidants. 2022;11(2):289. doi: https://doi.org/10.3390/antiox11020289
22. Soonthornsit N, Pitaksutheepong C, Hemstapat W, Utaisincharoen P, Pitaksuteepong T. In vitro anti-inflammatory activity of Morus alba L. stem extract in LPS-stimulated RAW 264.7 cells. Evidence-Based Complement Altern Med. 2017;2017:3928956. doi: https://doi.org/10.1155/2017/3928956
23. Liu Z-K, Ng C-F, Shiu H-T, Wong H-L, Wong C-W, Li K-K, et al. A traditional Chinese formula composed of Chuanxiong Rhizoma and Gastrodiae Rhizoma (Da Chuanxiong Formula) suppresses inflammatory response in LPS -induced RAW 264.7 cells through inhibition of NF-κB pathway. J Ethnopharmacol. 2017;196:20–8. doi: https://doi.org/10.1016/j.jep.2016.12.014
24. Suabjakyong P, Nishimura K, Toida T, Van Griensven LJLD. Structural characterization and immunomodulatory effects of polysaccharides from Phellinus linteus and Phellinus igniarius on the IL-6/IL-10 cytokine balance of the mouse macrophage cell lines (RAW 264.7). Food Funct. 2015;6:2834–44. doi: https://doi.org/10.1039/C5FO00491H
25. Chan CL, Wai HKF, Wu P, Lai SW, Chan OSK, Tun HM. A universal LC-MS/MS method for simultaneous detection of antibiotic residues in animal and environmental samples. Antibiotics. 2022;11(7):845. doi: https://doi.org/10.3390/antibiotics11070845
26. Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–9. doi: https://doi.org/10.1093/nar/gky1033
27. Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock vina 1.2.0: new docking methods, expanded force field, and python bindings. J Chem Inf Model. 2021;61:3891–8. doi: https://doi.org/10.1021/acs.jcim.1c00203
28. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: https://doi.org/10.1002/jcc.21334
29. Lestari AR, Batubara I, Wahyudi ST, Ilmiawati A. Phenolic Compound in Garlic (Allium sativum) and Black Garlic Potency as Antigout Using Molecular Docking Approach. J Kim Sains Dan Apl 2022;25:253–63. doi: https://doi.org/10.14710/jksa.25.7.253-263
30. Nurlela N, Awaluddin F, Batubara I, Wahyudi ST. Computational study of kaurene diterpenoids for antivirals against SARS-CoV-2. J Appl Pharm Sci. 2022;12:112–29. doi: https://doi.org/10.7324/JAPS.2022.120812
31. Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: https://doi.org/10.1038/srep42717
32. Bowers KJ, Chow DE, Xu H, Dror RO, Eastwood MP, Gregersen BA, et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. SC ’06 Proc. 2006 ACM/IEEE Conf. Supercomput; 2006, p 43. doi: https://doi.org/10.1109/SC.2006.54
33. López-García J, Lehocký M, Humpolí?ek P, Sáha P. HaCaT keratinocytes response on antimicrobial atelocollagen substrates: extent of cytotoxicity, cell viability and proliferation. J Funct Biomater. 2014;5:43–57. doi: https://doi.org/10.3390/jfb5020043
34. Wang Q, Liang J, Stephen Brennan C, Ma L, Li Y, Lin X, et al. Anti-inflammatory effect of alkaloids extracted from Dendrobium aphyllum on macrophage RAW 264.7 cells through NO production and reduced IL-1, IL-6, TNF-α and PGE2 expression. Int J Food Sci Technol. 2020;55:1255–64. doi: https://doi.org/10.1111/ijfs.14404
35. Wang S, Xu J, Zheng J, Zhang X, Shao J, Zhao L, et al. Anti-inflammatory and antioxidant effects of acetyl-L-carnitine on atherosclerotic rats. Med Sci Monit Int Med J Exp Clin Res. 2020;26:e920250. doi: https://doi.org/10.12659/MSM.920250
36. Li M, Dong L, Du H, Bao Z, Lin S. Potential mechanisms underlying the protective effects of Tricholoma matsutake singer peptides against LPS-induced inflammation in RAW264.7 macrophages. Food Chem. 2021;353:129452. doi: https://doi.org/10.1016/j.foodchem.2021.129452
37. Zhao X, Yi R, Zhou X, Mu J, Long X, Pan Y, et al. Preventive effect of Lactobacillus plantarum KSFY02 isolated from naturally fermented yogurt from Xinjiang, China, on d-galactose–induced oxidative aging in mice. J Dairy Sci. 2019;102:5899–912. doi: https://doi.org/10.3168/jds.2018-16033
38. Kim YA, Keogh JB, Clifton PM. Probiotics, prebiotics, synbiotics and insulin sensitivity. Nutr Res Rev. 2018;31:35–51. doi: https://doi.org/10.1017/S095442241700018X
39. Liu Y, Alookaran JJ, Rhoads JM. Probiotics in autoimmune and inflammatory disorders. Nutrients. 2018;10. doi: https://doi.org/10.3390/nu10101537
