INTRODUCTION
The mucus has several crucial functions. The first function is as a lubricant because it can keep the epithelium hydrated and help with the transport and mobility of food (Ali and Pearson, 2007). In addition, this type of mucus is also a permeable gel layer where gas and nutrient exchange occurs. Mucus is a slippery secretion that coats the epithelial surface in the middle ear, eyes, respiratory tract, digestive tract, and urogenital tract (Linden et al., 2008). The mucus consists of two layers. The first layer, with a depth of about 7 µm, is the periciliary fluid layer (also known as the solid/sol phase) which lies above the epithelial cilia (Rubin, 2010). The second layer covers the slime layer, known as the gel phase. Mucus gels generally have a neutral pH with a thickness varying from 5 µm to 50 µm. Mucus has a short lifespan as it is replaced every 10 to 20 minutes (Lai et al., 2009). Mucus is a complex consisting of ± 97% water (Lu and Zheng, 2013). The remaining 3% consists of enzymes, lysozyme, defensins, cytokines, inorganic salts, proteins, immunoglobulins, glycoproteins (known as mucin), lipids, and cellular debris (Vareille et al., 2011).
Mucins, which are included in 3% of the main components of mucous membranes, are a type of glycoprotein with a molecular weight ranging from 2 to 50 MDa. Mucin comprises 80% of carbohydrates, namely, N-acetylglucosamine, traces of mannose, fucose, N-acetylgalactosamine, galactose, sulfate, and sialic acid (N-acetylneuraminic acid). This carbohydrate component is a signaling mediator between mucins and external components such as pathogens and drugs. These oligosaccharide chains are attached to the protein core, especially serine and threonine hydroxyl side chains (Wagner et al., 2018).
The protein core itself is organized into different regions. A vital characteristic of mucins is a proline-threonine-serine (PTS) rich domain in the central glycosylation region, which contains residues that form glycosidic links with N-Acetylgalactosamine (GalNAc). PTS domain is expressed as a repeating tandem, resulting in a domain that carries large numbers of glycan. Furthermore, the second type region between the PTS repeats has a small number of N-glycosylation and O-glycosylation sites. This region is known as the cysteine-rich domain because the cysteine content in this region is more than 10%. Due to this structure, several interactions are present and define mucin properties, including hydrogen bonding, electrostatic bonding, and hydrophobic interactions (Yang et al., 2012). Molecular docking analysis as an in-silico approach helps explain how interactions are formed and the types of bonds.
Various ethnobotanical and ethnopharmaceutical studies have made a list of medicinal plants used by the Indonesian people and their formulation and use, mainly based on ethnicity and regional origin. Several herbal medicines that have been studied for their efficacy and are composed as polyherbal include asam jawa (Tamarindus indica), tembelekan (Lantana camara), rosella (Hibiscus sabdariffa), kencur (Kaempferia galanga), sirih merah (Piper crocatum), kelor (Moringa oleifera), meniran hijau (Phyllanthus niruri), temulawak (Curcuma xanthorrhiza), jeruk nipis (Citrus aurantifolia), and jahe (Zingiber officinale) (Aristya et al., 2018; Fatimah et al., 2017; Grosvenor et al., 1995; Jadid et al., 2020; Kristianto et al., 2022; Silalahi et al., 2015; Puspitarini et al., 2022; Wijayakusuma, 2008). This study investigated the interaction mechanism between Indonesian herbal medicine and mucin in the digestive tract. Herbs and drugs in solution for the treatment of pathological conditions are most often consumed orally; therefore, herbal preparations should support mucoadhesive properties to interact or initiate the penetration process with mucosal components, especially mucin. Mucoadhesive properties are strong adhesion binding to the mucin component that maintains an attractive interaction with each other for an extended period based on interfacial forces (Smart, 2005).
The pathological mechanisms associated with immune escape have also been widely discussed regarding metalloprotein action. Examples are the enzymes SOD (superoxide dismutase) and CAT (catalase) (Agoro and Mura, 2019; Liao et al., 2013). From the point of view of bioenergetics and electron transfer systems, this pathogenesis mechanism can occur due to uncontrolled sequestration and acquisition of metalloprotein cofactors in the transition metal ions form. The study of roles and mechanisms played by non-carbon (inorganic) elements in a life process (biochemistry) from a multidisciplinary point of view is called bioinorganic (Kaim et al., 2013). Thus, this study constructed bioactive compounds from herbs used as samples bioinorganically in complexes with transition metal ions. This bioinorganic form will be closer to the native condition of bioactive compounds from herbs, commonly called low molecular weight (LMW) (O’Brien and Nunn, 2001). As an antioxidant defense and natural scavenger, especially an indirect-acting and preventive mechanism, LMW needs to interact with transition metals as cofactors (Engwa, 2018).
In comparison, radical scavenging of a single LMW will form new radicals as LMW loses its electrons (Ahmadinejad et al., 2017). As a potential chelator, LMW complex bioinorganic has prospects as a pharmaceutical agent, especially in energy transfer systems (Widyarti et al., 2019; Sumitro and Sukmaningsih, 2018). Besides that, transition metals such as Fe, Cr, Zn, Co, Mn, Ni, Cu, Mg, and Ca are also required in small amounts to maintain the homeostasis of the human body, while either a deficiency or an excess of essential metals may result in various diseases (Bencini et al., 2010; Gupta, 2018; Jomova et al., 2022). A transition metal can be defined as any set of metallic elements occupying the central block in groups IVB, VIII, IB, and IIB, or 4–12 in the periodic table. The characteristics of this group exhibit variable valence and a strong tendency to form coordination compounds or complexes; many of their compounds are colored. Examples of the set of transition metals that are also the focus of this study are Fe (iron), Cu (copper), Mn (manganese), and Zn (zinc). The four metal elements are also included in the four elements that have crucial activities and roles in human biochemical metabolic processes and several pathological mechanisms. Metal ions are involved in catalytic mechanisms and play an active role in maintaining stability/complementing metalloproteins’ tertiary and quaternary structures. Iron or Fe is the most abundant essential transition metal in living things, with 2–6 g of Fe found in normal adult humans (Alexander and Kowdley, 2009). In the cell, Fe forms in the form of ions (Fe+2 or Fe+3), where the higher the oxidation state is, the more it will initiate acceleration or shorten the reaction in the enzyme catalytic cycle (Andreini et al., 2007). Fe binds to metalloproteins as a cofactor and is also found in many cells. Examples of metalloproteins with Fe cofactors are the most widely known heme or iron-sulfur clusters which have functions in oxygen transport. Furthermore, copper (Cu) is commonly found as a cofactor of various proteins in living systems (Andreini et al., 2008). The protein needs to bind Cu ions to reach its active form. However, the free ion concentration of Cu needs to be kept at the essential threshold, which is a maximum of 100 mg, because it is toxic (Cox, 1999). In addition, transition metals tend to catalyze the formation of free radicals that can easily damage cell membranes (Vulpe and Packman, 1995). Meanwhile, manganese (Mn), with average concentrations in the adult human body as much as 12–20 mg, negatively correlates with Fe because they are both transported by DMT1 (Brandt and Schramm, 1986; Tuschl et al., 2013). Manganese metal is also a cofactor of mitochondrial superoxide dismutase (SOD). Zinc metal (Zn) is a constituent of living systems other than Fe, with an average concentration of 1.4–2.3 g (Aggett and Harries, 1979). This metal also dominantly functions as an enzyme cofactor with regulatory, catalytic, and structural activities. An example of a metalloprotein that has a Zn metal cofactor is cytoplasmic SOD together with Cu metal (Ferreira and Gahl, 2017). Therefore, it is crucial to maintain complex bioinorganic forms. Thus, this study used bioactive compounds complex with transition metal ions.
The proper mucin conformation is affected by mucin source, ionic strength, pH, and purity (Balabushevich et al., 2018). Therefore, the target mucin used in this study is gel-forming mucin commonly found in the digestive tract, namely, MUC2 (Corfield, 2018; Johansson et al., 2013). Furthermore, this study also analyzed the stability of the bioactive compound in bioinorganic form with mucin, using molecular dynamics simulation in physiological pH (pH 7.4).
