INTRODUCTION
Background
Bacterial identification is generally accomplished through labor-intensive and time-consuming culture-based methods involving various biochemical tests. Such applications may be ineffective if the findings are needed for prompt emergency medical diagnosis. Furthermore, biochemical approaches may not always identify bacteria accurately and frequently provide incorrect conclusions [1,2]. Even though culture-based techniques are considered the gold standard for pathogen detection, these drawbacks include the need for trained personnel, standard reagents, labor intensiveness, time constraints, and limited specificity [3].
Developed countries or advanced hospital diagnostic facilities utilize analytical profile index assays, Vitek-2/mass spectroscopy (MS), nuclear magnetic resonance spectroscopy, or matrix-assisted laser desorption/ionization-time of flight MS as an alternative to culture-based approaches for pathogen detection [4–6]. In addition to these procedures, diagnostic facilities regularly utilize nucleic acid amplification techniques such as polymerase chain reaction (PCR), real-time PCR, isothermal amplification, and next-generation sequencing [7–10]. Though the diagnostic application has been implemented to identify signature sequences within species, these tests are not implemented outside reference laboratories due to their high cost and technical necessity [11]. Unlike conventional amplification methods, isothermal amplification methods such as loop-mediated isothermal amplification (LAMP) is a molecular approach known for being straightforward, fast, cost-effective, and highly reliable if adequately designed and suggested for usage in resource-constrained areas [12–15]. Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) are components of the commensal gut flora, and they are also the common opportunistic pathogens often associated with urinary tract and bloodstream infections [16]. Klebsiella pneumoniae is reported to be responsible for nearly one-third of all Gram-negative bacterium-related infections, such as urinary tract infections, cystitis, pneumonia, surgical wound infections, endocarditis, and septicemia [17]. On the other hand, although most E. coli species are harmless, few strains have been associated with various human infections categorized as diarrheagenic and extraintestinal pathogens, with various pathotypes and natural hybrid strains [18]. In addition, a high mortality risk is associated with patients infected with antibiotic-resistant strains of these microbes [19].
Importance of the study
Antibiotic resistance is a significant threat to healthcare systems. Annually, 700,000 people lose their lives worldwide due to infection with antibiotic-resistant bacteria. The fatality rate due to antibiotic resistance is estimated to be more than cancer by 2050 [20]. In line with such observations, the World Health Organization has designated antimicrobial resistance (AMR) as one of the top 10 worldwide public health problems confronting humanity and 12 families of bacteria posing a great threat, as treating infections against these is becoming quite tricky due to multidrug resistance [21]. Among these are the Enterobacteriaceae family members, such as E. coli and Klebsiella etiological agents, especially for urinary tract infections. Though AMR develops naturally, the misuse of antibiotics by humans accelerates the process. The factors contributing to AMR development are inappropriate prescription practices, inadequate patient literacy levels, unauthorized antibiotic sales, insufficient diagnostic facilities, and uncontrolled antimicrobial usage in animals [22,23].
In low- and middle-income countries (LMICs), there is often a lack of standardized national diagnostic protocols or monitoring policies. Diagnostic facilities may use inconsistent characterization methods or fail to adhere to protocols, leading to variable and inaccurate outcomes. This inconsistency in pathogen identification can result in improper treatment interventions, potentially contributing to AMR development. More studies are needed to monitor the accuracy of diagnoses in LMICs, as misidentification of clinical pathogens can promote incorrect antibiotic use, fueling AMR.
Aim of the study
This pilot study collected the previously characterized clinical E. coli and Klebsiella spp. isolates from two reputed hospital-based diagnostic facilities near Dhaka, Bangladesh. We attempted to determine their proper identity by applying different phenotypic, biochemical, and molecular techniques. Interestingly, we confirmed apparent misidentification in those diagnostic center-identified (DCI) clinical isolates, suggesting one of the possible routes of developing AMR in LMICs such as Bangladesh.
MATERIALS AND METHODS
Study samples
One hundred DCIs previously characterized by diagnostic facilities were collected from 2017 to 2018 [24]. The isolates were stored at −80°C and sub-cultured annually. Before working with the isolates, the cultures were thawed and grown in minimal media for recovery. To ensure the purity of the isolates, they were cultured on MacConkey agar (Scharlau, Spain, Catalog number: 01-118-500) and eosin methylene blue agar (Himedia, India, Catalog number: SKU M317) media.
Phenotypic characterization of the isolates
Immediately upon receiving the isolates, biochemical tests were carried out in our laboratory for characterization and identification. To cross-check this earlier characterization, we performed biochemical tests such as indole production, methyl red, Voges-Proskauer, citrate utilization, catalase, motility, triple sugar iron (TSI), and sugar utilization before starting this work.
Molecular detection
Boiling method for total DNA extraction
The chromosomal DNA of the bacterial isolates was extracted using the modified boiling DNA method [25]. Colonies isolated from the nutrient agar (Himedia, India, Catalog number: SKU M001) plate were grown overnight at 37°C in a test tube containing 5 ml of nutrient broth (Himedia, India, Catalog number: SKU M002). 1 ml culture was placed in a 1.5 ml microcentrifuge tube, and cells were separated by centrifugation at 12,000 rpm for 10 minutes. The pellets were resuspended in 200 μl of UltraPure™ DNase/RNase-Free Distilled Water (Invitrogen, USA, Catalog number: 10977015). Following this, each microcentrifuge tube was kept at 90°C–95°C for 10 minutes in a block heater (Thermo Fisher Scientific, USA, Catalog number: 88870003), then placed on ice for 10 minutes. The tubes were then centrifuged for 10 minutes at 10,000 rpm. The supernatant was collected into a new microcentrifuge tube and kept at −80°C for long-term storage.