40. Rahminiwati M, Trivadila, Iswantini D, Takemori H, Koketsu M, Sianipar RNR, et al. Indonesian medicinal plants with anti-inflammatory properties and potency as chronic obstructive pulmonary disease (COPD) herbal medicine. Pharmacogn J. 2022;14:432–44. doi: https://doi.org/10.5530/pj.2022.14.119
41. Le B, Anh PT, Yang SH. Enhancement of the anti-inflammatory effect of mustard kimchi on rAW 264.7 macrophages by the Lactobacillus plantarum fermentation-mediated generation of phenolic compound derivatives. Foods. 2020;9(2):181. doi: https://doi.org/10.3390/foods9020181
42. Cordingley DM, Cornish SM, Candow DG. Anti-inflammatory and anti-catabolic effects of creatine supplementation: a brief review. Nutrients. 2022;14(3):544. doi: https://doi.org/10.3390/nu14030544
43. Di Biase S, Ma X, Wang X, Yu J, Wang YC, Smith DJ, et al. Creatine uptake regulates CD8 T cell antitumor immunity. J Exp Med. 2019;216:2869–82. doi: https://doi.org/10.1084/jem.20182044
44. Masoumi-Ardakani Y, Fallah H, Shahouzehi B. Carnitine effects on serum and pancreas inflammatory response in diabetic rats. Ukr Biochem J 2019;91:59–66. doi: https://doi.org/10.15407/ubj91.06.059
45. Mathieson S, Lin C-WC, Underwood M, Eldabe S. Pregabalin and gabapentin for pain. BMJ. 2020;369:m1315. doi: https://doi.org/10.1136/bmj.m1315
46. Brosnan JT, da Silva RP, Brosnan ME. The metabolic burden of creatine synthesis. Amino Acids. 2011;40:1325–31. doi: https://doi.org/10.1007/s00726-011-0853-y
47. Bhandari SD, Gallegos-Peretz T, Wheat T, Jaudzems G, Kouznetsova N, Petrova K, et al. Amino acid fingerprinting of authentic nonfat dry milk and skim milk powder and effects of spiking with selected potential adulterants. Foods. 2022;11(18):2868. doi: https://doi.org/10.3390/foods11182868
48. Meléndez-Hevia E, De Paz-Lugo P, Cornish-Bowden A, Cárdenas ML. A weak link in metabolism: the metabolic capacity for glycine biosynthesis does not satisfy the need for collagen synthesis. J Biosci. 2009;34:853–72. doi: https://doi.org/10.1007/s12038-009-0100-9
49. Bessler H, Mendel C, Straussberg R, Gurary N, Aloni D, Sirota L. Effects of dexamethasone on IL-1beta, IL-6, and TNF-alpha production by mononuclear cells of newborns and adults. Biol Neonate. 1999;75:225–33. doi: https://doi.org/10.1159/000014099
50. Shah K, Mujwar S, Gupta JK, Shrivastava SK, Mishra P. Molecular docking and In Silico Cogitation validate mefenamic acid prodrugs as human cyclooxygenase-2 inhibitor. Assay Drug Dev Technol. 2019;17:285–91. doi: https://doi.org/10.1089/adt.2019.943
51. Desiraju G, Steiner T. The weak hydrogen bond: in structural chemistry and biology. Oxford: Oxford University Press; 2001. doi: https://doi.org/10.1093/acprof:oso/9780198509707.001.0001
52. Sari BL, Elfrieda NSAL, Marsuan K, Sapitri P, Hafidh A. Aktivitas antioksidan dan studi In Silico ekstrak buah pala (Myristica fragrans Houtt). J Farmamedika. 2022;7:28–40. doi: https://doi.org/10.47219/ath.v7i1.142
53. Tambunan USF, Nasution MAF, Parikesit AA, Noviardi H, Kerami D. Designing of disulfide cyclic peptide for inhibiting polymerase A and B1 (PAC-PB1N) in H1N1 virus using molecular simulation approach. Online J Biol Sci. 2016;16:122–9. doi: https://doi.org/10.3844/ojbsci.2016.122.129
54. Alnajjar R, Mostafa A, Kandeil A, Al-Karmalawy AA. Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin II receptor blockers to inhibit the COVID-19 main protease. Heliyon. 2020;6:e05641. doi: https://doi.org/10.1016/j.heliyon.2020.e05641
55. Pattar SV, Adhoni SA, Kamanavalli CM, Kumbar SS. In silico molecular docking studies and MM/GBSA analysis of coumarin-carbonodithioate hybrid derivatives divulge the anticancer potential against breast cancer. Beni-Suef Univ J Basic Appl Sci. 2020;9(1):1–0. doi: https://doi.org/10.1186/s43088-020-00059-7
56. Falcón-Cano G, Molina C, Cabrera-Pérez MÁ. ADME Prediction with KNIME: development and validation of a Publicly available workflow for the prediction of human oral bioavailability. J Chem Inf Model. 2020;60:2660–7. doi: https://doi.org/10.1021/acs.jcim.0c00019
57. Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1:337–41. doi: https://doi.org/10.1016/j.ddtec.2004.11.007