MATERIAL AND METHODS
Materials
Lipinski’s rule evaluation was performed using the Protox II database (Banerjee et al., 2018). Each ligand format was converted using the Open Babel (O’Boyle et al., 2011) into pdbqt. AutoDock Vina, as a plug-in in PyRx v.0.9.8 (Trott and Olson, 2010) used as a docking program. Post-docking results were analyzed using PyMOL v.2.4.1 (DeLano Scientific LLC, USA) and Discovery Studio 2021 Client (Accelrys, Inc., USA). Molecular dynamics simulations were performed using YASARA Dynamic v.20.10.4 (Krieger et al., 2002).
Preparation of MUC2 as a protein target
The three-dimensional crystal structure of Homo sapiens MUC2 was retrieved from Protein Data Bank (PDB ID: 7A5O) (Javitt et al., 2020). The MUC2 3D structure was prepared by removing water molecules and ions using Discovery Studio 2021 Client. Ligand NAG (N-Acetyl Glucosamine) was not removed to validate the ability of herbal bioactive compounds as competitors of NAG to form electrostatic bonds.
Ligand preparation for molecular docking
Marker or fingerprint compounds were obtained from Dr. Duke’s Phytochemical and Ethnobotanical Databases (US Department of Agriculture, 2016). The three-dimensional structures of the herbal medicine ligands, as well as Fe3+ (CID: 29936), Cu2+ (CID: 27099), and Mn2+ (CID: 27854) ions, were retrieved in SDF format from the PubChem database (Kim et al., 2016). These marker compounds of the herbal medicine were geraniol (CID: 637566) from the flower of tembelekan (Lantana camara), hibiscetin (CID: 15559735) from the flower of rosella (Hibiscus sabdariffa), kaempferol (CID: 5280863) from the rhizome of kencur (Kaempferia galanga), carvacrol (CID: 10364) from the leaf of sirih merah (Piper crocatum), moringyne (CID: 131751186) from the leaf of kelor (Moringa oleifera), phyllanthin (CID: 358901) from the leaf of meniran hijau (Phyllanthus niruri), curcumin (CID: 969516) from the rhizome of temulawak (Curcuma xanthorrhiza), epicatechin (CID: 72276) from the leaf of asam jawa (Tamarindus indica), citric acid (CID: 311) from the rind of jeruk nipis (Citrus aurantifolia), and gingerol (CID: 442793) from the rhizome of jahe (Zingiber officinale). Complexes of bioactive compounds with ions were made using Avogadro software v.1.2.0 (Marcus et al., 2012). Furthermore, the preparation process was also carried out on ligands using PyRx 0.9.8 software to lower or minimize the free energy of the ligands as well as convert compounds into AutoDock ligands. The pre-prepared ligands are then saved in PDB (.pdb) format.
QSAR analysis and drug-likeness properties
A quantitative structure–activity relationship or QSAR analysis was carried out using the Pass Online server from Way2Drug (Lagunin et al., 2010; Filimonov and Poroikov, 2008) to predict the potential bioactivity of each ligand. Drug-likeness properties were determined for each ligand with Lipinski’s rules of 5. These methods were done to ensure that all ligands meet the requirements of drug-likeness properties to be used as drug molecules. These properties are analyzed using the Protox II database (Banerjee et al., 2018; Drwal et al., 2014) and ADMETLab 2.0 database (Dong et al., 2018; Xiong et al., 2021). Input for both databases was the canonical SMILE of each ligand.
Molecular docking
All molecular docking was performed using AutoDock Vina v.1.1.2, integrated into PyRx v.09.8, operating under Microsoft Windows 10. A rigid docking method was used with 50 exhausted and 9 modes parameters. The size of the D3 backbone protein grid box was adjusted to the binding position of the NAG, with the center xyz (312.596; 123.177; 211.069) and dimensions xyz (14.757; 18.817; 20.541), while the CysD1 protein grid box uses maximized values with center (176,539; 182,435; 192,295) and dimensions (53,644; 31,481; 36,168). The program calculated the binding energy using the Vina scoring formula [1]. This formula can be interpreted as a sum of the intermolecular part of the lowest-scoring conformation and the intramolecular contributions (Trott and Olson, 2010). Once docking was completed, the results were saved in PDB (.pdb) format. Post-docking results were visualized and interpreted using PyMOL v.2.3.2.1 (Schrödinger and DeLano, 2020) and Discovery Studio 2021 Client (Biovia, 2020).
(1)
Molecular dynamics simulation
Molecular dynamics simulations were performed using YASARA Dynamic v.14.12.2 (Krieger et al., 2002), operating under Microsoft Windows 10. YASARA program makes preparation, minimization, equilibration, and production processes in one parameter file, namely, the md_run macro. Variables from the preparation step include the water solvent model TIP3P (transferable intermolecular potential) with a density of 0.997 g/l in cubic simulation cell (Sgrignani et al., 2018); periodic type of cell boundaries; water box cell size 20 Å larger than the protein; 0.9% ions of NaCl; forcefield AMBER03 12-6-4 (Duan et al., 2003; Panteva and Giambasu, 2015). The variables for the minimization steps include the number of steepest gradient descent minimization steps of 50 steps; pH and temperature were edited to physiological (pH 7.4 and 310K), and without any constrain (Cojocaru and Clima, 2019; Krieger et al., 2006). The variables of the equilibration step are carried out with an ensemble in the form of NVT and a protonation scheme where pKas are only predicted for the side-chains of Asp, Glu, His, and Lys residues, but neither for Tyr, Cys, and Arg residues nor for the N- and C-termini. Variables from the production step include a 2.5 fs timestep automatically saved as a .sim file every 25 ps and a running time of 20,000 ps (20 ns). After production, the step is an analysis run using two-parameter files, namely, the macro md_analyze and md_analyzeres. The analysis of root means square deviation (RMSD), the RG, and potential energy was obtained by running macro md_analyze. Furthermore, macro md_analyzeres was run to analyze root means square fluctuation (RMSF).
RESULTS AND DISCUSSION
Analysis was carried out between single compounds selected based on their tendencies as fingerprint compounds and their ability to bind transition metal ions. This chelating process can be determined by the presence or the absence of functional groups ROH (hydroxyl) and RCOOH (carboxyl) (Fig. 1). In addition, mucoadhesive properties were predicted by interpreting the interaction of the ligand with the two D3 and CysD1 chains of MUC2. The D3 chain is a part of the backbone protein rich in PTS (Pro-Thr-Ser) domains and interacts with carbohydrate groups, especially NAG. The CysD1 chain is a structure at the end of each mucin peptide containing many residues of the amino acid cysteine (Cys).
Lipinski’s rule of 5 helps to distinguish molecules of drugs from non-drugs. Lipinski can predict the probability of a compound’s success or failure metabolism based on its similarity to the drug. The criteria for compounds that meet Lipinski’s rules have at least 2 of the seven properties (Lipinski et al., 1997). The molecular mass of the compound is less than 500 Dalton. The compound is highly lipophilic (Log p value of less than 5). Then it has not more than ten hydrogen bond acceptors type, not more than five hydrogen bond donor type, and less than ten rotatable bonds. TPSA should also be between 0 and 140. At the same time, molar refractivity should be between 40 and 130. All variables used as Lipinski rule relate to the tendency of a drug/herbal compound to interact with macromolecules such as target proteins in cells as lipophilic environments, compared to its tendency to interact with solvents (Pollastri, 2010). Therefore, it is closely related to its absorption, distribution, metabolism, excretion, and toxicity (ADMET). All ligands meet Lipinski’s rule of 5 (Fig. 2a) and are included as safe compound because it has a toxicity class ≥3 (Fig. 2b). The results of the ADMET analysis showed that the bioactive metal complex would potentially increase bioavailability, decrease Lipinski’s rule violations, increase the L50 value, and reduce the level of toxicity of a compound compared to its single form.
The results of the QSAR analysis showed that almost all herbal fingerprint compounds, except moringyne, were predicted to have potential as mucomembranous protectors. However, carvacrol, phyllanthin, citric acid, and gingerol were predicted to have significant activity as mucositis treatment, while geraniol, carvacrol, moringyne, and gingerol compounds are also predicted to have activity as membrane permeability enhancers. Bioactivities such as steroid n-acetylglucosaminyl transferase inhibitor, TNF expression inhibitor, free radical scavenger, antioxidant, anti-inflammatory, lipid peroxidase inhibitor, peroxidase inhibitor, and MMP9 expression inhibitor are also likely to be possessed by almost all compounds. However, some bioactivity is predicted to show optimal values only for two bioactive compounds. These bioactivities are intestinal-specific anti-inflammatory (curcumin and gingerol), immunosuppressant, and immunostimulant (geraniol and moringyne).