Randomly amplified polymorphic DNA (RAPD) analysis
Extracted bacterial DNA was subjected to RAPD genotyping. Random primer (10 bp) 5’-AAGAGCCCGT-3’ was selected to observe their band pattern to classify them into different groups. The PCR reactions were performed in 25 μl volumes containing 12.5 μl GoTaq® G2 Green Master Mix (2×) master mix (Promega, USA, Catalog number: M7822), 2 μl of primer, 4 μl template DNA, and 6.5 μl nuclease-free water. PCR condition was set at 94°C for 30 seconds, followed by 36°C for 15 seconds, and 72°C for 30 seconds for 45 cycles in TaKaRa PCR Thermal Cycler Dice® Touch (Takara, Japan, Catalog number: TP350) [26]. Amplicons were characterized in 1% agarose gel. Binary data charts were produced based on the presence or absence of specific bands at specific locations for constructing dendrograms. The DendroUPGMA program (Universitat Rovira i Virgili (URV), Tarragona, Spain) was used to create dendrograms from binary data charts. The program calculates a similarity matrix, transforms similarity coefficients into distances, and makes a clustering using the unweighted pair group method with arithmetic mean (UPGMA) [27].
PCR of E. coli phoA gene
The alkaline phosphatase (phoA) gene was used to identify E. coli. 5’-AAGTTGAAGGTGCGTCAAT-3’-F3 and 5’-CTTGTGAATCCTCTTCGGAG-3’-B3 primers were used to identify E. coli isolate more accurately [28]. The PCR reactions were performed in 25 μl volumes containing 12.5 μl master mix, 2 μl each of the forward and reverse primers, 2 μl template DNA, and 8.5 μl nuclease-free water. PCR condition was set at 94°C for 30 seconds, followed by 50°C for 15 seconds and 72°C for 30 seconds for 45 cycles. Amplicon’s length of approximately 277 bp was initially confirmed using a molecular ladder to amplify phoA. Later, PCR bands were extracted from agarose gel and sequenced to confirm the specific amplification.
PCR of K. pneumoniae rcsA gene
The rcsA gene in K. pneumoniae produces capsular polysaccharides, an essential factor in its virulence. 5’-GGATATCTGACCAGTCGG-3’-KP27F3 and 5’- GGGTTTTGCGTAATGATCTG-3’-KP27B3 primer was used for the detection of K. pneumoniae [29]. The PCR reactions were performed in 25 μl volumes containing 12.5 μl master mix, 1 μl each of primer F and primer R, 4 μl template DNA, and 6.5 μl nuclease-free water. PCR condition was set at 94°C for 30 seconds, followed by 55°C for 15 seconds, and 72°C for 30 seconds for 45 cycles. Amplicon’s length of approximately 170 bp was initially confirmed using a molecular ladder to amplify rcsA and later confirmed by Sanger sequencing.
PCR of 16S rRNA gene
5’-AGAGTTTGATCCTGGCTCAG-3’ forward and 5’-CGGTTACCTTGTTACGACTT-3’ reverse primers were used to amplify the template DNA for 16S rRNA gene sequencing. For 25 μl PCR reactions, the PCR mixture contained 12.5 μl master mix, 2 μl each of the forward and reverse primers, 2 μl template DNA, and 8.5 μl nuclease-free water [30].
Phylogenetic analysis
The PCR products were analyzed on 1% agarose gel using an agarose gel electrophoresis Mupid-2plus system (Takara Bio, Japan, Catalog number: AD110). The amplicons were purified using the FavorPrep GEL/PCR Purification Kit (Favorgen Taiwan, Catalog number: FAGCK 001), as directed by the manufacturer. After purification, PCR products were used for Sanger dideoxy sequencing (3500 Series Genetic Analyzer, Applied Biosystems). The primary local alignment search tool (BLAST) was used to identify close phylogenetic relatives by comparing partial sequences to the GenBank database of the National Center for Biotechnology Information (NCBI) [31]. The phylogenetic tree was generated using BioEdit, ApE plasmid editor, and MEGA 11 software.
Loop-mediated isothermal amplification
LAMP reactions were conducted in 25 μl volumes containing 15 μl Bst 2.0 Polymerase Isothermal master mix (NEB, UK, Catalog number: M0537S), 5 μl (1×) Primers, and 5 μl Template DNA [32]. The list of primer sequences required to detect E. coli and K. pneumoniae is shown in Table 1. For detection by LAMP, the template DNA was boiled in a water bath at 95°C for 5 minutes before being immediately transferred to ice. Master mix, primer, and template DNA were mixed to prepare the reaction solution and incubated at 63°C temperature in a water bath for 30 minutes. The enzymatic reaction was stopped by incubating the tubes at 80°C temperature for 2 minutes. The LAMP products were visualized by subjecting them to gel electrophoresis in 1% agarose gel.
RESULTS
Isolation and characterization of E. coli and Klebsiella spp. isolates
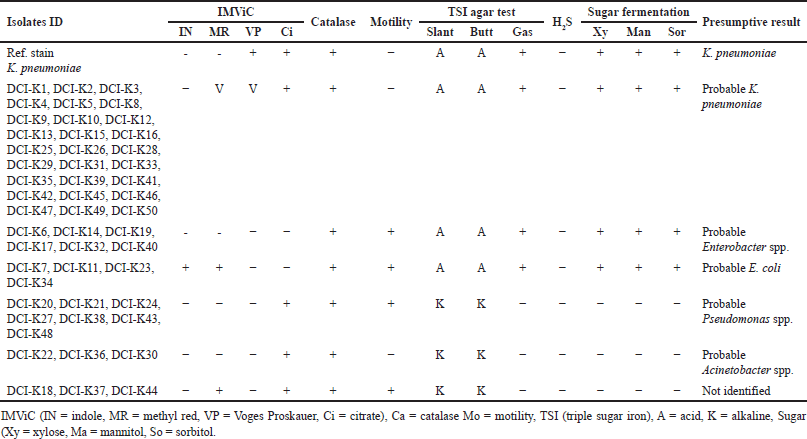
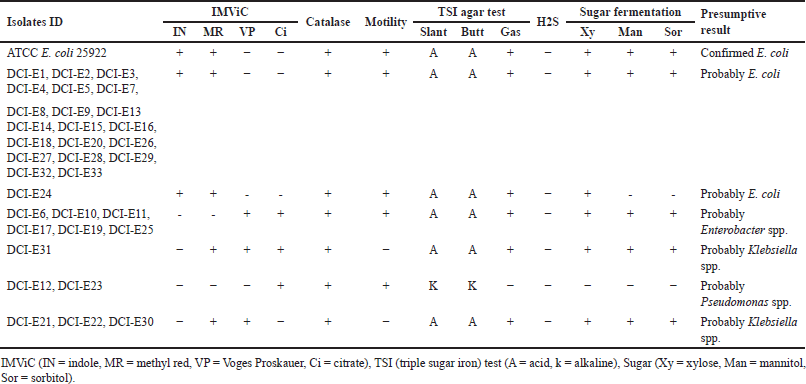
One hundred precharacterized bacterial isolates were collected from diagnostic facilities. Previously, 33 E. coli and 50 Klebsiella isolates were selected for further study due to their prevalence and clinical significance [24]. Presumptive re-identification through culturing on selective media and biochemical testing revealed 30% of presumed E. coli and 46% of suspected Klebsiella isolates were misidentified by the diagnostic centers (Tables 2 and 3). Based on the biochemical profile, the misidentified isolates belonged to genera including Enterobacter, Acinetobacter, and Pseudomonas.