Meanwhile, when viewed from the overall average bioactivity, compounds with potential Pa (probability to be active) values above the threshold include geraniol, hibiscetin, kaempferol, carvacrol, curcumin, epicatechin, and gingerol (Fig. 3). The value of Pa describes the potential bioactivity of a sample being tested. Pa value analyzed above 0.7 indicates that the compound has significant potential and high similarity with the compound in the database that has been proven as the treatment in the wet-lab experiment. We use a score of 0.5 as the cut-off score for the minimum potential for bioactivity. The Pa value means the accuracy of the bioactivity prediction. The better the level of accuracy displayed is, the higher the Pa value of the bioactivity will be (Lagunin et al., 2010; Filimonov and Poroikov, 2008).
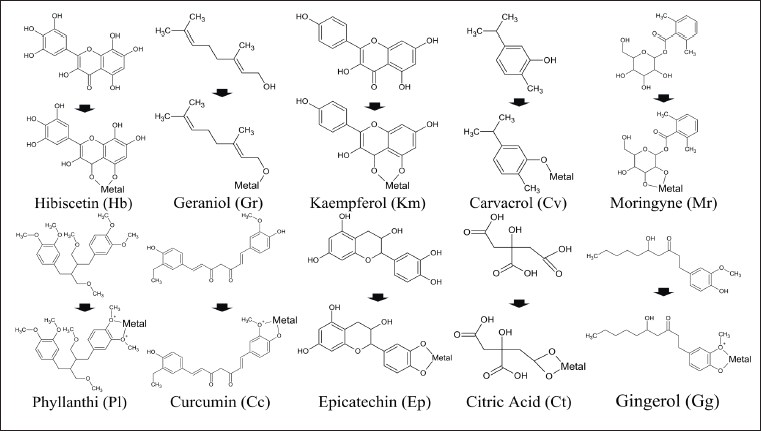 | Figure 1. 2D structure of compounds used as ligands in single and metal complexes form. [Click here to view] |
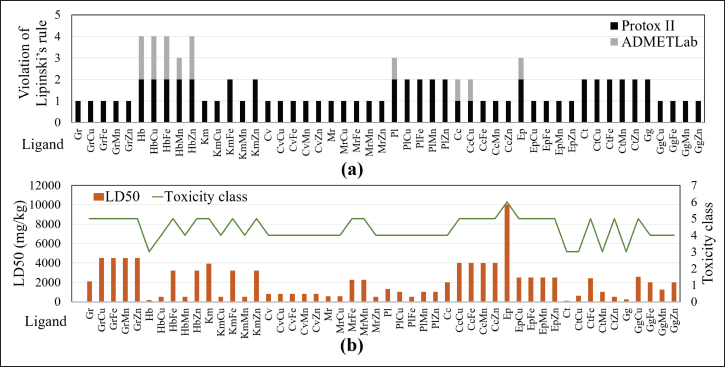 | Figure 2. Comparison of (a) Lipinski’s rules violations and (b) toxicity between single and complex metals for each ligand. [Click here to view] |
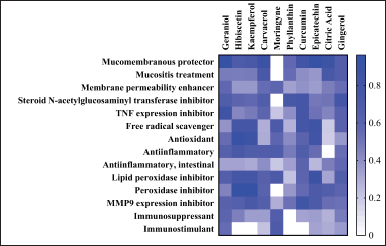 | Figure 3. Prediction of the bioactivity of each ligand using PASS online. [Click here to view] |
Molecular docking analysis
Molecular docking analysis was performed on two essential domains of MUC2, the D3 domain composed of PTS and NAG glycosylation and the CysD1 cysteine-rich domain (Yang et al., 2012). The molecular docking results show that almost all compounds that have formed complexes by binding to transition metal ions have a much lower binding affinity than a single compound when interacting with the two mucin chains. In addition, NAG used as a control compound required a relatively large binding affinity, videlicet −6.2 kcal/mol for the D3 chain and −4.6 kcal/mol for the CysD1 chain. The compound hibiscetin is predicted to have a stable interaction with the CysD1 chain but is less stable in interacting with D3. Meanwhile, kaempferol, moringyne, curcumin, and epicatechin showed the opposite results, with more stable binding to D3 than CysD1 based on the value of binding affinity (Fig. 4). This prediction is based on a minimum binding affinity value of −7 kcal/mol as the threshold (Trott and Olson, 2010).
Molecular dynamic analysis
Molecular dynamics simulation shows the dynamic stability of single and complex ion compounds when interacting with mucin in the human digestive tract. This molecular dynamic simulation was chosen as one of the drug discovery methods for predicting protein-ligand interactions by including the dynamic properties and flexibility of the target protein as the measured variables (Liu et al., 2018). The AMBER03 forcefield type used for simulating biological materials (Duan et al., 2003) is suitable for a secondary structure peptide, especially intrapeptide bonds (Smith et al., 2015), while the AMBER03 12-6-4 version is used because it has been optimized as a force field for monovalent, divalent, trivalent, and tetravalent ions, especially Mn2+, Zn2+, Cu2+, and Fe3+ (Panteva and Giambasu, 2015). The samples subjected to molecular dynamics analysis were the two most potent complexes, curcumin-metal ions, and kaempferol-metal ions. This simulation also uses 2.5 fs of timestep instead of 5 fs because when the timestep for nonbonded forces is larger, hydrogen angle vibrations become critical at 2.5 fs (Krieger and Vriend, 2015).
The total potential energy analysis is obtained after running the md_analyze macro. Furthermore, it generally shows that potential energy will experience a very significant increase from 0 to 0.6 ns running time. This pattern indicates an energy initiation process to achieve energy stability. However, fluctuations began at 0.25 ns of running time, indicating that both samples changed the molecular bond energy. Even though it has reached a stable potential energy range (equilibrium phase), this fluctuation continues. Up-fluctuations can be interpreted as a strengthening of bonds in molecules, while on the contrary a decrease in potential energy can be interpreted as a result of the relaxation of molecular bonds (Krieger et al., 2006).
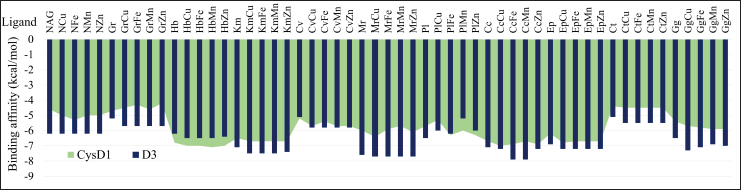 | Figure 4. Binding affinities result from molecular docking for every sample. [Click here to view] |
In addition to the potential energy, the complex’s stability can also be analyzed based on the RMSD value. RMSD is a score that provides information on conformational changes in a macromolecule that acts as a receptor after interacting with a particular ligand. RMSD can also be used as a standard deviation of conformational changes, with standards < 2 Å and > 2 Å generally being applied to docking results (Trott and Olson, 2010). The dynamic stability in question is the absence of significant conformational changes, better known as the unfolding process. The RMSD standard for a simulated protein receptor is 3 Å that the protein has undergone a conformational change (Smith et al., 2015). The pH treatment does not change the fluctuation pattern, and it only increases the average value of the distance in Å. The MUC2 chain D3 complex showed stability better if the control in the form of NAG binds to the PTS domain. Furthermore, the addition of curcumin-metal ions to the MUC2-NAG complex will significantly reduce the total RMSD value. However, the simulation results of the kaempferol compound bound with mucin showed the most unstable complex with average and maximum RMSD values of 5.413 Å and 9.976 Å (Fig. 5a). Meanwhile, the MUC2 chain CysD1 complex showed better stability if there was no interaction with curcumin or kaempferol although kaempferol showed a lower mean total RMSD value (Fig. 5b). However, all samples had a maximum total RMSD value not exceeding 3.5 Å. Therefore, it could be predicted that bonding between the mucin and the tethered compound would not be released.
The comparison of curcuminutes-metal ions complexes shows the order from the most stable to the most likely to be apart from the complex form with D3 chain after 20 ns simulation are three samples (single curcuminutes, CcMn, and CcFe) have almost the same stability, CcZn, and CcCu (Fig. 6a). Curcumin in a complex form with Fe and Mn has the best binding affinity (−7.9 kcal/mol) compared to curcumin without metal ions and other curcumin-metal complexes. Meanwhile, comparing the kaempferol-metal ions complex showed that the order KmCu, KmFe, KmZn, KmMn, and single kaempferol became the least stable complex (Fig. 6b). These results indicate that the complex kaempferol metals are stronger and more stable than kaempferol per se. Fluctuations values above 4 Å in the compound-ion complex are predicted to release the ions in the complex to find the most stable conformation. Next, the comparison of curcumin-metal ion complexes shows the order from the most stable to the most likely to be apart from the complex form with CysD1 after 20 ns of simulation are CcFe, CcMn, CcZn, single curcumin, and CcCu (Fig. 6c). Fluctuations in the curcumin-Cu complex that binds to CysD1 show an increase in the average number of RMSD close to 4 Å, indicating that Cu ions are no longer bound to curcumin. Meanwhile, comparing the kaempferol-metal ions complex showed the order KmCu, KmZn, KmFe, single kaempferol, and finally KmMn (Fig. 6d). These results show a consistent pattern of complex stability similar to the interaction between the kaempferol-metal ion and the CysD1 mucin chain.
The YASARA program’s RMSD value can also be used to see the RMSD of the ligand configuration, which in this study consisted of two single compounds (curcumin and kaempferol) and each complex structure with four types of transition metal ions. Ligand configuration RMSD was calculated by superposing the ligand with its reference structure as a function of the simulation period. The lower value of ligand configuration RMSD usually indicates proximity to the reference and hence better ligand binding affinity (Odhar et al., 2021). The RMSD ligand value above 4 Å indicates a release of ligand, especially the metal ion portion chelated by the compound (Smith et al., 2015). The RMSD of the curcumin ligand configuration was in line with the total RMSD values.
Meanwhile, in the kaempferol ligand configuration, different results between the RMSD ligand configuration and the total RMSD were seen in the KmCu (chain D3) and KmZn (CysD1 chain) complexes. This result indicates that the transition metal ion Cu tends to make the complex interactions of Cu compounds with the D3 MUC2 chain of the electrostatic type more unstable (Fig. 7a), while in the CysD1 chain, it can be predicted that the release of Zn ions from the kaempferol complex after molecular dynamic simulations was due to the RMSD value of the ligand configuration, which exceeded the value of 4 Å (Fig. 7b).
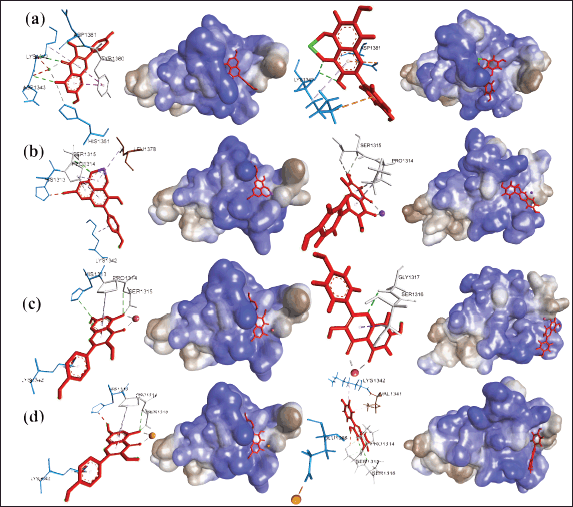
A comparison of the results before and after the simulation for 20 ns was performed by superimposing the complex at 0 ns (colored dark blue-red ligand) with the sample at 20 ns (colored dark green-white ligand). The single curcumin compound bonded to MUC2 chain D3 did not experience significant changes in position and conformation (Fig. 8a). The curcumin-Cu compound that bonds to the MUC2 chain D3 makes the NAG ligand shift position so that it successfully forms electrostatic bonds, which are also needed in the signaling process for exogenous compounds from the diet to the intestinal flora. The compound curcumin complexed with Fe, Mn, and Zn binding to the MUC2 chain D3 made the NAG ligand shift position but failed to form an electrostatic bond. Meanwhile, the single kaempferol compound that binds to the MUC2 chain D3 experienced a significant change in position and conformation, thus supporting the average total RMSD results (Fig. 8b). The kaempferol-metal ions compound attached to the MUC2 chain D3 causes the NAG ligand to shift its position and form an electrostatic bond (Fig. 8c–g). The types of electrostatic bonds formed include Pi-cation type (KmCu, KmFe, KmMn, and KmZn), Pi-anion (KmMn), and attractive charge (KmZn). The visualization results show that the arginine residue (ARG) becomes the dominant amino acid forming electrostatic bonds of Pi-cation, and GLU will form Pi-anion. Cationic environments in the Pi-cation bond are provided by the protonation of primary arginine, lysine, and histidine groups. At the same time, deprotonation of the carboxylic acid side chain will occur in GLU or aspartic acid residues to form an anionic environment (Schaeffer, 2008).
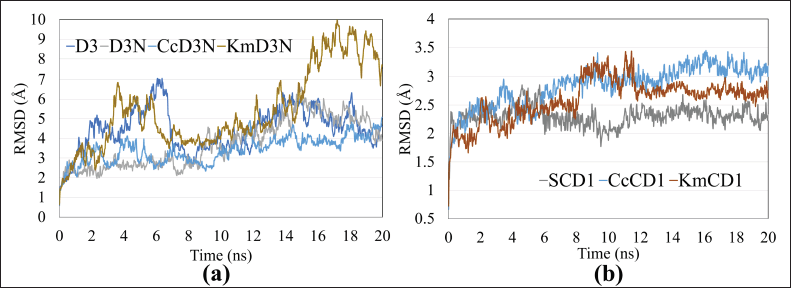 | Figure 5. Total RMSD of a single compound sample and compounds interacting with (a) the D3 chain and (b) the CysD1 chain of MUC2. [Click here to view] |
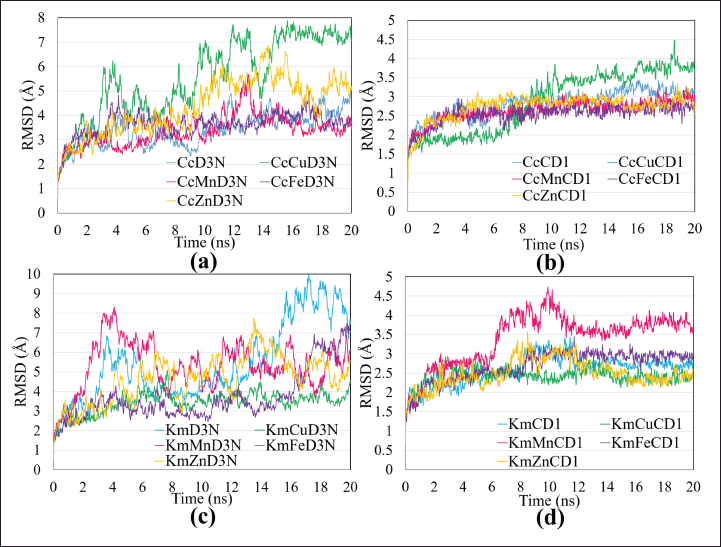 | Figure 6. Total RMSD of samples of single curcumin and curcumin-metal ions interacting with chains (a) D3 and (b) CysD1 of MUC2; single kaempferol and kaempferol-metal ions interacting with chains (c) D3 and (d) CysD1 of MUC2. Color codes of single compounds (blue), compounds-Cu (green), compounds-Mn (pink), compounds-Fe (Purple), and compounds-Zn (orange). [Click here to view] |
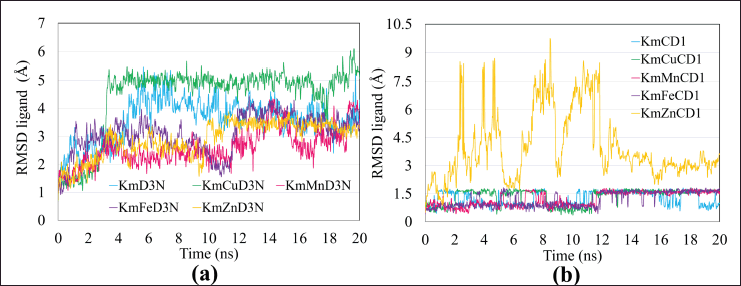 | Figure 7. Ligand configuration RMSD of samples of single kaempferol and kaempferol-metal ions interacting with chains (a) D3 and (b) CysD1 of MUC2. Color codes of single compounds (blue), compounds-Cu (green), compounds-Mn (pink), compounds-Fe (Purple), and compounds-Zn (orange). [Click here to view] |
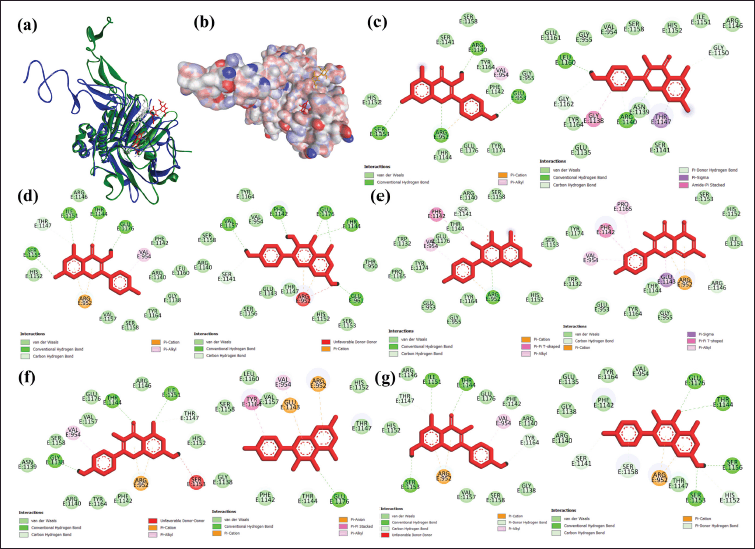 | Figure 8. Comparison of the conformational complexes of D3 mucin with (a) curcumin and (b) kaempferol and ligand interactions with (c) single kaempferol, (d) KmCu, (e) KmFe, (f) KmMn, and (g) KmZn, before (left) and after (right) 20 ns simulation. [Click here to view] |
The metal ion successfully initiates the formation of an electrostatic bond, which shows that the bioinorganic form of the bioactive compound is a functional form that naturally has more potent activity than the single form without metal ions. This natural form is a LMWC. LMW can be divided into primary and secondary metabolites based on molecular weight characteristics below 900 Da (Macielag, 2011). Secondary metabolites of LWM are a group of bioactive compounds commonly studied as medicinal compounds. Transition metals such as Mn, Cu, and Fe can catalyze the electron reduction process of molecular oxygen. Hence, LMWCs usually act as organic ligands in coordination complexes formed by various transition metals as central atoms (Hadacek and Bachmann, 2015).