Molecular characterization of the bacterial isolates
RAPD results
RAPD profiling revealed eight different band patterns among the DCI-K (Fig. 1A). All isolates belonging to the first seven groups displayed the same intra-group band pattern, whereas those in group 8 showed different banding patterns from each other. However, although K17 and K45 showed the same RAPD profile biochemically, they differed. On the contrary, based on the banding pattern of 16 of all 33 isolates, DCI-E isolates were represented by three groups. The other 17 isolates exhibited dissimilar band patterns, and they all were represented by group 4 (Fig. 1B). These banding patterns were used to select representative isolates for characterization through PCR and sequencing.
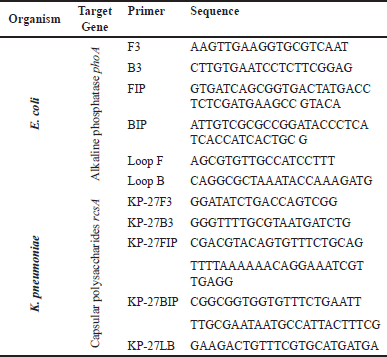 | Table 1. The primer sequence of the LAMP process for detecting E. coli and K. pneumoniae. [Click here to view] |
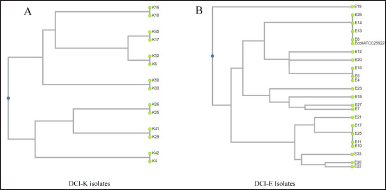 | Figure 1. UPGMA dendrograms of the representative DCI-K and DCI-E isolates, showing the formation of clusters based on their RAPD profiles. (A) DCI-K4 and DCI-K42 from group 1, DCI-K10 and DCI-K16 from group 2, DCI-K25 and DCI-K26 from group 3, DCI-K17 and DCI-K45 from group 4, DCI-K6 and DCI-K32 from group 5, DCI-K33 and DCI-K50 from group 6, and DCI-K29 and DCI-K41 from group 7 were selected as the representative isolates of the respective DCI-K groups. They were plotted as distinct clusters based on their similarities in the RAPD band pattern. (B) In the cases of DCI-E isolates, DCI-E8, DCI-E13, DCI-E14, and DCI-E26 from group 1 showed RAPD band patterns such as E. coli ATCC 25922 and were clustered together, whereas DCI-E4, DCI-E5, and DCI-E15 from group 2, DCI-E10, DCI-E11, DCI-E17, and DCI-E25 from group 3 were clustered together as seen in the dendrogram. On the contrary, E7DCI-E12, DCI-E18, DCI-E19, DCI-E20, DCI-E21, DCI-E22, DCI-E23 DCI-E27, and DCI-E30 showed different band patterns and were plotted distantly in the dendrogram. [Click here to view] |
 | Table 2. Biochemical test results of DCI-K (according to hospital record) pathogens. [Click here to view] |
Detection of E. coli and K. pneumoniae using phoA and rcsA genes specific PCR
The alkaline phosphatase gene was used to characterize 33 DCI-E isolates and 3 isolates biochemically positive for E. coli among the DCI-Klebsiella spp. samples. Vibrio cholerae ATCC (14035), K. pneumoniae reference strain, Micrococcus luteus ATCC [4698 (150307)], and Staphylococcus aureus ATCC (6538) were used as negative controls in the experiment. Twenty-three of the 33 DCI-E isolates and 2 from the DCI-K displayed a banding pattern similar to E. coli reference strains (JM109, V517, BL21, and DE3) and E. coli ATCC 25922 (Fig. 2A). Similarly, the K. pneumoniae strains amongst the DCI-K isolates that were biochemically determined to be Klebsiella spp. were identified using the K. pneumoniae-specific capsular polysaccharide gene rcsA. Reference strain was used as positive controls, while V. cholerae ATCC (14035), M. luteus ATCC (4698), S. aureus ATCC (6538), Salmonella typhi ATCC (14028), Bacillus cereus ATCC (14574), and E. coli ATCC (25922) were used as the negative controls. No bands were observed following PCR amplification and gel electrophoresis in negative control lanes. However, 13 of 27 isolates were biochemically positive for Klebsiella spp. gave a banding pattern similar to K. pneumoniae reference strains (Fig. 2B).