Furthermore, the interaction between LMW and inorganic transition metals as central ions can initiate charge-transfer complex formation. The formation of charge-transfer complexes occurs when LMW and protein mucins’ adjacent molecular orbitals as targets overlap (Szent-Györgyi, 1960; Szent-Györgyi, 1968). Charge-transfer complex formation in protein chemistry can be grouped as a variant of the dipole-dipole interactions (Silverman, 2002). Dipole–dipole interaction is a form of the electrostatic force between two permanent molecular dipoles (Pal, 2020). Hydrogen bonding is a dipole–dipole interaction formed between the proton of a group electronegative atom (X) and atom hydrogen (H), while there are also other electronegative atoms (Y) that contain a pair of nonbonded electrons (Silverman, 2004). Another example of an electrostatic interaction is Pi-anion and Pi-cation. Pi-Anion interactions are favorable non-covalent between an anion and an electron-deficient (Pi-acidic) aromatic system (Schottel, 2008). Opposite to Pi-anion, Pi-cation also forms favorable interaction between the cation and an electron-rich aromatic system. Many studies have analyzed Pi interactions, especially Pi-cation, which can enhance binding energies by 2–5 kcal/mol. Hence, hydrogen bonds and attractive charge interaction (ion pairs) are crucial factors in drug-receptor and protein-protein interactions (Dougherty, 2013). Pi interactions also play an essential role in nature as enzyme catalysis, protein structure, and molecular recognition. Molecular recognition also includes the chemical signaling of exogenous compounds and a NAG assignment on the MUC2 PTS domain. Based on these interpretations, it can be predicted that the kaempferol complex is more stable and has better mucoadhesive properties than the D3 chain because it has succeeded in forming electrostatic bonds for all types of ligand complexes than the curcumin complex. However, due to the tendency of metal ions to change the binding affinity of the compound, several types of unfavorable bonds were also formed from the ligand interaction analysis of the KmCu complex (unfavorable bump and unfavorable donor-donor), KmMn complex (unfavorable donor-donor), and KmZn complex (unfavorable donor-donor and unfavorable positive-positive). This type of bond can reduce the stability of the interaction between the ligand and macromolecules according to the results reflected in the total RMSD average value and the ligand configuration RMSD.
Furthermore, comparisons on the CysD1 chain focused on the hydrophobicity of the positions of the amino acid residues interacting with the ligands. Interpretation results show that adding transition metal ions Zn, Mn, and Cu will increase the tendency of curcumin to interact with hydrophobic amino acid residues, while Fe ions tend to interact with hydrophilic amino acid residues lysine (Fig. 9b). However, Cu (Fig. 9a) and Zn (Fig. 9c) ions are unstable and tend to be apart from the complex after simulation. The addition of transition metal ions Mn, Cu, and especially Zn will increase the tendency of kaempferol compounds to interact with mainly hydrophilic amino acid residues. In contrast, Fe ions interact with hydrophobic amino acid residues such as leucine (Fig. 10b). In addition, similar to the curcumin complex, the Zn ion is unstable and tends to escape after simulation (Fig. 10c). However, Cu ions form covalent bonds with kaempferol, which align with the total RMSD and RMSD ligand configurations (Fig. 10a). Thus, curcumin generally forms more bonds with hydrophobic amino acids in the CysD1 chain than kaempferol to form hydrophobic pockets. This hydrophobic pocket makes the compound environment more hydrophobic and protects the compound to maintain its bioavailability.
The prediction that the kaempferol-metal ions have a higher binding affinity for the D3 domain was also confirmed by the results of RMSF (Fig. 11a) and RG (Fig. 11c). The RMSF showed that kaempferol binding could significantly maintain the bound amino acid residue (in the dotted box), especially in the results of the kaempferol-metal ions complex, which was used as a ligand. This stability can be seen in the average RMSF, which is still below 5 Å as the cut-off value (Krieger et al., 2006). These results align with RG results. The kaempferol-metal ions complex significantly maintains the consistency of the RG pattern of MUC2, which is much more stable than the MUC2-kaempferol complex. RMSF is a score that provides information on conformational changes in more detail because it is associated with fluctuations in the level of amino acid residues. High RMSF values indicate ligand mobility, while low RMSF suggests the interacted ligand stability with MUC2 (Dash et al., 2017). The value of RG and RMSF analysis helps explain that the formation of an unfavorable bond in the kaempferol-metal ions complex with MUC2 only affects the instability of the ligand bond, according to the results of RMSD and RMSD ligand configurations. However, interactions from hydrogen, electrostatic, hydrophobic, and van der Waals help maintain the complex’s stability, especially the amino acid residues of MUC2 that bind the ligands.
Meanwhile, curcumin, predicted to have a better binding affinity for the CysD1 domain, maintained conformational stability with an RMSF mean value below 4 Å and a consistent RG pattern (Fig. 10b and d). Furthermore, the RG is a parameter that describes the conformation equilibrium for all the various simulations performed. The RG value, which can also be explained as the radius of rotation of the dynamic movement of a complex, both proteins and protein compounds to the solvent, thus becomes one way of predicting the simulation of the solubility of the sample in a liquid solution or solvent (Lobanov et al., 2008). The lowest value indicates the folded protein condition, while the highest value indicates the protein conformational condition when unfolded (Yamamoto et al., 2021). Therefore, RG is commonly used to predict the dynamics pattern in a solvent environment.