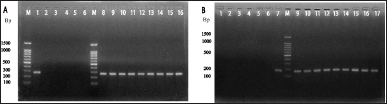 | Figure 2. Gel electrophoresis analysis of PCR amplified (A) phoA gene and (B) rcsA gene. Agarose gel electrophoresis (on 1.5% agarose gel) image showing the amplified products by PCR, which was done by using phoA primer (~270 bp) and rcsA primer (~176 bp). Lane M represents the molecular marker spanning 100–1,500 base pairs (bp). (A) Lane 1 corresponds to E. coli ATCC (25922) as a positive control. Lanes 2–6 represent negative control, with lane 2 designated as general negative control, lane 3 as K. pneumoniae reference stain, lane 4 as V. cholerae ATCC (14035), lane 5 as M. luteus ATCC (4698), and lane 6 as S. aureus ATCC (6538). The sample lanes (8–16) contain the amplified phoA products (located between 200 and 300 bp sized band of DNA marker) showing positive results in the form of distinct bands. (B) Lanes 1–6 corresponds to negative control with as lane 1 as V. cholerae ATCC (14035), lane 2 as E. coli ATCC (25922), lane 3 as M. luteus ATCC (4698), lane 4 as S. typhi ATCC (14028), lane 5 as S. aureus ATCC (6538), and lane 6 as B. cereus ATCC (14574). Lane 7 represents K. pneumoniae reference strain as the positive control, and lanes 9–17 show positive results (bands) for rcsA gene-positive clinical isolates. These band sizes are ~176 bp and located just below 200 bp. [Click here to view] |
Sequencing and homology alignment of amplified phoA and rcsA genes
phoA and rcsA amplicons were found in 25 and 17 isolates of presumptively characterized E. coli and K. pneumoniae, respectively. Three amplicons from each group were subjected to sequencing using the forward primers. phoA and rcsA gene sequences from E. coli (Accession No. MG1655) and K. pneumoniae (Accession No. NNP58345) were retrieved for homology alignment. Multiple sequence alignments (MSAs) of these gene sequences were performed using ClustalW multiple alignment tools. MSA found the amplicons specific to their respective genes (Fig. 3).
16S rRNA PCR, sequencing, and phylogenetic analysis
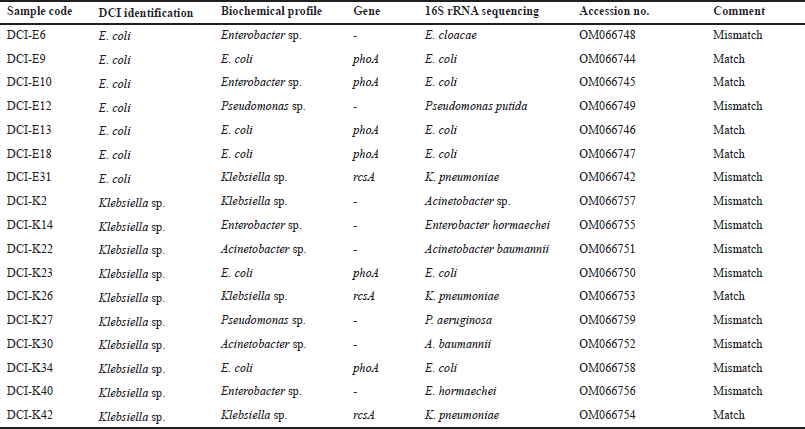
Representative isolates from the RAPD pattern and biochemical tests were subjected to 16S rRNA PCR for further confirmation of the match and mismatches of bacterial identification by diagnostic centers. The PCR products were gel extracted and sequenced. The sequences were submitted to Genebank. Sequencing confirms the misidentification of clinical pathogens by diagnostic centers (Table 4).
Closely related reference 16S rRNA sequences were downloaded from the NCBI database, and sequences were aligned with the ClustalW MSA tool. The alignments were subjected to phylogenetic tree development using the maximum likelihood method and the Tamura-Nei model (Mega 11). A bootstrap value of 1,000 was set to minimize the error of distant measurement. The phylogenetic tree generated with selected DCI-E and DCI-K shows matched and mismatched isolates in different branches (Fig. 4).
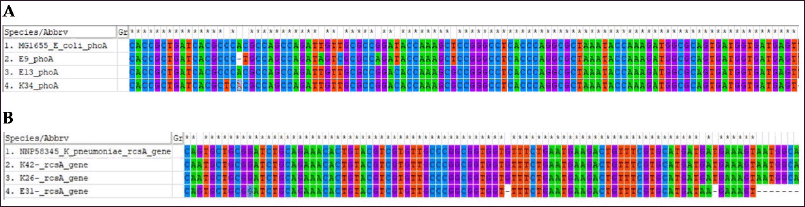 | Figure 3. Homology alignment of phoA and rcsA gene. (A) phoA amplicon sequences from three isolates, two from DCI-E (DCI-E9 and DCI-E13) and one DCI-K (DCI-K34), were aligned with the phoA gene sequence of reference E. coli (MG1655), and the amplicons matched the reference sequence. (B) rcsA amplicon sequences from three isolates, two from DCI-K (DCI-K26 and DCI-K42) and one DCI-E (DCI-E31), were aligned with the rcsA gene sequence of reference K. pneumoniae (NNP58345), and the amplicons matched the reference sequence. [Click here to view] |
 | Table 3. Biochemical test results of selected DCI-E (according to hospital record) pathogens. [Click here to view] |
 | Table 4. Comparative identification of bacterial pathogens by the diagnostic centers, biochemical profiling, phoA, and rcsA gene-specific PCR, and 16S rRNA sequencing. [Click here to view] |
Rapid molecular identification of clinical isolates by LAMP method
Biochemical, phoA, and rcsA gene PCR-positive E. coli (DCI-E9, DCI-E10, DCI-E13, DCI-K23, and DCI-K34) (Fig. 5A) and K. pneumoniae (DCI-E31, DCI-K26, and DCI-K42) were subjected to LAMP analysis (Fig. 5B). All phoA and rcsA gene-positive isolates showed multiple band patterns in 1% agarose gel.