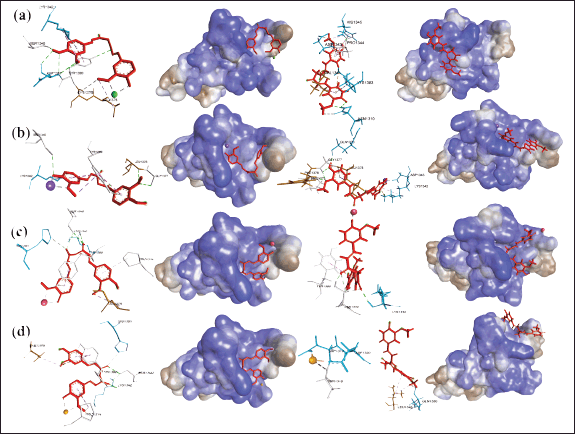 | Figure 9. Comparison of the ligand interaction and hydrophobicity diagram complexes of CysD1 mucin with (a) CcCu, (b) CcFe, (c) CcMn, and (d) CcZn, before (left) and after (right) 20 ns simulation. [Click here to view] |
 | Figure 10. Comparison of the ligand interaction and hydrophobicity diagram complexes of CysD1 mucin with (a) KmCu, (b) KmFe, (c) KmMn, and (d) KmZn, before (left) and after (right) 20 ns simulation. [Click here to view] |
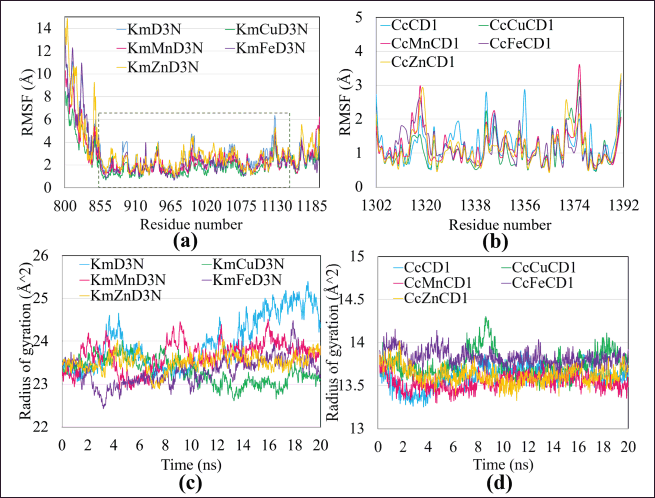 | Figure 11. Confirmation of the binding effect of the compound on the conformational stability of MUC2, based on (a, b) RMSF and (c, d) radius of gyration. [Click here to view] |
Based on the overall results, it is predicted that kaempferol and curcumin tend to interact with MUC2 in the D3 and the CysD1 chain, respectively. The interaction of both will strengthen and synergize polyherbal interactions with MUC2. In addition, the bioactive compound kaempferol tends to interact with the D3 chain of MUC2 compared to the CysD1 chain, reducing the possibility of crosslinking (Menchicchi et al., 2015). Chain D3 also has a more hydrophilic surface than chain CysD1 to maintain the viscosity of MUC2. Besides having high affinity, both bioactive compounds can significantly increase or maintain conformational stability and dynamic pattern in the bioinorganic form. The conclusion of this study was obtained from only one fingerprint compound, which represented the complex compounds contained in herbs. Therefore, it is necessary to do an in-vitro study of the mucoadhesive properties of Indonesian herbs.
Kaempferol is widely distributed in different genera like Berberis, Camellia, Allium, Citrus, Delphinium, Brassica, and Malus (Devi et al., 2015). Kaempferol is naturally bonded to different glycoside moieties (Montano et al., 2011). This study shows that kaempferol has potential as a mucoadhesive component aided by its ability to form interaction with glucan groups such as NAG. Kaempferol is also commonly known as a flavonol-derived and fingerprint compound from Kaempferia galanga L. rhizome (Khairullah et al., 2020). Kencur (Kaempferia galanga L.) is one of the Indonesian medical plants from the family Zingiberaceae, distributed throughout the world (Mitra et al., 2007; Ling, 2009; Techaprasan et al., 2010; Sukenti et al., 2016; Purba et al., 2018). Kencur also has been mentioned to have many bioactivities such as a stimulant, analgesic, expectorant, diuretic, antipyretic, antioxidant, anti-inflammatory, anticholelithiatic, and antimicrobial (Umar et al., 2011). This provides the results of the QSAR analysis. Compounds with choleretic effects, which are also equipped with potential anti-inflammatory, antioxidant, and antimicrobial activities, will interact with gel-forming mucin to treat intestinal wounds (Nazar et al., 2008). However, no research still evaluates the mucoadhesive properties of kaempferol in compound form and in complex form as kencur in more detail. Thus, conducting a wet lab study is necessary to explore the mucoadhesive properties of kaempferol both as pure extracts and herbal components. Meanwhile, curcumin, a fingerprint compound of curcuminoid from Curcuma longa extract, has been investigated for its mucoadhesive properties (Ortiz et al., 2019). However, the binding mechanism of curcumin to the mucin structure is not precise.
Several studies have also proven that plant herbal medicine contains transition metals as one of the micronutrients absorbed with groundwater. Jahe (Zingiber officinale) and meniran (Phyllanthus niruri L.) are proven to contain 6-gingerol and shogaol as phenolic compounds that bond with Fe and Mn (Kristianto et al., 2022). Zaitun (Olea europaea) is also proven to contain oleuropein-Cu complex (Capo et al., 2017). Brotowali (Tinospora crispa) contains various metals such as Fe, Cu, and Mn, significantly predicted in choline-Fe and DL-Carnitine-Fe complex molecules (Widodo et al., 2021). Duwet (Syzygium cumini) is proven to contain flavonoid-Fe complexes, especially in the cyanidin-Fe complex molecule (Sukmaningsih et al., 2018). Flavonoid complexes with transition metals Fe and Cu were of more vigorous radical scavenging activity than their single form (Jabeen et al., 2017; Sukmaningsih et al., 2018).
CONCLUSION
Herbs containing one or both bioactive compounds, namely, curcumin and kaempferol, are suggested to be composed into a polyherbal composition to obtain optimal mucoadhesive properties for MUC2. One of the suggested compositions based on this research is polyherbal composition formed from Kencur (Kaempferia galanga) with Temulawak (Curcuma xanthorrhiza). In addition, the preparation or manufacture of herbal medicine that maintains the availability of transition metal content is recommended because it is predicted to have much better bioactivity. It is crucial to preserve the natural form of LMW compounds in complexes formed with inorganic transition metals as the central ion. The complex form of these metal-ionic compounds can initiate charge transfers, which are crucial in signaling processes, functional activities, mucoadhesive properties, and other metabolic processes.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
The Research Grant Hibah Penelitian Profesor Universitas Brawijaya 2022 No. 3084.24/UN10.F09/PN/2022 funded this research.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Aggett PJ Harries JT. Current status of zinc in health and disease states. Arch Dis Childh, 1979; 54(12):909. CrossRef
Agoro R, Mura C. Inflammation-induced up-regulation of hepcidin and down-regulation of ferroportin transcription are dependent on macrophage polarization. BCMD, 2016; 61:16–25. CrossRef
Ahmadinejad F, Geir Møller S, Hashemzadeh-Chaleshtori M, Bidkhori G, Jami MS. Molecular mechanisms behind free radical scavengers function against oxidative stress. Antioxidants, 2017; 6(3):51. CrossRef
Alexander J, Kowdley KV. HFE-associated hereditary hemochromatosis. Genet Med, 2009; 11(5):307–13. CrossRef
Ali MS, Pearson JP. Upper airway musin gene expression: a review. Laryngoscope, 2007; 117(5):932–8. CrossRef
Andreini C, Banci L, Bertini I, Elmi S, Rosato A. Non-heme iron through the three domains of life. Proteins, 2007; 67(2):317–24. CrossRef
Andreini C, Banci L, Bertini I, Rosato A. Occurrence of copper proteins through the three domains of life: a bioinformatic approach. J Proteome Res, 2008; 7(01):209–16. CrossRef
Aristyani S, Widyarti S, Sumitro, SB. Network analysis of indigenous Indonesia medical plants for treating tuberculosis. Pharmacogn J, 2018; 10(6):1159–64. CrossRef
Balabushevich NG, Kovalenko EA, Mikhalchik EV, Filatova LY, Volodkin D, Vikulina AS. Mucin adsorption on vaterite CaCO3 microcrystals for the prediction of mucoadhesive properties. J Colloid Interface Sci, 2019; 545:330–9. CrossRef
Banerjee P, Dehnbostel FO, Preissner R. Prediction is a balancing act: importance of sampling methods to balance sensitivity and specificity of predictive models based on imbalanced chemical data sets. Front. Chem, 2018a; 362. CrossRef
Banerjee P, Eckert OA, Schrey AK, Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res, 2018b; 46(W1):W257–63. CrossRef
Bencini A, Failli P, Valtancoli B, Bani D. Low molecular weight compounds with transition metals as free radical scavengers and novel therapeutic agents. Cardiovasc Hematol Agents Med Chem, 2010; 8(3):128–46. CrossRef
Biovia. Dassault Systèmes, Discovery studio visualizer, V21.1.0.20298. Dassault Systèmes, San Diego, CA, 2020.