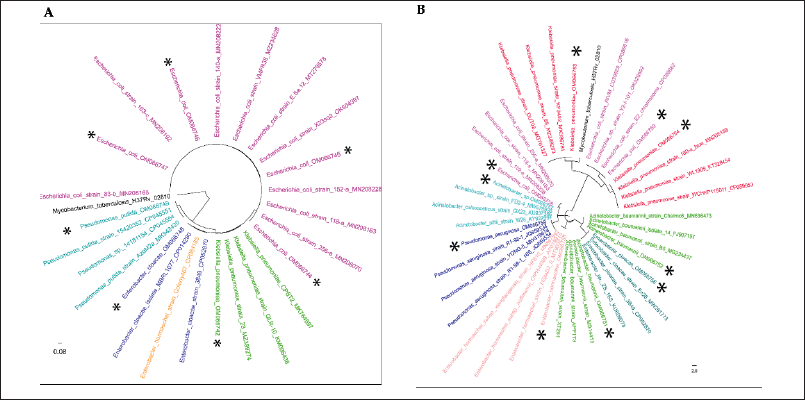 | Figure 4. Evolutionary analyses were conducted in MEGA11 using the maximum likelihood method and the Tamura-Nei model. The study isolates are marked with the * sign. (A) Tree with the highest log likelihood (−4,590.91) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura-Nei model and then selecting the topology with a superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. There were a total of 1,558 positions in the final dataset. Four of the phoA gene-positive isolates, DCI-E9, DCI-E10, DCI-E13, and DCI-E18 (OM066744, OM066745, OM066746, and OM066747), closely clustered with reference E. coli isolates. phoA PCR negative isolate DCI-E6 (OM066748), DCI-E31 (OM066742), and DCI-E12 (OM066749), closely clustered with Enterobacter cloacae, K. pneumoniae, and Pseudomonas spp. (B) The tree with the highest log likelihood (−6,340.37) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura-Nei model and then selecting the topology with a superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 41 nucleotide sequences. Codon positions included were first + second + third + noncoding. There were a total of 1,288 positions in the final dataset. Two of the rcsA gene-positive isolates, DCI-K26 and DCI-K42 (OM066753 and OM066754), are closely clustered with reference K. pneumoniae isolates. rcsA PCR negative isolate DCI-K23 (OM066750) and DCI-K34 (OM066758) closely clustered with E. coli reference strains; isolates DCI-K2 (OM066757), DCI-K22(OM066751), and DCI-K30 (OM066752) closely clustered with Acinetobacter spp. reference strains; isolate DCI-K14 (OM066755) and DCI-K40 (OM066756) closely clustered with Enterobacter reference strains, and isolate DCI-K27 (OM066759) closely clustered Pseudomonas aeruginosa reference strains. [Click here to view] |
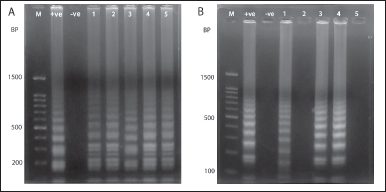 | Figure 5. Agarose gel electrophoresis (1% agarose gel) image of LAMP method. A. From left, M indicates molecular marker (100–1,500) bp, E. coli ATCC 25922, and K. pneumoniae reference strain as the positive and negative control, respectively. Lanes 1–5 represent positive bands of clinical isolates (DCI-E9, DCI-E10, DCI-E13, DCI-K23, and DCI-K34) positive for the phoA gene. B. Lane 1 (+ve) was positive control of K. pneumoniae reference strain, lane 2 (−ve) was negative control [E. coli ATCC (25922)], lanes 1, 3, and 4 were rcsA positive isolates (DCI-E31, DCI-K26, and DCI-K42), and lanes 2 and 5 were blanks. [Click here to view] |
DISCUSSION
This study suggests that diagnostic tests’ accuracy, precision, analytical sensitivity, analytical specificity, reportable range, and internal reference should be validated regularly against gold standards and monitored by proper regulatory authorities [33]. Like other departments of a diagnostic facility, the microbiology department should also strictly follow standards set by the Clinical and Laboratory Standards Institute (CLSI), European Committee on Antimicrobial Susceptibility Testing, and so on, while performing culture and sensitivity assays [34–36]. The false-negative results generated by these centers can increase mortality and morbidity, whereas false-positive results influence wrong interventions, misuse of drugs, and economic burden [2,37].
Previously, there were some reports published on misidentification by diagnostic centers. However, those reports implemented the identification of organisms using biochemical tests [1,24,38]. In this pilot study, we have used molecular techniques in conjunction with biochemical tests to identify the extent of misidentification, suggesting that rapid and cost-effective assay for clinical diagnostics is suitable for LMICs such as Bangladesh. After the biochemical test, among the DCI-E isolates, 70%, 12%, 9%, 6%, and 3% were found to be E. coli, Enterobacter spp., Klebsiella spp., Pseudomonas spp., and unknown, respectively, whereas, among all DCI-K isolates, 54%, 12%, 6%, 16%, 6%, and 6% were identified as Klebsiella spp., Enterobacter spp., E. coli, Pseudomonas spp., Acinetobacter spp., and unknown, respectively (Tables 2 and 3). This high rate of discrepancies in pathogen identification prompted us to use molecular tests to justify our claims.
Molecular techniques are the more sensitive and fast diagnostic tools for detecting pathogens in clinical samples. RAPD is a PCR method requiring no prior knowledge of the genome since the amplified fragments are random. This makes the technique well-regarded for comparing biological systems’ DNA and diversity analysis of pathogens in environmental and clinical samples [39–43]. We tried to utilize RAPD to cluster the isolates based on band patterns to reduce the number of tests. UPGMA-based dendrograms showed the representative Klebsiella spp., and E. coli isolates in distinct clusters based on the similarities among their RAPD band patterns. When DCI-E and DCI-K were analyzed, the former isolates were clustered into four groups, whereas the latter were into eight groups (Fig. 1). The last groups of each DCI isolate contained isolates with unique band patterns that could not be clustered into a specific group. Thus, RAPD helped us to shorten the number of samples that needed further analysis. However, multiple clustering of the same genus makes RAPD unsuitable for diagnostic use in pathogen identification.