Brandt M, Schramm VL. Mammalian manganese metabolism and manganese uptake and distribution in rat hepatocytes. Manganese in metabolism and enzyme function, 1986; 50:3–16. CrossRef
Capo CR, Pedersen JZ, Falconi M, Rossi L. Oleuropein shows copper complexing properties and noxious effect on cultured SH-SY5Y neuroblastoma cells depending on cell copper content. J Trace Elem in Med Biol, 2017; 44:225–32. CrossRef
Coates J. Interpretation of infrared spectra, a practical approach. Encyclopedia Anal Chem, 2000; 12:10815–37.
Cojocaru C, Clima L. Binding assessment of methylene blue to human serum albumin and poly (acrylic acid): experimental and computer-aided modeling studies. J Molecular Liquids, 2019; 285:811–21. CrossRef
Corfield P. The interaction of the gut microbiota with the mucus barrier in health and disease in human. Microorganisms, 2018; 6(3): 78–135. CrossRef
Cox DW. Disorders of copper transport. Br Med Bull, 1999; 55(3):544–55. CrossRef
Dash R, Das R, Junaid M, Akash MFC, Islam A, Hosen SZ. In silico-based vaccine design against Ebola virus glycoprotein. AABC, 2017; 10:11–28. CrossRef
Devi KP, Malar DS, Nabavi SF, Sureda A, Xiao J, Nabavi SM, Daglia M. Kaempferol and inflammation: from chemistry to medicine. Pharmacol Res, 2015; 99:1–10. CrossRef
Dong J, Wang NN, Yao ZJ, Zhang L, Cheng Y, Ouyang D, Lu A, Cao D. ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J Cheminform, 2018; 10:29. CrossRef
Dougherty DA. The cation-π interaction. Acc Chem Res, 2013; 46(4):885–93. CrossRef
Drwal MN, Banerjee P, Dunkel M, Wettig MR, Preissner R. ProTox: a web server for the insilico prediction of rodent oral toxicity. Nucleic Acids Res, 2014; 42(W1):W53–8. CrossRef
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R and Lee T. A point-charge force field for molecular mechanics simulations of proteins. J Comput Chem, 2003; 24:1999–2012. CrossRef
Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: new docking methods, expanded force field, and python bindings. J Chem Inf Model, 2021; 61(8):3891–8. CrossRef
Engwa GA. Free radicals and the role of plant phytochemicals as antioxidants against oxidative stress-related diseases. Phytochemicals: source of antioxidants and role in disease prevention. BoD–Books on Demand, 2018; 7:49–74. CrossRef
Fatimah C, Rosidah HU, Suryanto D. Anti-tuberculosis assay of nanoherbal and ethanolic extract of Lantana camara Linn flos in vitro and in vivo. IIOABJ, 2017; 8(2):92–7.
Ferreira CR, Gahl WA. Disorders of metal metabolism. Transl Sci Rare Dis, 2017; 2(3–4):101–39. CrossRef
Filimonov D, Poroikov V. Probabilistic approaches in activity prediction Royal Society of Chemistry, Cambridge, UK, pp. 182–216, 2008. CrossRef
Gupta SP. Roles of metals in human health. MOJ Bioorg Org Chem, 2018; 2(5):221–4. CrossRef
Hadacek F, Bachmann G. Low-molecular-weight metabolite systems chemistry. Front Environment Sci, 2015; 3:1–2. CrossRef
Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform, 2012; 4(1):1–7. CrossRef
Jabeen E, Janjua NK, Ahmed S, Murtaza I, Ali T, Hameed S. Radical scavenging propensity of Cu2+, Fe3+ complexes of flavonoids and in-vivo radical scavenging by Fe3+-primuletin. Spectrochim Acta Part A Mol biomol spectr, 2017; 171:432–8. CrossRef
Jadid N, Kurniawan E, Himayani CES, Prasetyowati I, Purwani KI, Muslihatin W, Hidayati D, Tjahjaningrum ITD. An ethnobotanical study of medicinal plants used by the Tengger tribe in Ngadisari village, Indonesia. PLos One, 2020; 15(7):e0235886. CrossRef
Javitt G, Khmelnitsky L, Albert L, Bigman LS, Elad N, Morgenstern D, Ilani T, Levy Y, Diskin R, Fass D. Assembly mechanism of mucin and von willebrand factor polymers. Cell, 2020; 183(3):717–29. CrossRef
Jayaram B, Singh T, Mukherjee G, Mathur A, Shekhar S, Shekhar V. Sanjeevini: a freely accessible web-server for target directed lead molecule discovery. BMC Bioinformat. BioMed Central, 2012; 13(17):1–3. CrossRef
Johansson MEV, Sjövall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol, 2013; 10(6):352–61. CrossRef
Jomova K, Makova M, Alomar SY, Alwasel SH, Nepovimova E, Kuca K, Rhodes CJ, Valko M. Essential metals in health and disease. Chem Biol Interactions, 2022; 22:110173. CrossRef
Kaim W, Schwederski B, Klein A. Bioinorganic chemistry: inorganic elements in the chemistry of life—an introduction and guide. 2nd edition, John Wiley & Sons, New York, NY, 2013.
Khairullah AR, Solikhah TI, Ansori ANM, Fadholly A, Ram SC, Ansharieta R, Anshori A. A review of an important medicinal plant: alpinia galanga (L.) willd. Syst Rev Pharm, 2020; 11(10):387–95.
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH. PubChem: substance and compound databases. Nucleic Acids Res, 2016; 44(D1):D1202–13. CrossRef
Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins, 2002; 47(3):393–402. CrossRef
Krieger E, Nielsen JE, Spronk CAEM, Vriend G. Fast empirical pKa prediction by Ewald summation. J Molecular Graph Mod, 2006; 25(4):481–6. CrossRef
Krieger E, Vriend G. New ways to boost molecular dynamics simulations. J Comput Chem, 2015; 36(13):996–1007. CrossRef
Kristianto S, Widyarti S, Santjojo DJ, Sumitro SB. Identification and characterization of Shogaol And 6-Gingerol complex from Madurese Herbal medicine. Egypt J Chem, 2022; 65(3):1–2.
Lagunin A, Filimonov D, Poroikov V. Multi-targeted natural products evaluation based on biological activity prediction with PASS. Curr Pharm Des, 2010; 16(15):1703–17. CrossRef
Lai SK, Wang YY, Wirtz D, Hanes J. Micro- and macrorheology of mucus. Advanced Drug Delivery Reviews, 2009; 61(2):86–100. CrossRef
Liao D, Fan Q, Bao L. The role of superoxide dismutase in the survival of mycobacterium tuberculosis in Macrophages. Jpn J Infect Dis, 2013; 66(6):480–8. CrossRef
Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Musins in the mucosal barrier to infection. Mucosal Immunol, 2008; 1(3):183–97. CrossRef
Ling KH. 2009. Guide to medicinal plants: an illustrated scientific and medicinal approach. World Scientific, Singapore.
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ, An Experimental and computational approach to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev, 1997; 23:3–25. CrossRef
Liu X, Shi D, Zhou S, Liu H, Liu H, Yao X. Molecular dynamics simulations and novel drug discovery. Expert Opin Drug Discov, 2018; 13(1):23–37. CrossRef
Lobanov MY, Bogatyreva NS, Galzitskaya OV. Radius of gyration as an indicator of protein structure compactness. Molecular Biology, 2008; 42(4):623–8. CrossRef
Lu W, Zheng J. The function of musins in the COPD airway. Curr Respirator Care Rep, 2013; 2(3):155–66. CrossRef
Macielag MJ. Chemical properties of antimicrobials and their uniqueness. Antibiot Discov Devel, 2011; 1:793–820. CrossRef
Menchicchi B, Fuenzalida JP, Hensel A, Swamy MJ, David L, Rochas C, Goycoole FM. Biophysical analysis of the molecular interactions between polysaccharides and mucin. Biomacromolecules, 2015; 16(3):924–35. CrossRef
Mitra R, Orbell J, Muralitharan MS. Agriculture — medicinal plants of Malaysia. Asia Pac. Biotech. News, 2007; 11:105–10. CrossRef
Montano MCJ, Burgos-Morón E, Pérez-Guerrero C, López-Lázaro M. A review on the dietary flavonoid kaempferol. Mini Rev Med Chem, 2011; 11(4):298–344. CrossRef
Nazar S, Ravikumar S, Williams GP. Ethnopharmacological survey of medicinal plants along the Southwest coast of India. J. Herbs, Spices, Med. Plants, 2008; 14(3–4):219–39. CrossRef
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Chem, 2011; 3(1):1–4. CrossRef
O’Brien RJ, Nunn PP. The need for new drugs against tuberculosis. Obstacles, opportunities, and next steps. Am J Respir Crit Care Med, 2001; 163(5):1055–8. CrossRef
Odhar HA, Ahjel SW, Odhar ZA. Computational screening of FDA approved drugs from ZINC database for potential inhibitors of Zika Virus NS2B/NS3 protease: a molecular docking and dynamics simulation study. J Pharm Res Int, 2021; 33(39B):308–19. CrossRef
Ortiz AO, Torres LM, Moyado JAV, Peña EAP, López JLB, Bernad MJ, Carvalho JCT, Navarrete A. Mucoadhesive efect of Curcuma longa extract and curcumin decreases the ranitidine efect, but not bismuth subsalicylate on ethanol-induced ulcer model. Sci Rep, 2019; 9:16622. CrossRef
Pal S. Fundamentals of molecular structural biology. Chapter 4: chemical basis of biology. Academic Press, New York, NY, pp61–82, 2020. CrossRef
Panteva MT, Giambasu GM, York DM. Force field for Mg2+, Mn2+, Zn2+, and Cd2+ ions that have balanced interactions with nucleic acids. J Phys Chem B, 2015; 119(50):15460–70. CrossRef
Pollastri MP. Overview on the rule of five. Curr Protocol Pharmacol, 2010; 49(1):9–12. CrossRef
Purba EC, Silalahi M, Nisyawati N. Gastronomic ethnobiology of “terites”—a traditional Batak Karo medicinal food: a ruminant’s stomach content as a human food resource. J. Ethn. Foods, 2018; 5(2):114–20. CrossRef
Puspitarini S, Widodo N, Widyarti S, Jatmiko YD, Rifa’i M. Polyherbal effect between Phyllanthus urinaria and Curcuma longa as an Anticancer and Antioxidant. RJPT, 2022; 15(2):671–8. CrossRef
Rubin BK. Mucus and musins. Otolaryngol Clin North Am, 2010; 43(1):27–34. CrossRef
Schaeffer L. 2008. The role of functional groups in drug–receptor interactions. In: the practice of medicinal chemistry. New York: Academic Press 464–80. CrossRef
Schottel BL, Chifotides HT, Dunbar KR. Anion-π interactions. Chem Soc Rev, 2008; 37(1):68–83. CrossRef
Schrödinger L, DeLano W. PyMOL. 2020. Available via http://www.pymol.org/pymol
Sgrignani J, Chen J, Alimonti A, Cavalli A. How phosphorylation influences E1 subunit pyruvate dehydrogenase: a computational study. Sci Rep, 2018;8(1):1–1. CrossRef
Silalahi M, Walujo EB, Supriatna J, Mangunwardoyo W. The local knowledge of medicinal plants trader and diversity of medicinal plants in the Kabanjahe traditional market, North Sumatra, Indonesia. J Ethnopharmacol, 2015; 175:432–43. CrossRef
Silverman RB. The organic chemistry of drug design and drug action. Chapter 3: receptors. 2nd edition, Academic Press, New York, NY, pp 121–72l, 2004. CrossRef
Smart JD. The basics and underlying mechanisms of mucoadhesion. Adv Drug Deliv Rev, 2005; 57(11):1556–68. CrossRef
Smith MD, Rao S, Segelken E, Cruz L. Force-field induced bias in the structure of Aβ21−30: a comparison of OPLS, AMBER, CHARMM, and GROMOS force fields. J Chem Inf Model, 2015; 55:2587−95. CrossRef
Sukenti K, Hakim L, Indriyani S, Purwanto Y, Matthews PJ. Ethnobotanical study on local cuisine of the Sasak tribe in Lombok Island, Indonesia. J Ethn Foods, 2016; 3:189–200. CrossRef
Sukmaningsih AASA, Permana S, Santjojo DJDH, Wardoyo AYP, Sumitro SB. Investigating natural transition metal coordination anthocyanin complex in java plum (Syzygium cumini) fruit as free radical scavenging. RASAYAN J Chem, 2018; 11(3):1193–03. CrossRef
Sumitro SB, Sukmaningsih A. Herbal medicine, radical scavenger and metal detoxification: bioinorganic, complexity and nano science perspectives. IOP Conf Ser Earth Environ Sci, 2018; 130:012003. CrossRef
Szent-Györgyi A. Introduction to a submolecular biology. Academic Press, New York, NY, 1960. CrossRef
Szent-Györgyi A. Bioelectrics. Academic Press, New York, NY, 1968.
Techaprasan J, Klinbunga S, Ngamriabsakul C, Jenjittikul T. Genetic variation of Kaempferia (Zingiberaceae) in Thailand based on chloroplast DNA (psbA-trnH and petA-psbJ) sequences. Genet. Mol. Res. 2010; 9:1957–73. CrossRef
Trott O, Olson AJ. Autodock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem, 2010; 31:455–61. CrossRef
Tuschl K, Mills PB, Clayton PT. Manganese and the brain, Int Rev Neurobiol. 2013; 110:277–312. CrossRef
Umar MI, Bin Asmawi MZ, Sadikun A, Altaf R, Iqbal MA. Phytochemistry and medicinal properties of Kaempferia galanga L. (Zingiberaceae) extracts. Afr J Pharm Pharmacol, 2011; 5(14):1638–47. CrossRef
US Department of Agriculture, Agricultural Research Service. 1992-2016. Dr. Duke’s phytochemical and ethnobotanical databases. Available via http://phytochem.nal.usda.gov/ (Accessed 13 May 2021).
Vareille M, Kieninger E, Edwards MR, Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin Microbiol Rev, 2011; 24(1):210–29. CrossRef
Vulpe CD, Packman S. Cellular copper transport. Ann Rev Nutr, 1995; 15(1):293–22. CrossRef
Wagner CE, Wheeler KM, Ribbeck K. Mucins and their role in shaping the functions of mucus barriers. Annu Rev Cell Dev Biol, 2018; (34):189–215. CrossRef
Widodo WT, Santjojo DJDH, Widyarti S, Sumitro SB. Exploring and proving complex compounds in brotowali (Tinospora crispa). RASAYAN J. Chem, 2021; 14(4):2666–70. CrossRef
Widyarti S, Kamaruddin M, Aristyani S, Elvina M, Subagjo S, Rahayu T, Sumitro SB. Bioinorganic chemistry and computational study of herbal medicine to treatment of tuberculosis. Intechopen, 2019; 1(1):e90155. CrossRef
Wijayakusuma H. Ramuan lengkap herbal taklukkan penyakit. Pustaka Bunda Depok, Depok, Indonesia, 2008.
Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Chen X, Hou T, Cao D. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res, 2021; 49:W5–14. CrossRef
Yamamoto E, Akimoto T, Mitsutake A, Metzler R. Universal relation between instantaneous diffusivity and radius of gyration of proteins in aqueous solution. Phys Rev Lett, 2021; 126(12):128101. CrossRef
Yang X, Forier K, Steukers L, Vlierberghe SV, Dubrue P, Braeckmans K, Glorieux S, Nauwynck HJ. Immobilization of pseudorabies virus in porcine tracheal respiratory mucus revealed by single particle tracking. PLoS One, 2012; 7(12):e51054. CrossRef