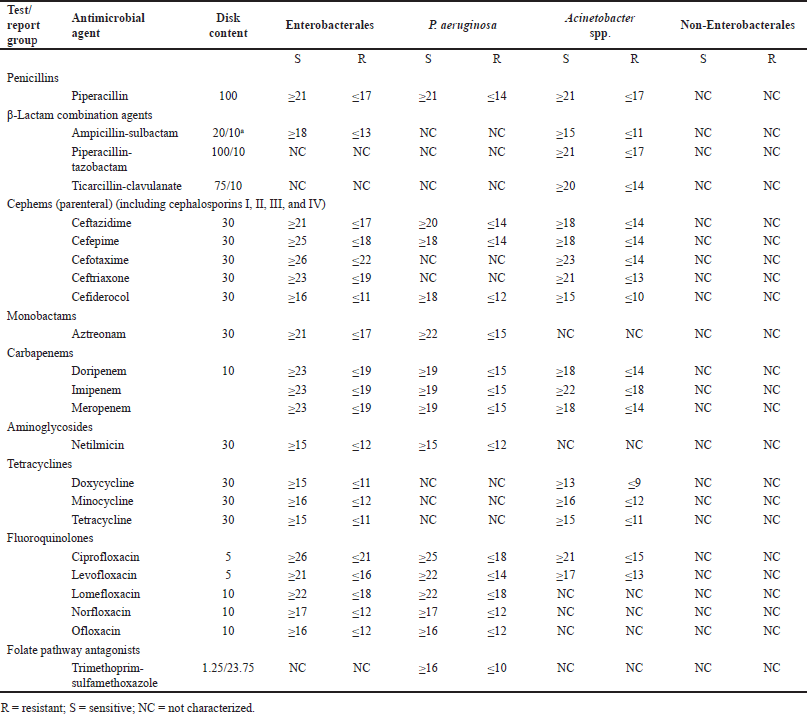 | Table 5. Variation of antibiotic susceptibility pattern in different groups of Gram-negative bacterial pathogens according to CLSI. [Click here to view] |
 | Table 6. List of antibiotics for which no standard is set for P. aeruginosa, Acinetobacter spp., and non-Enterobacterales according to CLSI. [Click here to view] |
phoA gene encodes bacterial alkaline phosphatase. Under low phosphate conditions, E. coli alkaline phosphatase is synthesized and secreted across the inner membrane to the periplasmic space, which plays a critical role in the breakdown of organic phosphate esters [44,45]. Among 33 DCI-E, 23 (70%) isolates and 2 isolates from DCI-K were phoA positive (Fig. 2A). Sequencing the phoA amplicons confirmed their proper amplification and identification (Fig. 3A). On the other hand, rcsA is involved in expressing the K antigen capsule [46] of K. pneumoniae. Among Klebsiella spp., K. pneumoniae is mainly responsible for infectious diseases. rcsA gene was selected for molecular detection of K. pneumoniae because of its clinical significance [2]. Thirteen (48%) isolates from 27 biochemically positive Klebsiella spp. among the 50 DCI-K were rcsA gene positive (Fig. 2B). Identities were confirmed by sequencing the rcsA amplicons (Fig. 3B).
The absence of phoA and rcsA genes in 30% DCI-E and 46% DCI-K were further characterized by 16S rRNA PCR followed by sequencing. Misidentification was confirmed in both groups, where Enterobacter sp., Klebsiella sp., and Pseudomonas sp. were misidentified as E. coli (Table 4, Fig. 4A). On the other hand, Enterobacter sp., E. coli, Pseudomonas sp., and Acinetobacter spp. were misidentified as Klebsiella, not only by the diagnostic centers (Table 4, Fig. 4B).
Later, we employed LAMP, a rapid, simple, specific, and cost-effective nucleic acid amplification method developed by Eiken Chemical Co., Ltd., LAMP amplifies DNA with high specificity, efficiency, and rapidly under the isothermal condition where two to three sets of primers (outer, loop, and inner) are used. It is a cost-effective detection method that can be performed with simple instruments, such as a heater or water bath [47]. The LAMP method can detect E. coli and K. pneumoniae [27,28]. We used the rcsA gene and phoA gene primer set (Table 1) to detect K. pneumoniae and E. coli from clinical samples within 60 minutes (Fig. 5). Due to the high-sensitivity nature of LAMP in the case of K. pneumoniae, we observed some cross-reacting nonspecific bands with negative controls. Such discrepancies were reported previously, emphasizing the importance of validating these methods before clinical use and overcoming the misidentification of clinical pathogens [48,49].
The impact of the misidentification of clinical pathogens on AMR development is yet to be understood. However, Table 5 shows that, according to CLSI, antibiotic suggestion and susceptibility patterns in the disk-diffusion method vary among bacterial groups. Although the pattern or suggested antibiotics are the same for the genera belonging to Enterobacterales, the largest group of pathogenic bacteria [50], the situation varies outside this order. For non-Enterobacterales, no standards are set for the disk-diffusion method. Moreover, sensitivity patterns of many antibiotics (Table 6) are only known for Enterobacterales. Therefore, we hypothesized that misidentification followed by improper antibiotic suggestions may lead to AMR development.
LIMITATION OF THE STUDY
This pilot study works with a limited number of clinical pathogens; however, the scenario of misidentification was observed. Further experimental proof is required to identify the prospective route of antibiotic resistance development, which is beyond the scope of this study.
CONCLUSION
Here, we found that traditional culture and biochemical processes are not always accurate, whereas molecular techniques are more precise and ensure proper identification. Since antibiotic susceptibility varies among genera, misidentification by the diagnostic facilities/laboratories may lead to the wrong antibiotic prescribing and, consequently, may result in antibiotic resistance development. Therefore, healthcare authorities of a country, especially in the LMICs, should develop policies for the healthcare and diagnostic institutes to provide standard services and regularly monitor the practices. Policies should be included in a country’s antibiotic stewardship program to mitigate diagnostic centers’ misidentification of clinical pathogens. Diagnostic centers and healthcare facilities should follow international and national guidelines for correctly identifying clinical pathogens and their susceptibility to antibiotics. Moreover, healthcare workers should be adequately trained to perform and interpret the test results.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
This research was conducted with funds from the Ministry of Science and Technology (MOST), Bangladesh, and the University Grant Commission (UGC), Bangladesh.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
The study protocol was approved by the Biosafety, Biosecurity, and Ethic Committee of Jahangirnagar University Bangladesh [Ref No: BBEC, JU/M-2022(5), dated 02-02-2022].
DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
PREPRINT
A major part of this article was previously posted to the medRxiv preprint server on January 19, 2022. https://doi.org/10.20944/preprints202201.0225.v2. The authors undertake that they will inform the preprint server once the article is accepted.
REFERENCES
1. Frederiksen W, Tønning B. Possible misidentification of Haemophilus aphrophilus as Pasteurella gallinarum. Clin Infect Dis. 2001;32(6):987–9. CrossRef
2. Filice GA, Drekonja DM, Thurn JR, Hamann GM, Masoud BT, Johnson JR. Diagnostic errors that lead to inappropriate antimicrobial use. Infect Control Hosp Epidemiol. 2015;36(8):949–56. CrossRef
3. Abayasekara LM, Perera J, Chandrasekharan V, Gnanam VS, Udunuwara NA, Liyanage DS, et al. Detection of bacterial pathogens from clinical specimens using conventional microbial culture and 16S metagenomics: a comparative study. BMC Infect Dis. 2017;17:631. CrossRef
4. Abrar S, Ain NU, Liaqat H, Hussain S, Rasheed F, Riaz S. Distribution of blaCTX - M, blaTEM, blaSHV and blaOXA genes in extended-spectrum-β-lactamase-producing clinical isolates: a three-year multi-center study from Lahore, Pakistan. Resist Infect Control. 2019;8:80. CrossRef
5. Rodrigues C, Passet V, Rakotondrasoa A, Brisse S. Identification of Klebsiella pneumoniae, Klebsiella quasipneumoniae, Klebsiella variicola and related phylogroups by MALDI-TOF mass spectrometry. Front Microbiol. 2018;9:3000. CrossRef
6. Mitchell SL, Alby K. Performance of microbial identification by MALDI-TOF MS and susceptibility testing by VITEK 2 from positive blood cultures after minimal incubation on solid media. Eur J Clin Microbiol Infect Dis. 2017;36(11):2201–6. CrossRef
7. Leach L, Zhu Y, Chaturvedi S. Development and validation of a real-time PCR assay for rapid detection of Candida auris from surveillance samples. J Clin Microbiol. 2018;56(2):e01223–17. CrossRef
8. Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341–55. CrossRef
9. Zhong LL, Zhou Q, Tan CY, Roberts AP, El-Sayed Ahmed MAE, Chen G, et al. Multiplex loop-mediated isothermal amplification (multi-LAMP) assay for rapid detection of mcr-1 to mcr-5 in colistin-resistant bacteria. Infect Drug Resist. 2019;12:1877–87. CrossRef
10. Jiang M, Pan W, Arasthfer A, Fang W, Ling L, Fang H, et al. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. Front Cell Infect Microbiol. 2020;10:331. CrossRef
11. Kai S, Matsuo Y, Nakagawa S, Kryukov K, Matsukawa S, Tanaka H, et al. Rapid bacterial identification by direct PCR amplification of 16S rRNA genes using the MinION™ nanopore sequencer. FEBS Open Bio. 2019;9(3):548–57 CrossRef
12. Kapalamula TF, Thapa J, Akapelwa ML, Hayashida K, Gordon SV, Hang’Ombe BM, et al. Development of a loop-mediated isothermal amplification (LAMP) method for specific detection of Mycobacterium bovis. PLoS Negl Trop Dis. 2021;15(1):e0008996. CrossRef
13. Schoepp NG, Schlappi TS, Curtis MS, Butkovich SS, Miller S, Humphries RM, et al. Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Sci Transl Med. 2017;9(410):eaal3693. CrossRef
14. Tian Y, Zhang T, Guo J, Lu H, Yao Y, Chen X, et al. A LAMP-based microfluidic module for rapidly detecting pathogen in cryptococcal meningitis. Talanta. 2022;236:122827. CrossRef
15. Ramezani R, Kardoost Parizi Z, Ghorbanmehr N, Mirshafiee H. Rapid and simple detection of Escherichia coli by loop-mediated isothermal amplification assay in urine specimens. Avicenna J Med Biotechnol. 2018;10(4):269–72.
16. Hyun M, Lee JY, Kim HA, Ryu SY. Comparison of Escherichia coli and Klebsiella pneumoniae acute pyelonephritis in Korean patients. Infect Chemother. 2019;51(2):130–41. CrossRef
17. Effah CY, Sun T, Liu S, Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. 2020;19(1):1. CrossRef
18. Braz VS, Melchior K, Moreira CG. Escherichia coli as a multifaceted pathogenic and versatile bacterium. Front Cell Infect Microbiol. 2020;10:548492. CrossRef
19. Sianipar O, Asmara W, Dwiprahasto I, Mulyono B. Mortality risk of bloodstream infection caused by either Escherichia coli or Klebsiella pneumoniae producing extended-spectrum β-lactamase: a prospective cohort study. BMC Res Notes. 2019;12(1):719. CrossRef
20. Mallah N, Badro DA, Figueiras A, Takkouche B. Association of knowledge and beliefs with the misuse of antibiotics in parents: a study in Beirut (Lebanon). PLoS One. 2020;15(7):e0232464. CrossRef
21. World Health Organization. [cited 2023 Nov 15]. Available from: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed
22. Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. 2017;6:47. CrossRef
23. Nepal A, Hendrie D, Selvey LA, Robinson S. Factors influencing the inappropriate use of antibiotics in the Rupandehi district of Nepal. Int J Health Plann Manage. 2021;36(1):42–59. CrossRef
24. Bristy FS, Uddin MR, Mahmud SA, Uddin MJ, Talukder AZ, Khondoker M, et al. Reassessment of clinical pathogens diagnosed by sub-urban facilities of Dhaka: necessity of comprehensive techniques to manage antibiotic resistance. Microbial Bioactives. 2018;1:51–8. CrossRef
25. Queipo-Ortuño MI, De Dios Colmenero J, Macias M, Bravo MJ, Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin Vaccine Immunol. 2008 Feb;15(2):293–6. CrossRef
26. Khan A, Das SC, Ramamurthy T, Sikdar A, Khanam J, Yamasaki S, et al. Antibiotic resistance, virulence gene, and molecular profiles of Shiga toxin-producing Escherichia coli isolates from diverse sources in Calcutta, India. J Clin Microbiol. 2002 Jun;40(6):2009–15. CrossRef
27. Garcia-Vallvé S, Palau J, Romeu A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Biol Evol. 1999 Sep 1;16(9):1125–34. CrossRef
28. Stratakos AC, Linton M, Millington S, Grant IR. A loop-mediated isothermal amplification method for rapid direct detection and differentiation of nonpathogenic and verocytotoxigenic Escherichia coli in beef and bovine faeces. J Appl Microbiol. 2017;122(3):817–28. CrossRef
29. Dong D, Liu W, Li H, Wang Y, Li X, Zou D, et al. Survey and rapid detection of Klebsiella pneumoniae in clinical samples targeting the rcsA gene in Beijing, China. Front Microbiol. 2015;6:519. CrossRef
30. Vasco K, Nohomovich B, Singh P, Venegas-Vargas C, Mosci RE, Rust S, et al. Characterizing the cattle gut microbiome in farms with a high and low prevalence of Shiga toxin-producing Escherichia coli. Microorganisms. 2021;9(8):1737. CrossRef
31. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. CrossRef
32. Bhadra S, Jiang YS, Kumar MR, Johnson RF, Hensley LE, Ellington AD. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of middle east respiratory syndrome coronavirus (MERS-CoV). PLoS One. 2015;10(4):e0123126. CrossRef
33. Burd EM. Validation of laboratory-developed molecular assays for infectious diseases. Clin Microbiol Rev. 2010;23(3):550–76. CrossRef
34. Humphries RM, Ambler J, Mitchell SL, Castanheira M, Dingle T, Hindler JA, et al. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol. 2018;56:e01934–17. CrossRef
35. Melvin P, Weinstein MD. Performance standards for antimicrobial susceptibility testing, M100. 31st ed. Wayne, NJ: Clinical and Laboratory Standards Institute; 2022. p 352. Available from: https://clsi.org/standards/products/microbiology/documents/m100/
36. Giske CG, Turnidge J, Cantón R, Kahlmeter G; EUCAST Steering Committee. Update from the European Committee on Antimicrobial Susceptibility Testing (EUCAST). J Clin Microbiol. 2022;60(3):e0027621. CrossRef
37. Rodziewicz TL, Houseman B, Hipskind JE. medical error reduction and prevention [Updated 2023 May 2]. [Internet]. Treasure Island, FL: StatPearls Publishing; 2023. Available from: https://www.ncbi.nlm.nih.gov/books/NBK499956/
38. Fateh R, Jalal M, Maryam D. Misidentification of common pathogenic bacteria in a University Hospital Laboratory. Iran J Infect Dis Trop Med. 2016;21:49–54. Available from: https://www.sid.ir/en/journal/ViewPaper.aspx?id=523967
39. Nielsen KL, Godfrey PA, Stegger M, Andersen PS, Feldgarden M, Frimodt-Møller N. Selection of unique Escherichia coli clones by random amplified polymorphic DNA (RAPD): evaluation by whole genome sequencing. J Microbiol Methods. 2014;103:101–3. CrossRef
40. Zare S, Derakhshandeh A, Haghkhah M, Naziri Z, Broujeni AM. Molecular typing of Staphylococcus aureus from different sources by RAPD-PCR analysis. Heliyon. 2019;5(8):e02231. CrossRef
41. Mobasherizadeh S, Shojaei H, Havaei SA, Mostafavizadeh K, Davoodabadi F, Khorvash F, et al. Application of the random amplified polymorphic DNA (RAPD) fingerprinting to analyze genetic variation in community associated-methicillin resistant Staphylococcus aureus (CA-MRSA) isolates in Iran. Glob J Health Sci. 2016;8(8):53822. CrossRef
42. Feruke-Bello YM, Babalola G, Odeyemi O. Genetic variability of Klebsiella variicola by RAPD-PCR technique and bioremoval of Pb2+ and Cd2+ from simulated contaminated soils. Soil Sediment Contamin Int J. 2021;31:770–84. CrossRef
43. Auda IG, Al-Kadmy IM, Kareem SM, Lafta AK, A’Affus MH, Khit IA, et al. RAPD- and ERIC-based typing of clinical and environmental Pseudomonas aeruginosa isolates. J AOAC Int. 2017;100(2):532–6. CrossRef
44. Zhou Y, Zhang T, Jin S, Chen S, Zhang Y. Effects of Escherichia coli alkaline phosphatase phoA on the mineralization of dissolved organic phosphorus. Water. 2021;13:3315. CrossRef
45. Kriakov J, Lee Sh, Jacobs WR Jr. Identification of a regulated alkaline phosphatase, a cell surface-associated lipoprotein, in Mycobacterium smegmatis. J Bacteriol. 2003;185:4983–91. CrossRef
46. McCallum KL, Whitfield C. The rcsA gene of Klebsiella pneumoniae O1:K20 is involved in expression of the serotype-specific K (capsular) antigen. Infect Immun. 1991;59(2):494–502. CrossRef
47. Hema M, Konakalla NC. Recent developments in detection and diagnosis of plant viruses. In: Viswanath B. Recent developments in applied microbiology and biochemistry. Academic Press; 2021. pp 163–80. CrossRef
48. Senarath K, Usgodaarachchi R, Navaratne V, Nagahawatte A, Wijayarathna C, Alvitigala J, et al. Non-specific amplification with the LAMP technique in the diagnosis of tuberculosis in Sri Lankan settings. J Tuberculosis Res. 2014;2:168–72. CrossRef
49. Hardinge P, Murray JAH. Reduced false positives and improved reporting of loop-mediated isothermal amplification using quenched fluorescent primers. Sci Rep. 2019;9(1):7400. CrossRef
50. Ordonez AA, Wintaco LM, Mota F, Restrepo AF, Ruiz-Bedoya CA, Reyes CF, et al. Imaging Enterobacterales infections in patients using pathogen-specific positron emission tomography. Sci Transl Med. 2021;13(589):eabe9805. CrossRef